 |
 |
| Plant Pathol J > Volume 32(1); 2016 > Article |
Abstract
Bacterial wilt of tomatoes caused by Ralstonia solanacearum is a devastating disease that limits the production of tomato in Korea. The best way to control this disease is using genetically resistant tomato plant. The resistance degree to R. solanacearum was evaluated for 285 tomato accessions conserved in the National Agrobiodiversity Center of Rural Development Administration. These accessions of tomato were originated from 23 countries. Disease severity of tomato accessions was investigated from 7 days to 14 days at an interval of 7 days after inoculation of R. solanacearum under greenhouse conditions. A total of 279 accessions of tomato germplasm were susceptible to R. solanacearum, resulting in wilt and death in 70 to 90% of these plants. Two tomato accessions were moderately resistant to R. solanacearum. Only four accessions showed high resistance against R. solanacearum. No distinct symptom of bacterial wilt appeared on the resistant tomato germplasms for up to 14 days after inoculation of R. solanacearum. Microscopy of resistant tomato stems infected with R. solanacearum revealed limited bacterial spread with thickening of pit membrane and gum production. Therefore, these four resistant tomato germplasms could be used in tomato breeding program against bacterial wilt.
Wild tomatoes (Solanum L. section Lycopersicon [Mill.] Wettst. subsection Lycopersicon) are native to South western America (from Ecuador south to northern Chile) and the Galapagos Islands (Darwin et al., 2003). The putative progenitor of cultivated tomatoes Solanum lycopersicum is widespread throughout warm regions of the world. Tomatoes include cultivated tomato (Lycopersicon esculentum = Solanum lycopersicum) and its wild relatives such as S. pimpinellifolium, S. cheesmaniae, S. chmielewskii, S. chilense, S. neorickii, S. peruvianum, S. habrochaites, and S. pennellii (Peralta and Spooner, 2007).
Tomato is one of the most consumed vegetables in the world. It provides dietary source of vitamins, minerals and fiber. Tomato plants are grown worldwide in the field or greenhouse (Peralta and Spooner, 2007). Tomatoes belong to Solanaceae. Thay contain lycopene and β-carotene that have anti-cancer and antioxidant properties. Tomatoes are recognized as healthy food. Therefore, cultivation acreage, production, and consumption of tomatoes have been substantially increased in worldwide.
Bacterial wilt caused by Ralstonia solanacearum is one of the most important and widespread bacterial disease. It limits the production of solanaceous crops in tropical, subtropical, and some warm temperate regions worldwide (Buddenhagen and Kelman, 1964; Hayward, 1991). Using genetically resistant cultivars is the most effective and environmental-friendly method to control bacterial wilt. Several resistant rootstocks have been developed and used to control bacterial wilt. However, bacterial wilt has been reported in commercial tomato cultivars grafted on resistant rootstocks in Korea and Japan (Lim et al., 2008; Nakaho et al., 2000). Therefore, selecting new resistant source from tomato germplasm is essential to control bacterial wilt.
This study was carried out to evaluate resistance of tomato germplasm lines against bacterial wilt disease. This will provide useful information on resistant sources of tomatos to improve the efficiency of tomato breeding. A total of 285 accessions of tomatoes were planted for the evaluation of resistance to bacterial wilt. Ten seeds of each accession were disinfected with 1% NaOCl for one minute. Disinfected seed were washed with distilled water twice. Tomato seeds were sowed into 50 commercial plug nursery filled with commercial peat mix soil (Baroker, Seoul Agriculture Materials Co., Seoul, Korea). Tomato seedlings were grown for 25 days in the green-house. The 285 tomato germplasms included 6 accessions of Solanum corneliomulleri, 1 accession of S. galapagense, 14 accessions of S. habrochaites, 170 accessions of S. lycopersicum, 48 accessions of S. peruvianum, 46 accessions of S. pimpinellifolium originated from 23 countries (Table 1). Resistant tomato cultivar (cv.) Support (Sakata Korea Seed) and susceptible tomato cv. Hoyong (Sakata Korea Seed) were used as control to compare disease severity.
