 |
 |
| Plant Pathol J > Volume 37(5); 2021 > Article |
|
Abstract
Pseudomonas cichorii secretes effectors that suppress defense mechanisms in host plants. However, the function of these effectors, including avirulence protein E1 (AvrE1), in the pathogenicity of P. cichorii, remains unexplored. In this study, to investigate the function of avrE1 in P. cichorii JBC1 (PcJBC1), we created an avrE1-deficient mutant (JBC1ΔavrE1) using CRISPR/Cas9. The disease severity caused by JBC1ΔavrE1 in tomato plants significantly decreased by reducing water soaking during early infection stage, as evidenced by the electrolyte leakage in infected leaves. The disease symptoms caused by JBC1ΔavrE1 in the cabbage midrib were light-brown spots compared to the dark-colored ones caused by PcJBC1, which indicates the role of AvrE1 in cell lysis. The avrE1-deficient mutant failed to elicit cell death in non-host tobacco plants. Disease severity and cell death caused by JBC1ΔavrE1 in host and non-host plants were restored through heterologous complementation with avrE1 from Pseudomonas syringae pv. tomato DC3000 (PstDC3000). Overall, our results indicate that avrE1 contributes to cell death during early infection, which consequently increases disease development in host plants. The roles of PcJBC1 AvrE1 in host cells remain to be elucidated.
Pseudomonas cichorii is a Gram-negative rod-shaped bacterial pathogen that produces effectors, toxins, and cyclic lipopeptides to infect a wide range of host plants (Cottyn et al., 2009; Pauwelyn et al., 2013; Ramkumar et al., 2015). In many Gram-negative plant-pathogenic bacteria, the hypersensitive reaction and pathogenicity (hrp) and hrp conserved (hrc) genes that encode a type 3 secretion system (T3SS) are essential determinants of disease development in compatible hosts and for the elicitation of the hypersensitive response (HR) on non-host plants (Hojo et al., 2008; Nomura et al., 2011). The effectors secreted via T3SS contribute to pathogen fitness in the host plants.
Avirulence protein E (AvrE) family effectors, including AvrE1 from Pseudomonas syringae pv. tomato DC3000 (PstDC3000), water-soaking E (WtsE) from Pantoea stewartii, disease-specific protein A (DspA) from Erwinia amylovora, and DspE from Pectobacterium carotovorum are strong inducers of water soaking and/or cell death when expressed in host or non-host plants (Boureau et al., 2006; Degrave et al., 2008; Frederick et al., 2001; Ham et al., 2006, 2008, 2009; Hogan et al., 2013; Kim et al., 2011). In addition, AvrE family effectors have been shown to suppress plant defense responses, such as callose deposition and expression of the defense gene PR-1 (Boureau et al., 2006; DebRoy et al., 2004; Ham et al., 2008, 2009). Xin et al. (2015) reported that AvrE1 localizes to the host plasma membrane and downregulates the expression of the nonrace-specific disease resistance1/harpin-induced1-like13 (NHL13) gene required for antibacterial immunity in Arabidopsis. Based on previous reports, the roles of avrE1 in the virulence of bacterial pathogens are diverse and versatile. Despite many studies, the avrE1 functions in P. cichorii JBC1 (PcJBC1) remain to be elucidated.
The AvrE family of effector proteins such as AvrE1, DspA/E, and WtsE are conserved in many plant-pathogenic bacteria and plant growth-promoting rhizobacteria (Preston et al., 2001). To analyze evolutionary diversity, the amino acid sequence of AvrE1 from PcJBC1 was compared with AvrE1 from PstDC3000 and Pseudomonas viridiflava UASWS0038 (PvUASWS0038), DspE of Erwinia amylovora 01SFR-BO (Ea01SFR-BO), and WtsE of Pantoea stewartii subsp. stewartii (Pss). The deduced amino acid sequence (1,818 aa) of the avrE1 gene (5,457 bp) from PcJBC1 was retrieved from the National Center for Biotechnology Information (NCBI GenBank accession no. CP007039) (Ramkumar et al., 2015) and aligned with those of PstDC3000 (WP_011103532.1), PvUASWS0038 (NCBI accession no. WP_004885164.1), Ea01SFR-BO (WP_004155375.1), and Pss (AAG01467.2). Multiple sequence alignments were performed using the Clustal O (Larkin et al., 2007) tool in the European Bioinformatics Institute (EMBL-EBI) web server; conserved and diverged amino acids were analyzed. The AvrE1 from PcJBC1 showed 52.2% and 42.8% identity with that of PvUASWS0038 and PstDC3000, respectively, and AvrE1 from PcJBC1 was far more different from DspE of Ea01SFR-BO (28.7%) or WtsE from Pss (30.8%) (Supplementary Fig. 1).