Bacterial wilt pathogen, R. solanacearum (WR-1) was kindly provided by E. Y. Yang (National Institute of Horticultural and Herbal Science, RDA). The isolate was stored in 20% glycerol at â70°C until use. The bacterial isolate was pure-cultured by repeated streaking of single conoly on triphenyl tetrazolium chloride (TTC) agar medium (Kelman, 1954) before use. To prepare the inoculum, bacterial cells were grown in nutrient broth (Difco, USA) with shaking at 28°C for 2 days, suspended in distilled water, and adjusted to 108 cfu/ml (OD600=0.8). The roots of the tomato germplasms were immersed in bacterial suspension in a plastic container (56 cm Ă 35 cm Ă 14 cm) until saturation after artificial wounding along two sides of the plant by inserting a blade in the soil at 1 cm from the collar. After inoculation, plants were grown in a green house at 25 to 30°C under natural light. Symptom development was recorded at 7 and 14 days after inoculation of R. solanacearum.
The development of disease was evaluated using index 0-4 according to Roberts et al. (1988), with index of 0 standing for no leaves wilted, index 1 for 25% of leaves wilted, index of 2 for 26 to 50% of leaves wilted, index of 3 for 51 to 75% of leaves wilted, index of 4 for 76 to 100% of leaves wilted. Symptom development was examined by assigning symptom grades based on visual inspection at 7 and 14 days after inoculation. Tomatoes with a mean disease index of less than 2 was graded as resistant (R). Mean disease index between 2 and 3 was graded was moderate resistant (MR). If mean disease index was greater than 3, it was grades as susceptible (S).
Hypocotyls of resistant (IT 201664) and susceptible (cv. Hoyong) tomato plants were cut into small pieces (5 mm in length) with a sterile razor blade at 3 and 7 days after inoculation. Specimens were fixed with modified Karnovskyâs fixative consisting of 2% (vol/vol) glutaraldehyde and 2% (vol/vol) paraformaldehyde in 0.05 M sodium cacodylate buffer (pH 7.2) They were vacuum infiltrated. Prefixed and infiltrated specimens were placed at 4°C overnight and washed three times with the same buffer for 10 min each wash. Specimens were postfixed with 1% (w/v) osmium tetroxide in the same buffer at 4°C for 2 hr, and washed briefly twice with distilled water. These postfixed specimens were en bloc stained with 0.5% uranyl acetate at 4°C overnight. They were dehydrated in a graded ethanol series (30, 50, 70, 90 and 100%) followed by three times of washing in 100% ethanol for 10 min each. These specimens were further treated with propylene oxide as a transitional fluid twice (15 min each) and embedded in Spurrâs medium (Spurr, 1969). Infected tomato tissues embedded in Spurr resin blocks were sectioned with a glass knife on an ultramicrotome (Ultracut UCT, Leica, Austria) for light microscopy. Sections (800 nm in thickness) of these tomato samples were placed on a glass slide and heated (40°C) on a slide warmer to adhere these sections to the slide. These sections were stained with 1% toluidine blue O for a few seconds at 40°C, washed with distilled water, and examined under a light microscope (Leica DM5500B Research Microscope, Wetzlar, Germany) equipped with a Leica DFC450 digital camera (Leica Microsystems, Heerbrug, Germany) and Leica Application Suite vision V4.0 acquisition software.
Ultrathin sections (approximately 80 nm thick) were mounted onto copper grid, followed by double-staining with 2% (w/v) uranyl acetate and Reynoldâs lead citrate for 7 min. Sections were examined under a transmission electron microscope (LEO912AB, Carl Zeiss, Germany) at an accelerating voltage of 120 kV. Digitalized images were recorded with charge couple device (CCD) camera.
In this study, 285 accessions of tomato germplasm were inoculated with 108 cfu/ml of R. solanacearum at seedling stages in green house to evaluate their resistance to bacterial wilt.
Typical symptoms of bacterial wilt on tomato plant began to appear in highly susceptible tomato germplasms at 5 days after inoculation of R. solanacearum. Initial symptoms of bacterial wilt appeared on the leaves and progressed with time after inoculation. The wilt reached the entire plant of susceptible tomato germplasm. In resistant tomato germplasms, although theirfirst leaves turned yellow with age, they did not have wilt for 14 days after inoculation (Fig. 1A). Some leaves were wilted in moderately resistant tomato germplasms at 14 days after inoculation (Fig. 1B). In the susceptible control (cv. Hoyong), the entire plant wilted and died (Fig. 1C). Some leaves of resistant control (cv. Support) wilted at 14 days after inoculation (Fig. 1D).