The AvrE1 effectors possess W/YxxxE/D motifs at the N-terminus, and LKKEG [F/L]E[L/M]KS, a putative endoplasmic reticulum membrane targeting/retention signal (ERMRS), at the C-terminus, which are important for host defense suppression and HR elicitation in non-host plants (Ham et al., 2009; Kim et al., 2011). Substitution of amino acids W (tryptophan), Y (tyrosine), F (phenylalanine), E (glutamic acid), or D (aspartic acid) within the WxxxE motif may alter or disrupt AvrE1 virulence function (Ham et al., 2009). AvrE1 proteins from PcJBC1, PvUASWS0038, and PstDC3000 possess one WxxxE motif at positions 402, 336, and 392, respectively, and another at positions 639, 575, and 635, respectively (Supplementary Fig. 1). In the first location (W402E) of the motif, E was conserved in PvUASWS0038 and PstDC3000 but substituted by P (proline) in PcJBC1. In the second location of the motif, E was substituted by G (glycine) in all three Pseudomonas strains. The DspE of Ea01SFR-BO and WtsE of PsM009 have only one WxxxE motif at positions 706 and 693, respectively. The E in this motif is conserved in Pss and Ea01SFR-BO but substituted with G in PcJBC1, PvUASWS0038, and PstDC3000. The ERMRS, which is important for endoplasmic reticulum targeting, is conserved in all strains except PvUASWS0038. In PcJBC1, the K (lysine) present in the LKKEG [F/L]E[L/M]KS motif is replaced by Q (glutamine) and S (serine). Xin et al. (2015) reported that AvrE1 from PstDC3000 has two functional domains (AvrE1-N1-995aa and AvrE1-C990-1795aa) and requires most of the full length AvrE1200-1795aa to retain the necrosis-inducing ability in plants, and AvrE1-N contains a plasma membrane targeting signal. Collectively, the variations and low identity in amino acid composition among these strains suggest that AvrE1 may have diverse roles in PcJBC1 virulence.
In P. syringae pv. tomato, E. amylovora, and P. agglomerans pv. gypsophilae, loss of AvrE1 or DspE effectors reduced the virulence of the strain (Badel et al., 2006; Bogdanove et al., 1998; Gaudriault et al., 1997; Lorang et al., 1994; Mor et al., 2001). AvrE1 from PstDC3000 induced water-soaking (Xin et al., 2016), lesion formation (Badel et al., 2006), suppressed salicylic acid-mediated basal immunity and promoted necrosis in tomato and Arabidopsis plants (DebRoy et al., 2004; Jin et al., 2016). In the bacterial canker disease causing P. syringae pv. actinidiae, the cooperation of AvrE1 and HopR1 was required to foster in planta bacterial growth and lesion production in kiwi fruits (Jayaraman et al., 2020).
In this study, to determine the role of avrE1 in the virulence of P. cichorii, we created an avrE1-deficient PcJBC1 mutant (JBC1ΔavrE1) strain and complemented strains with avrE1 genes from the PcJBC1 (JBC1ΔavrE1 + pavrE1JBC1) and PstDC3000 (JBC1ΔavrE1 + pavrE1DC3000). The avrE1 gene (from start to stop codon) from PcJBC1 was knocked out using the pCasPA/pACRISPR system developed by Chen et al. (2018) with minor modifications (Supplementary Fig. 2). The single-guide RNA (sgRNA) design tool Benchling (Benchling, Inc., San Francisco, CA, USA; https://benchling.com/) was used to find a high-efficiency gRNA target sequence followed by the protospacer adjacent motif nucleotide sequence NGG. To construct a plasmid with the appropriate gRNA sequence for the target sequence, primers were designed with 5′-ends annealing back-to-back using the NEBaseChanger Tool (http://nebasechanger.neb.com/). The gRNA sequence was replaced with the designed primers (Supplementary Table 1) using Q5 Site-Directed Mutagenesis kit (New England BioLabs). Briefly, 5 μl 2× KLD reaction buffer, 1 μl PCR reaction, 3 μl distilled water (DW), and 1 μl KLD enzyme mix were mixed and incubated at room temperature for 5 min and used for the transformation of Escherichia coli DH5α using standard techniques. The transformants with the pACRISPR-sgRNA-avrE1 plasmid were screened on carbenicillin plates (150 μg/ml) and confirmed through PCR using primers AvrE1-sgRNA/Amp F and sequencing. Simultaneously, the upstream and downstream regions of avrE1 (500 bp) were amplified individually through PCR. The 5′-forward primer upstream AvrE1.JBC1-Us-F and 3′-reverse primer downstream AvrE1.JBC1-Ds-R contained pACRISPR flanking regions (20-40 bp), while the 3′-primer upstream AvrE1.JBC1-Us-R and the-primer downstream AvrE1. JBC1-Ds-F were flanked (30-40 bp) with each other. Equal amounts of the digested pACRISPR-sgRNA-avrE1 with XbaI and XhoI was assembled with upstream and downstream fragments through Gibson assembly and cloned into E. coli DH5α. The construct was confirmed through PCR using primer sets AvrE1-Us-F/AvrE1-Ds-R primers and sequencing. PcJBC1 cells were washed twice with 10% glycerol to produce electrocompetent cells, and pCasPA plasmids (Chen et al., 2018) were transferred into PcJBC1 electrocompetent cells (PcJBC1-pCasPA). The colonies were verified through PCR using CasPA-C-F/CasPA-C-R primers. The pACRISPR-sgRNA-avrE1-Us-Ds plasmids were electroporated into PcJBC1-pCasPA cells. One percent of PcJBC1-pCasPA overnight culture was reinoculated into fresh Luria-Bertani (LB) broth containing 0.2% L-arabinose to induce the expression of Red system at 30°C until OD600 reached 0.5. Cells were made electrocompetent and transformed with pACRISPR-sgRNA-avrE1-Us-Ds through electroporation. The electroporated cells were recovered by adding 1 ml LB broth, incubated at 30°C for 2 h, and then plated on LB agar plates containing 100 μg/ml tetracycline and 150 μg/ml carbenicillin. The defective mutant (JBC1ΔavrE1) was verified for correct deletion through PCR and sequencing with the AvrE1-Us-F/AvrE1-Ds-R primers. The plasmids were cured by streaking the mutants on LB plates supplemented with 5% (w/v) sucrose.
To complement the JBC1ΔavrE1 mutant using homologous and heterologous avrE1 genes, the gene including its own promoter region was amplified from PcJBC1 and PstDC3000 gDNA through PCR (Hung et al., 2014) using the primer set AvrE1-J.EcoRI-F/AvrE1-J.BamHI-R and AvrE1-DC.KpnI-F/AvrE1-DC.BamHI.R, respectively (Supplementary Table 1). PCR products were purified using a PCR purification kit, cloned into pGEM-T Easy vector, subcloned into pUCP18 vector (West et al., 1994) using EcoRI and BamHI restriction sites, and then transformed into electrocompetent E. coli DH5α cells. Positive colonies were selected on LB plates containing ampicillin (100 μg/ml) and kanamycin (50 μg/ml). Recombinant plasmids containing avrE1 from PcJBC1 (pavrE1JBC1) and PstDC3000 (pavrE1DC3000) were transformed into JBC1ΔavrE1 electrocompetent cells to create a homologous (JBC1ΔavrE1 + pavrE1JBC1) and heterologous (JBC1ΔavrE1 + pavrE1DC3000) complemented strain, respectively. The complemented strains were confirmed through PCR using AvrE1-J.Eco-RI-F/AvrE1-J.BamHI-R and AvrE1-DC.KpnI-F/AvrE1-DC.BamHI.R for JBC1ΔavrE1 + pavrE1JBC1 and JBC1ΔavrE1+ pavrE1DC3000, respectively.
To investigate the role of avrE1 in PcJBC1 virulence, the wild-type (PcJBC1), avrE1-deficient mutant (JBC1ΔavrE1), and complemented strains (JBC1ΔavrE1 + pavrE1JBC1 and JBC1ΔavrE1 + pavrE1DC3000) were cultured in LB broth and prepared for inoculation in 10 mM MgCl2 (1 × 108 cfu/ml) with 0.025% Silwet L-77. Four-week-old tomato plants were infected by dipping the whole plants into the bacterial suspensions for 2 min, and were then kept under high humidity (approximately 95% relative humidity) for a day. Disease severity was observed 7 days after inoculation (dai) at 25°C with a 16 h light/8 h dark photoperiod. A disease index scale of 0-3, where 0 = 0-25%, 1 = 26-50%, 2 = 51-75%, and 3 = 76-100% was used to calculate the disease severity based on the size of the diseased area of the infected leaves. The infected area on each leaf was estimated by evaluating the pixel area from leaf pictures using the magnetic lasso and histogram tool of ImageJ software (https://imagej.nih.gov/ij/). Disease severity in each leaf was calculated using the following formula: (pixel of infected area/pixel of total leaf) × 100. The average disease index was calculated using the following formula: Disease severity = (∑ disease index number × number of leaves with similar disease index)/total leaves surveyed. Three individual plants were used for each treatment, and three independent experiments were conducted. Disease severity was significantly reduced in leaves infected with JBC1ΔavrE1 (6.38%) compared to those infected with PcJBC1 (67.22%), JBC1ΔavrE1 + pavrE1JBC1 (39.19%), and JBC1ΔavrE1 + pavrE1DC3000 (21.23%) (Fig. 1). These results indicate that AvrE1 plays a determinative role in PcJBC1 virulence in tomato plants.