Of the 285 accessions tested, 17 had a mean disease index of less than 2, while 24 accessions scored a mean index of less than 3 for bacterial wilt at 7 days after inoculation. Of the 285 accessions, 244 exhibited more than 76% disease severity (disease index greater than 3) at 7 days after inoculation (data not shown) before they reach maximum severity (plant death). Four accessions were resistant while two accessions were moderately resistant to bacterial wilt at 14 days after inoculation (Table 2). Three S. lycopersicum and one S. peruvianum were resistant to bacterial wilt with mean disease index of less than 2 at 14 days after inoculation. One S. lycopersicum and one S. peruvianum were moderately resistant to bacterial wilt at 14 days after inoculation. The rest 279 accessions of these tomato germplasms were susceptible to bacterial wilt (Table 2). All S. corneliomulleri, S. galapagense, S. habrochaites, and S. pimpinellifolium were susceptible to bacterial wilt at 14 days after inoculation of R. solanacearum (Table 2).
At 14 days after inoculation, four accessions of tomato germplasms (IT 201664, IT 201662, IT 201659, and IT 173773) were resistant to R. solanacearum (mean disease index of less than 2 after high inoculums density at 108 cfu/ml, Table 3). IT 201664 was the most resistant one with a mean disease index of 1.11. K177647 and IT 173830 showed moderate resistance to R. solanacearum with a mean disease index of less than 3 (Table 3). IT 201664, IT 201662, IT 201659, and IT 173773 were more resistant than cv. Support, a commercially available resistant rootstock against bacterial wilt.
In hypocotyl tissues inoculated with R. solanacearum, the primary and secondary xylem tissues differentiated in both susceptible and resistant tomato plants (Fig. 2). In a transverse section of a wilting plant of susceptible cv. Hoyong, numerous bacteria (arrows) were scattered in the vessels of both primary and secondary xylem tissues under light microscope (Fig. 2A). However, in resistant tomato (IT 201664) hypocotyl, bacterial colonization was not observed in the xylem tissues under light microscope (Fig. 2B). It has been reported that the bacterial population is reduced significantly in stems compared to taproot or collar region after root inoculation in resistant plant compared to that in susceptible plant (Dahal et al., 2009; Dannon and Wydra, 2004; Diogo and Wydra, 2007).
In the hypocotyl of infected susceptible plants, numerous bacteria were scattered in the vessels of xylem tissues under electron microscope (Fig. 3). Fewer bacteria were observed in resistant plants than in susceptible plants. Many bacteria cells had electron translucent spots on the central portions. Those electron translucent spots of bacterial cell might be storage carbon such as poly-β-hydroxybutyrate (PHB). PHB is produced in response to conditions of stress (Kang et al., 2008). Parenchyma cells next to the vessels that were colonized with bacteria were degenerated. Some degenerated parenchyma cells contained bacteria. A degenerated parenchyma cell and xylem vessel colonized with bacteria showed partial or total degradation of pit membrane (Fig. 3A). Some vessel colonized with bacteria and parenchyma cell were degenerated by infection in resistant tomato plant. Some pit membrane loosened by bacteria were still connected (Fig. 3B). Bacteria can move through openings of vessel or pit membrane upward or horizontally in vascular bundles. Degeneration and breakdown of the pit membranes have been observed in the stem and roots of susceptible tomato cultivars infected with R. solanacearum (Grimault et al., 1994; Vasse et al., 1995; Wallis and Truter, 1978).
The secondary walls and pit membrane are structurally loosened in susceptible plants (Fig. 3C). Pit membrane (arrow) between two connecting vessels were thicker and darker in electron density (Fig. 3D). Generally, the pit membrane of parenchyma cells and vessels were thicker and more conspicuous in the xylem tissue of resistant tomato plants than susceptible tomato plants after inoculation. Resistant tomato (IT 201664) showed the production of gums (g) in the vessel lumen (Fig. 3D).
The typical symptoms of R. solanacearum in tomatoes are wilting of the upper leaves during the hottest part of the day. When temperatures are cooler, the infected plants may recover temporarily. However, the plant will wither within a few days. In the lower stem, brown discoloration of vascular tissues can be observed in wilted plants (Anith et al., 2004). In this study, the symptoms of bacterial wilt began to appear on tomato seedling 4 days after inoculation of R. solanacearum in the susceptible accessions, including the susceptible variety of commercial tomato cv. Hoyong.
Resistance to bacterial wilt was first reported in PI 127805A (Acosta et al., 1964). Numerous resistant sources have been found until now. Among those, Hawaii 7996 was evaluated the highest resistance to evaluated resistance by international collaboration (Wang et al., 1998). The resistance of tomato against bacterial wilt is known to be controlled by several QTL (Quantitative trait loci). QTL on chromosome 6 plays the important role controlling resistance and strain-specific locus on chromosome 12 were founded from Hawaii 7996 (Wang et al., 2000). Studies examining the progeny of a cross between Hawaii 7996 and a susceptible line revealed QTLs on chromosomes 3, 6, 8, 10 and 11, resistant cultivar L285 on chromosomes 6, 7 and 10. Therefore, the bacterial wilt resistance in tomato is generally known under polygenic control (Ishihara et al., 2012). In this study, three S. lycopersicum and one S. peruvianum were found to be resistant to bacterial wilt, two accessions were moderately resistant, and 279 accessions were found to be susceptible to bacterial wilt at 14 days after inoculation. The four tomato germplasms were found to be more resistant than the commercial resistant rootstock cv. Support against high inoculums density of R. solanacearum containing 108 cfu/ml. Of the four resistant tomato germplasms, three (S. lycopersicum) are native to Philippine landrace, one (S. peruvianum) is a relative to Peru wild. These four tomato germplasms might be used as breeding resources for bacterial wilt resistance breeding program.
Salicylic acid (SA), jasmonic acid (JA) and ethylene could be involved in the signaling for defense response in plants (Lee et al., 2014). Many genes including pathogenesis-related, hormone signaling and lignin biosynthesis increased in only resistant tomato LS-89 (Ishihara et al., 2012). SA, JA and Ethylene dependant pathway may be engaged in the defense response of tomato to R. solanacearum. This pathogenesis related gene expression and genetic study should be needed to verify our results of providing information. To breed stable resistant plant more effectively, understanding the progress of pathogen infection in resistant genetic resources is essential. Light microscopy on infected tissue from the hypocotyls showed the presence of bacterial masses in the vessel lumen of susceptible tomatoes at 3 days after inoculation. Bacterial colonization was also observed in the xylem tissues under electron microscope in resistant tomatoes (Fig. 3). The same frequency and density of bacteria have been detected in the taproot and collar of resistant and susceptible cultivars, regardless of the resistant level (Grimault and Prior, 1993). The spread of this pathogen in the aerial parts is reduced more in resistant cultivars than in susceptible cultivars (Grimault et al., 1994; Prior et al., 1996). Mechanical barrier may hamper the spread of this pathogen in resistant cultivar. The thickenings in the pit membrane and cell wall might be a response to bacterial invasion to provide a stronger barrier against bacteria. Limited bacterial spreading in resistant cultivar CaraĂŻbo has been correlated to the production of tyloses, gums, and deposits (Grimault et al., 1994). Accumulation of electron dense materials such as the production of tyloses and gums have been reported to be a general mechanism of resistance to vascular fungal pathogens (Beckman, 1987). Histological finding suggest that the resistance of IT 201664 to bacterial wilt was associated with limited bacterial spreading by the thickening of pit membrane and the production of gums (g).
Acknowledgments
This study was supported by a grant (Project No: PJ01015302) funded by National Academy of Agricultural Science, Rural Development Administration, and by a 2015 Postdoctoral Fellowship Program of the National Academy of Agricultural Science, Rural Development Administration, Republic of Korea.
Fig. 1
The symptoms of resistant (A, IT 201664) and moderately resistant (B, K177647) accessions of tomato germplasms to bacterial wilt at 14 days after inoculation of Ralstonia solanacearum at concentration of 1Ă108 cfu/ml. Susceptible (C, cv. Hoyong) and resistant control (D, cv. Support) showed wilt or non-wilt symptom, respectively.