The in planta bacterial growth was evaluated following the method described by Xin et al. (2016). Briefly, leaves of 4-week-old tomato plants were dip-inoculated with 0.2 OD600 (1 × 108 cfu/ml) cell suspension of each strain with 0.025% Silwet L-77 and kept under high humidity (approximately 95%) for disease to develop. After 72 hour after inoculation (hai), three infected leaves were collected and surface-sterilized with 2% NaClO for 3 min followed by washing 3 times with sterile DW. Leaf disk samples collected using a cork borer were homogenized with 1 ml sterile water and serially diluted. For bacterial enumeration, each diluted sample was spread plated on LB media containing vancomycin for PcJBC1 and JBC1ΔavrE1, vancomycin and ampicillin for JBC1ΔavrE1 + pavrE1JBC1 and JBC1ΔavrE1 + pavrE1DC3000 strains. The bacterial colonies were counted 48 hai at 30°C and the cfu was normalized as cfu/cm2. Experiments were repeated three times. The cell growth of JBC1ΔavrE1 was significantly reduced compared to those infected with PcJBC1, JBC1ΔavrE1 + pavrE1JBC1, and JBC1ΔavrE1 + pavrE1DC3000, which corresponds to the results of disease severity assay.
Water soaking is important for providing a suitable environment for nutrient mobilization, buffering temperature, movement, and multiplication of the pathogen to increase colonization of host cells (Aung et al., 2018; Beattie, 2011). In previous report, AvrE1 played important roles in producing water-soaked conditions, which are the most common symptoms of bacterial diseases (Xin et al., 2016). To investigate the role of avrE1 in P. cichorii during the early stages of infection, we inoculated PcJBC1, JBC1ΔavrE1, JBC1ΔavrE1 + pavrE1JBC1, and JBC1ΔavrE1 + pavrE1DC3000 cells (1 × 108 cfu/ml) into the abaxial sides of tomato leaves using a needleless syringe and incubated them in a growth chamber at 25°C and 16 h light under high humidity. Symptom development was recorded at 2, 4, 6, 12, and 24 hai. Water-soaking symptoms started to appear in PcJBC1, JBC1ΔavrE1 + pavrE1JBC1 and JBC1ΔavrE1 + pavrE1DC3000 infected tomato leaves 4 hai; lesion formation was observed from 6 to 12 hai, followed by cell death (Fig. 2). However, no water soaking symptoms, lesion formation, or cell death were observed in tomato leaves infected with JBC1ΔavrE1 12 hai, and disease development was significantly reduced compared to that induced by WT and complemented strains. The results indicated that avrE1 from PcJBC1 was essential for inducing water soaking, which promoted the growth of bacterial cells in plant tissues.
P. cichorii induced distinctive symptoms in different parts of host plants, including varnish spots on lettuce, midrib rot on butterhead lettuce, bacterial blight on celery, leaf rot on pepper and tomato, leaf spot, bud blight, and stem necrosis on chrysanthemum, and stem melanosis on spring wheat (Hung et al., 2014; Mirik et al., 2011; Pauwelyn et al., 2011; Yu and Lee, 2012). In this study, we investigated the role of AvrE1 in the virulence of PcJBC1 on the midrib of cabbage. The midribs of Kimchi cabbage were surface disinfected with 70% ethanol and washed twice with sterile DW. A sterile toothpick was used to make a 0.5 mm deep wound on the surface-disinfected midribs, and a 15 μl cell suspension (1 × 108 cfu/ml) was applied to the wound. The inoculated midribs were incubated in a growth chamber at 25°C in closed plastic containers (>90% relative humidity). Midribs inoculated with 10 mM MgCl2 served as the negative control. The diseased area [(distance from the center of the symptom to its farthest edge) × (distance from the center of the symptom to its closest edge) × 3.14] was measured 3 dai. Ten wounds were used for each treatment with three replicates. The size of the rot area in plants inoculated with JBC1ΔavrE1 (4.63 mm2) was significantly reduced in comparison to that of plants inoculated with PcJBC1 (14.54 mm2), JBC1ΔavrE1 + pavrE1JBC1 (12.46 mm2), and JBC1ΔavrE1 + pavrE1DC3000 (10.46 mm2). The rot symptoms were characterized by dark color with soft rot caused by PcJBC1 and by the complemented strains JBC1ΔavrE1 + pavrE1JBC1 and JBC1ΔavrE1 + pavrE1DC3000, whereas JBC1ΔavrE1 strain caused a light-brown color with dry rotting symptoms (Fig. 3). The avrE1 from PstDC3000 restored symptom size and color, indicating proper function of the PcJBC1 type III secretion system. The results indicated that AvrE1 significantly contributed to cell lysis and rot of the cabbage midrib, which consequently influenced the symptoms and disease development.
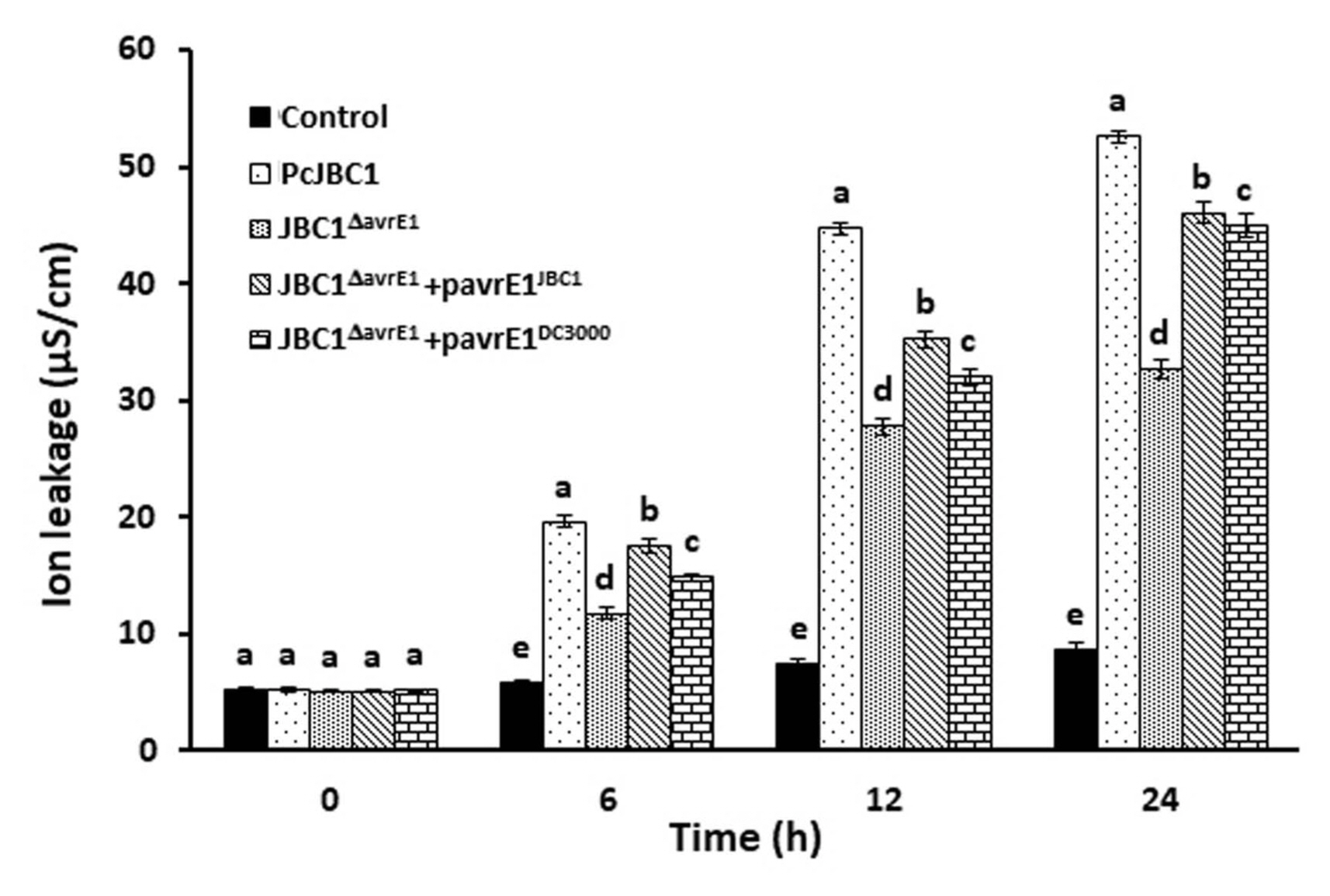