Fig. 2
Light micrographs of transverse sections from the hypocotyls of a susceptible (A) and resistant (B) plant at 3 days after inoculation of Ralstonia solanacearum. A large number of bacteria (arrows) were present in the vessel lumen of susceptible tomato (cv. Hoyong) stem at 3 days after inoculation (A). Transverse section of resistant tomato (IT 201664) stem showed the absence of colonization or vessel reaction (B).
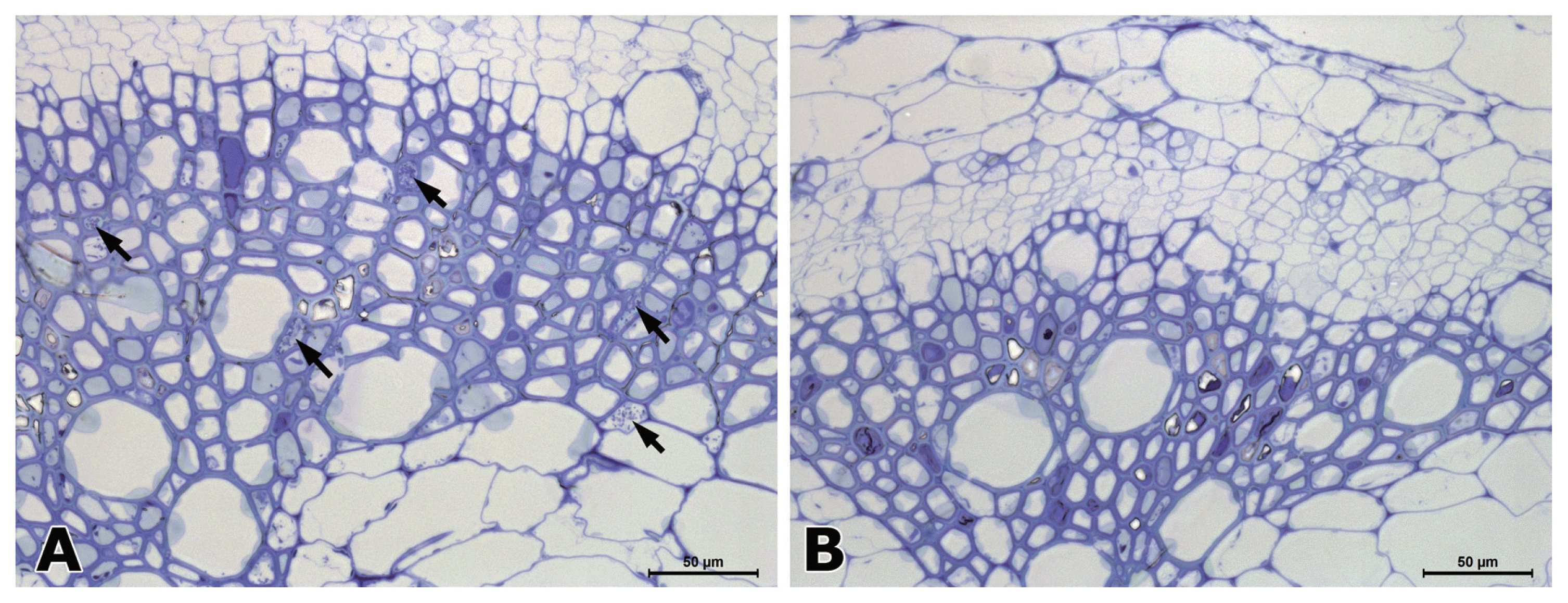
Fig. 3
Electron micrographs of transverse (A and B) and longitudinal (C and D) sections from the xylem tissues in the hypocotyls of susceptible (A and C) and resistant (B and D) plants at 3 days after inoculation of Ralstonia solanacearum. (A) Bacterial cells conolized in companion cell (cc) and intercellular space. Secondary cell walls (CW) adjacent to bacteria are structurally loosened by bacteria. The Primary cell walls were disconnected (arrows). (B) A vessel colonized with bacteria showing degeneration of parenchyma cell. V: vessel; PC: parenchyma cell; P: connecting pit membrane. (C) The secondary cell walls (CW) and pit membrane (P) are structurally loosened in susceptible plants. (D) Two vessels with secondary cell wall (CW) connecting pit membrane (P) and gums (g). Pit membranes that are thick and high in electron density are indicated by arrow.