The symptom development during the infection process indicated that AvrE1 from PcJBC1 influences water soaking and cell lysis of tomato tissues. To study the influence of AvrE1 during the infection stage, tomato leaves were inoculated with bacterial suspensions and the ion leakage was measured according to Hatsugai et al. (2017). Briefly, three leaf discs per leaf with a diameter of 7.5 mm were cut using a cork borer at 0, 12, and 24 h after bacterial infiltration (1 × 106 cfu/ml) and placed in 10 ml sterilized ultra-pure water in 50 ml falcon tubes for 90 min with simple agitation. Electrolyte leakage was determined by measuring the conductivity of three independent plants with an electrical conductivity meter (HQ14d, HACH, USA). Leaf discs inoculated with DW were used as negative controls. The electro-resistance was higher in the leaves inoculated with PcJBC1, JBC1ΔavrE1, JBC1ΔavrE1 + pavrE1JBC1, and JBC1ΔavrE1 + pavrE1DC3000 than in those inoculated with JBC1ΔavrE1 at 6, 12, and 24 hai (Fig. 4). The results of this study indicate the role of AvrE1 in cell lysis in the early stages of disease development.
AvrE family effectors strongly induce either water soaking, cell death, or both, once expressed in host or non-host plants (Boureau et al., 2006; Ham et al., 2008, 2009). The influence of avrE1 from PcJBC1on HR was determined using tobacco (Nicotiana tabacum cv. Samsun) plants. PcJBC1, JBC1ΔavrE1, JBC1ΔavrE1 + pavrE1JBC1, and JBC1ΔavrE1 + pavrE1DC3000 cells were diluted to a concentration of 1 × 108 cfu/ml in 10 mM MgCl2, and then infiltrated in tobacco leaves using a needleless syringe. The inoculated leaves were incubated at 25°C with high humidity (≥90%) and HR was visually estimated 3 dai. HR-associated necrosis was elicited by wild-type and both complemented strains within 3 dai, but no HR-associated necrosis was induced in the tobacco leaves infiltrated with the JBC1ΔavrE1 strain (Supplementary Fig. 3).
Overall, AvrE1-deficiency significantly suppressed PcJBC1 virulence by reducing water soaking in tomato leaves during the infection process, which was supported by electrolyte leakage because of cell lysis in the infected leaves. The disease symptoms in the cabbage midrib induced by JBC1ΔavrE1 were characteristically light-brown compared to the dark discoloration with soft rotting induced by PcJBC1, which also indicates the role of AvrE1 in cell lysis. Cell death in non-host tobacco plants was not induced by the AvrE1-deficient mutant. The disease severity and cell death in the host and non-host plants were restored through complementation with avrE1 from Pst-DC3000, which indicates that avrE1 from PstDC3000 can function in a PcJBC1 background with similar functions. Our results indicate that avrE1 is essential for PcJBC1 virulence during the early infection stage, which consequently determines disease severity in host plants. The results of this study contribute to the understanding of disease progression and symptom development induced by the bacterial pathogen P. cichorii on its host plants.
Acknowledgments
We gratefully acknowledge a grant from the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science, and Technology (No. 2017R1A2B2002221). This research was also partly supported by the Cooperative Research Program for Agriculture Science & Technology Development (Project No. PJ015566), and the Rural Development Administration.
Electronic Supplementary Material
Supplementary materials are available at The Plant Pathology Journal website (http://www.ppjonline.org/).
Fig. 1
Disease severity and symptoms in tomato plants. (A) Four-week-old tomato plants were dip-inoculated with Pseudomonas cichorii JBC1 (PcJBC1), avrE1-deficient (JBC1ΔavrE1), and complemented (JBC1ΔavrE1 + pavrE1JBC1 and JBC1ΔavrE1 + pavrE1DC3000) strains at a concentration of 1 × 108 cfu/ml and incubated in a growth chamber at 25°C with a photoperiod of 16 h light and 8 h dark. In planta bacterial growths were recorded and calculated as cfu/cm2 at 72 hours after inoculation. The results are presented as the means ± SD from three independent experiments in three replicates. Means with the same letter are not significantly different by Tukey’s studentized range (HSD) test (P ≤ 0.05). (B) The photo was taken 3 days after inoculation.
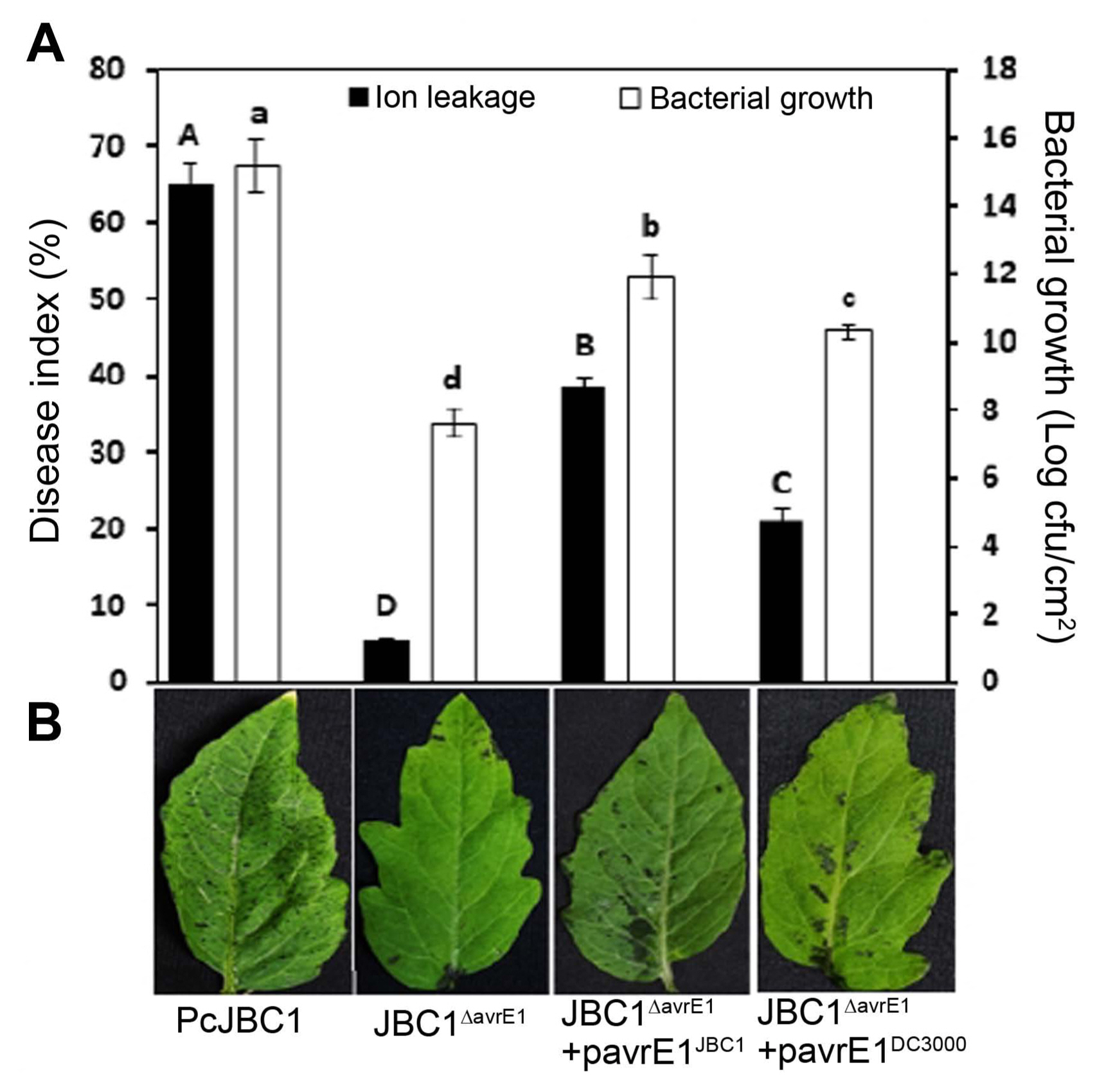
Fig. 2
Development of disease symptoms in tomato plants. Four-week-old tomato leaves were inoculated with wild-type (PcJBC1), avrE1-deficient (JBC1ΔavrE1), and complemented (JBC1ΔavrE1 + pavrE1JBC1, JBC1ΔavrE1 + pavrE1DC3000) strains using needleless syringes at a concentration of 1 × 108 cfu/ml and incubated in a growth chamber at 25°C with a photoperiod of 16 h light and 8 h dark. Symptom development was recorded 2, 4, 6, 12, and 24 hours after inoculation (hai). The results are presented as the means ± SD from three independent experiments in three replicates. Means with the same letter are not significantly different by Tukey’s studentized range (HSD) test (P ≤ 0.05).

Fig. 3
Disease severity and symptom on the midrib of cabbage. Each bacterial suspension (1 × 108 cfu/ml) of the wildtype (PcJBC1), avrE1-deficient (JBC1ΔavrE1), and complemented (JBC1ΔavrE1 + pavrE1JBC1 and JBC1ΔavrE1 + pavrE1DC3000) strains was placed in wounds made with a sterile toothpick on the midrib of Kimchi cabbage and incubated at 25°C with 90% relative humidity. Photographs were taken 3 days post inoculation. The results are presented as the means ± SD from three independent experiments in three replicates. Means with the same letter are not significantly different by Tukey’s studentized range (HSD) test (P ≤ 0.05).
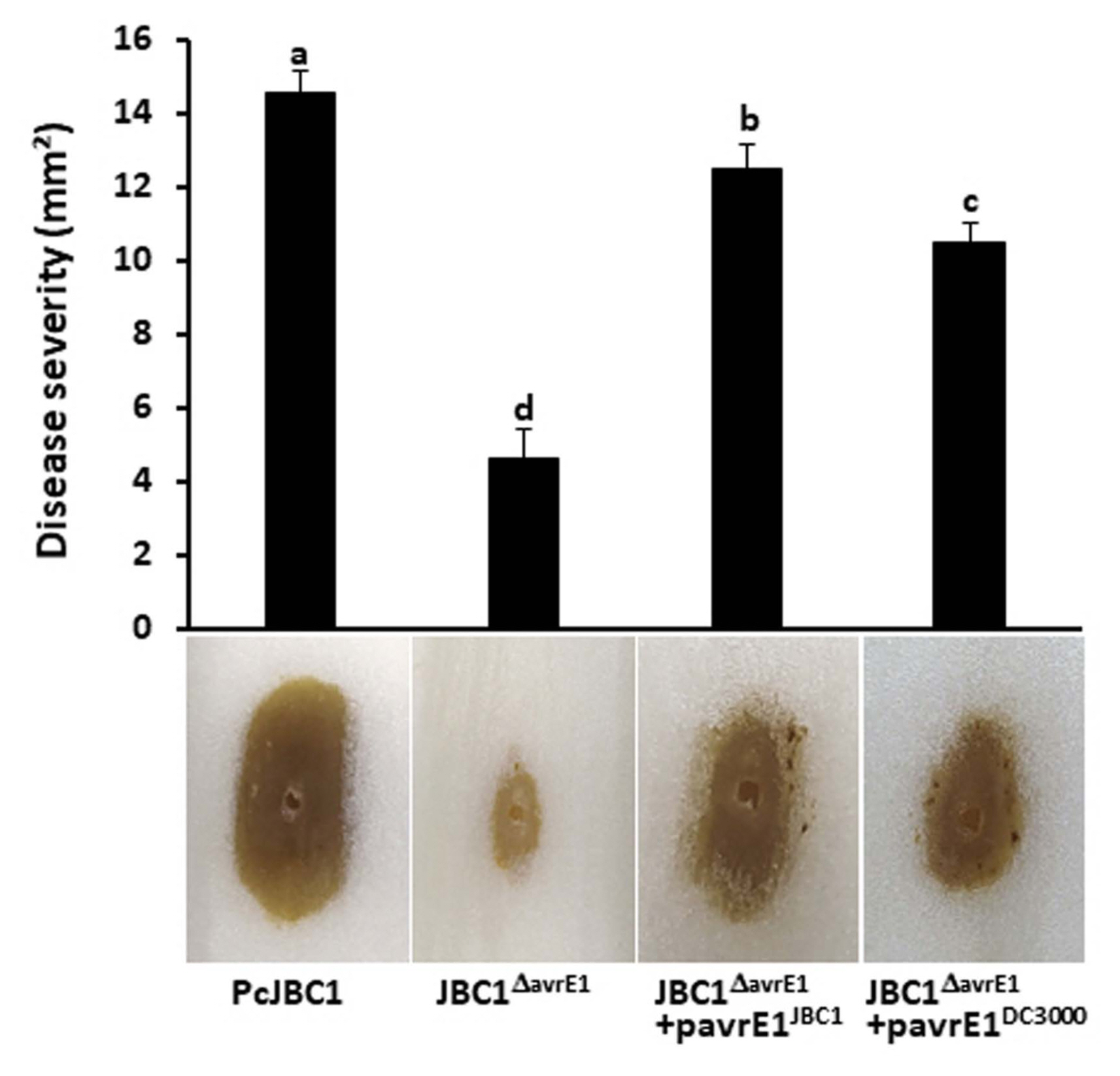
Fig. 4
Ion leakage by mutants of Pseudomonas cichorii JBC1 in tomato plants. Four-week-old tomato plant leaves were needle-less syringe-inoculated with bacterial suspensions of wild-type (PcJBC1), avrE1-deficient (JBC1ΔavrE1), and complemented (JBC1ΔavrE1 + pavrE1JBC1 and JBC1ΔavrE1 + pavrE1DC3000) strains at a concentration of 1 × 106 cfu/ml. Ion leakage was measured at 0, 12, and 24 hours after inoculation. Results are the average of three replications and ± SD. Different letters indicate significant difference at each time point at 0.5 significance level according to the Tukey Krammer HSD test (P ≤ 0.05).
References
Aung, K, Jiang, Y and He, SY 2018. The role of water in plant-microbe interactions. Plant J. 93:771-780.