Table 1
Geographical distribution of tomato germplasms used in this study to evaluate their resistance to bacterial wilt
| Species | Origin1 (Number of Accessions) |
|---|---|
| Solanum corneliomulleri | PER(6) |
| Solanum galapagense | ECU(1) |
| Solanum habrochaites | ECU(9), PER(4), SLV(1) |
| Solanum lycopersicum | BGR(84), BLR(1), CHN(2), GTM(2), KOR(2), MEX(2), MMR(3), NPL(1), PER(1), PHL(6), RUS(43), TUR(7), TWN(10), USA(2), UZB(4) |
| Solanum peruvianum | CHL(14), ECU(2), PER(26), POL(2), USA(4) |
| Solanum pimpinellifolium | CAN(1), ECU(4), JPN(1), MEX(4), NLD(1), PAN(1), PER(26), USA(8) |
Table 2
Resistance of 285 accessions of tomato germplasms to bacterial wilt at 14 days after inoculation of Ralstonia solanacearum
| Resistant | Moderate resistant | Susceptible | |
|---|---|---|---|
| Solanum corneliomulleri | 6 | ||
| Solanum galapagense | 1 | ||
| Solanum habrochaites | 14 | ||
| Solanum lycopersicum | 3 | 1 | 166 |
| Solanum peruvianum | 1 | 1 | 463 |
| Solanum pimpinellifolium | 46 |
Table 3
Disease severity of resistant tomato germplasms tested against Ralstonia solanacearum at 14 days after inoculation
| Accession No. | Replication | Disease severitya | Evaluationb |
|---|---|---|---|
| IT 201664 | 9 | 1.11Âą1.54a | R |
| IT 201662 | 8 | 1.50Âą1.31ab | R |
| IT 201659 | 3 | 1.67Âą2.08ab | R |
| IT 173773 | 9 | 1.78Âą1.92ab | R |
| K177647 | 7 | 2.86Âą1.46bc | MR |
| IT 173830 | 9 | 2.89Âą1.69bc | MR |
| Support (Resistant Control) | 10 | 2.10Âą1.10ab | MR |
| Hoyong (Susceptible control) | 9 | 4.00Âą0.00c | S |
References
Acosta, JC, Gilbert, JC and Quinon, VL 1964. Heritability of bacterial wilt resistance in tomato. Proc Am Soc Hort Sci. 84:455-462.
Anith, KN, Momol, MT, Kloepper, JW, Marois, JJ, Olson, SM and Jones, JB 2004. Efficacy of plant growth-promoting rhizobacteria, acibenzolar-S-methyl, and soil amendment for integrated management of bacterial wilt on tomato. Plant Dis. 88:669-673.