Badel, JL, Shimizu, R, Oh, H-S and Collmer, A 2006. A Pseudomonas syringae pv. tomato avrE1/hopM1 mutant is severely reduced in growth and lesion formation in tomato. Mol Plant-Microbe Interact. 19:99-111.


Beattie, GA 2011. Water relations in the interaction of foliar bacterial pathogens with plants. Annu Rev Phytopathol. 49:533-555.


Bogdanove, AJ, Kim, JF, Wei, Z, Kolchinsky, P, Charkowski, AO, Conlin, AK, Collmer, A and Beer, SV 1998. Homology and functional similarity of an hrp-linked pathogenicity locus, dspEF, of Erwinia amylovora and the avirulence locus avrE of Pseudomonas syringae pathovar tomato
. Proc Natl Acad Sci U S A. 95:1325-1330.



Boureau, T, ElMaarouf-Bouteau, H, Garnier, A, Brisset, M-N, Perino, C, Pucheu, I and Barny, M-A 2006. DspA/E, a type III effector essential for Erwinia amylovora pathogenicity and growth in planta, induces cell death in host apple and nonhost tobacco plants. Mol Plant-Microbe Interact. 19:16-24.


Chen, W, Zhang, Y, Zhang, Y, Pi, Y, Gu, T, Song, L, Wang, Y and Ji, Q 2018. CRISPR/Cas9-based genome editing in Pseudomonas aeruginosa and cytidine deaminase-mediated base editing in Pseudomonas species. iScience. 6:222-231.



Cottyn, B, Heylen, K, Heyrman, J, Vanhouteghem, K, Pauwelyn, E, Bleyaert, P, van Vaerenbergh, J, Höfte, M, de Vos, P and Maes, M 2009.
Pseudomonas cichorii as the causal agent of midrib rot, an emerging disease of greenhouse-grown butterhead lettuce in Flanders. Syst Appl Microbiol. 32:211-225.


DebRoy, S, Thilmony, R, Kwack, Y-B, Nomura, K and He, SY 2004. A family of conserved bacterial effectors inhibits salicylic acid-mediated basal immunity and promotes disease necrosis in plants. Proc Natl Acad Sci U S A. 101:9927-9932.



Degrave, A, Fagard, M, Perino, C, Brisset, MN, Gaubert, S, Laroche, S, Patrit, O and Barny, M-A 2008.
Erwinia amylovora type three-secreted proteins trigger cell death and defense responses in Arabidopsis thaliana
. Mol Plant-Microbe Interact. 21:1076-1086.


Frederick, RD, Ahmad, M, Majerczak, DR, Arroyo-Rodríguez, AS, Manulis, S and Coplin, DL 2001. Genetic organization of the Pantoea stewartii subsp. stewartii hrp gene cluster and sequence analysis of the hrpA, hrpC, hrpN, and wtsE operons. Mol Plant-Microbe Interact. 14:1213-1222.


Gaudriault, S, Malandrin, L, Paulin, JP and Barny, MA 1997. DspA, an essential pathogenicity factor of Erwinia amylovora showing homology with AvrE of Pseudomonas syringae, is secreted via the Hrp secretion pathway in a DspB-dependent way. Mol Microbiol. 26:1057-1069.



Ham, JH, Majerczak, DR, Arroyo-Rodriguez, AS, Mackey, DM and Coplin, DL 2006. WtsE, an AvrE-family effector protein from Pantoea stewartii subsp. stewartii, causes disease-associated cell death in corn and requires a chaperone protein for stability. Mol Plant-Microbe Interact. 19:1092-1102.


Ham, JH, Majerczak, DR, Ewert, S, Sreerekha, M-V, Mackey, D and Coplin, D 2008. WtsE, an AvrE-family type III effector protein of Pantoea stewartii subsp. stewartii, causes cell death in non-host plants. Mol Plant Pathol. 9:633-643.