Beckman, CH 1987. The nature of wilt diseases of plants. APS press, St. Paul. MN.
Buddenhagen, I and Kelman, A 1964. Biological and physiological aspects of bacterial wilt caused by Pseudomonas solanacearum. Annu Rev Phytopathol. 2:203-230.

Dahal, D, Heintz, D, Van Dorsselaer, A, Braun, H-P and Wydra, K 2009. Pathogenesis and stress related, as well as metabolic proteins are regulated in tomato stems infected with Ralstonia solanacearum. Plant Physiol Biochem. 47:838-846.


Dannon, EA and Wydra, K 2004. Interaction between silicon amendment, bacterial wilt development and phenotype of Ralstonia solanacearum in tomato genotypes. Physiol Mol Plant Pathol. 64:233-243.

Darwin, SC, Knapp, S and Peralta, IE 2003. Taxonomy of tomatoes in the GalĂĄpagos Islands: native and introduced species of Solanum section Lycopersicon (Solanaceae). Syst Biodivers. 1:29-53.

Diogo, RVC and Wydra, K 2007. Silicon-induced basal resistance in tomato against Ralstonia solanacearum is related to modification of pectic cell wall polysaccharide structure. Physiol Mol Plant Pathol. 70:120-129.

Grimault, V, GĂŠlie, B, Lemattre, M, Prior, P and Schmit, J 1994. Comparative histology of resistant and susceptible tomato cultivars infected by Pseudomonas solanacearum. Physiol Mol Plant Pathol. 44:105-123.

Grimault, V and Prior, P 1993. Bacterial wilt resistance in tomato associated with tolerance of vascular tissues to Pseudomonas solanacearum. Plant Pathol. 42:589-594.

Hayward, AC 1991. Biology and epidemiology of bacterial wilt caused by Pseudomonas solanacearum. Annu Rev Phytopathol. 29:65-87.


Ishihara, T, Mitsuhara, I, Takahashi, H and Nakaho, K 2012. Transcriptome analysis of quantitative resistance-specific response upon Ralstonia solanacearum infection in tomato. PLoS One. 7:e46763



Kang, Z, Wang, Q, Zhang, H and Qi, Q 2008. Construction of a stress-induced system in Escherichia coli for efficient polyhydroxyalkanoates production. Appl Microbiol Biotechnol. 79:203-208.



Kelman, A 1954. The relationship of pathogenicity of Pseudomonas solanacearum to colony appearance in a tetrazolium medium. Phytopathology. 44:693-695.
Lee, YH, Kim, SH, Yun, B-W and Hong, JK 2014. Altered cultivar resistance of kimchi cabbage seedlings mediated by salicylic acid, jasmonic acid and ethylene. Plant Pathol J. 30:323-329.



Lim, Y-S, Lee, M-J, Cheung, J-D, Rew, Y-H and Kim, B-S 2008 Occurrence and biovar classification of bacterial wilt caused by Ralstonia solanacearum in eggplant (Solanum melongena). Res Plant Dis. 14:10-14 in Korean.

Nakaho, K, Hibino, H and Miyagawa, H 2000. Possible mechanisms limiting movement of Ralstonia solanacearum in resistant tomato tissues. J Phytopathol. 148:181-190.

Peralta, IE and Spooner, DM 2007. History, origin and early cultivation of tomato (Solanaceae). In: Genetic improvement of solanaceous crops, eds. by MK Razdan and AK Mattoo, 1-24. Science Publishers, Enfield, NH, USA.

Prior, P, Bart, S, Leclercq, S, Darrasse, A and Anais, G 1996. Resistance to bacterial wilt in tomato as discerned by spread of Pseudomonas (Burholderia) solanacearum in the stem tissues. Plant Pathol. 45:720-726.
Rivard, CL and Louws, FJ 2008. Grafting to manage soilborne diseases in heirloom tomato production. HortScience. 43:2104-2111.

Rivard, CL, Oâconnell, S, Peet, MM, Welker, RM and Louws, FJ 2012. Grafting tomato to manage bacterial wilt caused by Ralstonia solanacearum in the southeastern United States. Plant Dis. 96:973-978.


Roberts, DP, Denny, TP and Schell, MA 1988. Cloning of the egl gene of Pseudomonas solanacearum and analysis of its role in phytopathogenicity. J Bacteriol. 170:1445-1451.




Son, S-H, Khan, Z, Kim, SG and Kim, YH 2009. Plant growth-promoting rhizobacteria, Paenibacillus polymyxa and Paenibacillus lentimorbus suppress disease complex caused by root-knot nematode and fusarium wilt fungus. J Appl Microbiol. 107:524-532.


Spurr, AR 1969. A low-viscosity epoxy resin embedding medium for electron microscopy. J Ultrastruct Res. 26:31-43.


Vasse, J, Frey, P and Trigalet, A 1995. Microscopic studies of intercellular infection and protoxylem invasion of tomato roots by Pseudomonas solanacearum. Mol Plant-Microbe Interact. 8:241-251.

Wallis, FM and Truter, SJ 1978. Histopathology of tomato plants infected with Pseudomonas solanacearum, with emphasis on ultrastructure. Physiol Plant Pathol. 13:307-317.

Wang, J-F, Hanson, PM and Barens, JA 1998. Worldwide evaluation of international set of resistance sources to bacterial wilt in tomato. In: Bacterial wilt disease: Molecular and Ecological Aspects, eds. by P Prior, C Allen and J Elphinstone, 269-275. Springer, Berlin, Germay.



 PDF Links
PDF Links PubReader
PubReader Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Print
Print