Ham, JH, Majerczak, DR, Nomura, K, Mecey, C, Uribe, F, He, S-Y, Mackey, D and Coplin, DL 2009. Multiple activities of the plant pathogen type III effector proteins WtsE and AvrE require WxxxE motifs. Mol Plant-Microbe Interact. 22:703-712.



Hatsugai, N, Igarashi, D, Mase, K, Lu, Y, Tsuda, Y, Chakravarthy, S, Wei, H-L, Foley, JW, Collmer, A, Glazebrook, J and Katagiri, F 2017. A plant effector-triggered immunity signaling sector is inhibited by pattern-triggered immunity. EMBO J. 36:2758-2769.




Hogan, CS, Mole, BM, Grant, SR, Willis, DK and Charkowski, AO 2013. The type III secreted effector DspE is required early in Solanum tuberosum leaf infection by Pectobacterium carotovorum to cause cell death, and requires Wx(3-6)D/E motifs. PLoS ONE. 8:e65534.



Hojo, H, Koyanagi, M, Tanaka, M, Kajihara, S, Ohnishi, K, Kiba, A and Hikichi, Y 2008. The hrp genes of Pseudomonas cichorii are essential for pathogenicity on eggplant but not on lettuce. Microbiology. 154:2920-2928.



Hung, NB, Ramkumar, G and Lee, YH 2014. An effector gene hopA1 influences on virulence, host specificity, and lifestyles of Pseudomonas cichorii JBC1. Res Microbiol. 165:620-629.


Jayaraman, J, Yoon, M, Applegate, ER, Stroud, EA and Templeton, MD 2020. AvrE1 and HopR1 from Pseudomonas syringae pv. actinidiae are additively required for full virulence on kiwifruit. Mol Plant Pathol. 21:1467-1480.




Jin, L, Ham, JH, Hage, R, Zhao, W, Soto-Hernández, J, Lee, SY, Paek, S-M, Kim, MG, Boone, C, Coplin, DL and Mackey, D 2016. Direct and indirect targeting of PP2A by conserved bacterial type-III effector proteins. PLoS Pathog. 12:e1005609.



Kim, H-S, Thammarat, P, Lommel, SA, Hogan, CS and Charkowski, AO 2011.
Pectobacterium carotovorum elicits plant cell death with DspE/F but the P. carotovorum DspE does not suppress callose or induce expression of plant genes early in plant-microbe interactions. Mol Plant-Microbe Interact. 24:773-786.


Larkin, MA, Blackshields, G, Brown, NP, Chenna, R, Mc-Gettigan, PA, McWilliam, H, Valentin, F, Wallace, IM, Wilm, A, Lopez, R, Thompson, JD, Gibson, TJ and Higgins, DG 2007. ClustalW and ClustalX version 2. Bioinformatics. 23:2947-2948.


Lorang, JM, Shen, H, Kobayashi, D, Cooksey, D and Keen, NT 1994. AvrA and AvrE in Pseudomonas syringae pv. tomato PT23 play a role in virulence on tomato plants. Mol Plant-Microbe Interact. 7:508-515.

Mirik, M, Aysan, Y and Sahin, F 2011. Characterization of Pseudomonas cichorii isolated from different hosts in Turkey. Int J Agric Biol. 13:203-209.
Mor, H, Manulis, S, Zuck, M, Nizan, R, Coplin, DL and Barash, I 2001. Genetic organization of the hrp gene cluster and dspAE/BF operon in Erwinia herbicola pv. gypsophilae
. Mol Plant-Microbe Interact. 14:431-436.


Nomura, K, Mecey, C, Lee, Y-N, Imboden, L, Chang, JH and He, SY 2011. Effector-triggered immunity blocks pathogen degradation of an immunity-associated vesicle traffic regulator in Arabidopsis
. Proc Natl Acad Sci U S A. 108:10774-10779.



Pauwelyn, E, Huang, C-J, Ongena, M, Leclère, V, Jacques, P, Bleyaert, P, Budzikiewicz, H, Schäfer, M and Höfte, M 2013. New linear lipopeptides produced by Pseudomonas cichorii SF1-54 are involved in virulence, swarming motility, and biofilm formation. Mol Plant-Microbe Interact. 26:585-598.


Pauwelyn, E, Vanhouteghem, K, Cottyn, B, de Vos, P, Maes, M, Bleyaert, P and Höfte, M 2011. Epidemiology of Pseudomonas cichorii, the cause of lettuce midrib rot. J Phytopathol. 159:298-305.

Preston, GM, Bertrand, N and Rainey, PB 2001. Type III secretion in plant growth-promoting Pseudomonas fluorescens SBW25. Mol Microbiol. 41:999-1014.


Ramkumar, G, Lee, SW, Weon, H-Y, Kim, B-Y and Lee, YH 2015. First report on the whole genome sequence of Pseudomonas cichorii strain JBC1 and comparison with other Pseudomonas species. Plant Pathol. 64:63-70.

West, SE, Schweizer, HP, Dall, C, Sample, AK and Runyen-Janecky, LJ 1994. Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa
. Gene. 148:81-86.


Xin, X-F, Nomura, K, Aung, K, Velásquez, AC, Yao, J, Boutrot, F, Chang, JH, Zipfel, C and He, SY 2016. Bacteria establish an aqueous living space in plants crucial for virulence. Nature. 539:524-529.




Xin, X-F, Nomura, K, Ding, X, Chen, X, Wang, K, Aung, K, Uribe, F, Rosa, B, Yao, J, Chen, J and He, SY 2015.
Pseudomonas syringae effector avirulence protein E localizes to the host plasma membrane and down-regulates the expression of the NONRACE-SPECIFIC DISEASE RESISTANCE1/HARPIN-INDUCED1-LIKE13 gene required for antibacterial immunity in Arabidopsis. Plant Physiol. 169:793-802.



- TOOLS
-
METRICS

- ORCID iDs
-
Yong Hoon Lee

https://orcid.org/0000-0001-9921-3871 - Related articles



 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Supplement
Supplement Print
Print



