 |
 |
| Plant Pathol J > Volume 32(4); 2016 > Article |
Abstract
Grapevine Algerian latent virus (GALV) is a member of the genus Tombusvirus in the Tombusviridae and infects not only woody perennial grapevine plant but also herbaceous Nicotiana benthamiana plant. In this study, we developed GALV-based gene expression and virus-induced gene silencing (VIGS) vectors in N. benthamiana. The GALV coat protein deletion vector, pGMG, was applied to express the reporter gene, green fluorescence protein (GFP), but the expression of GFP was not detected due to the necrotic cell death on the infiltrated leaves. The p19 silencing suppressor of GALV was engineered to inactivate its expression and GFP was successfully expressed with unrelated silencing suppressor, HC-Pro, from soybean mosaic virus. The pGMG vector was used to knock down magnesium chelatase (ChlH) gene in N. benthamaina and the silencing phenotype was clearly observed on systemic leaves. Altogether, the GALV-derived vector is expected to be an attractive tool for useful gene expression and VIGS vectors in grapevine as well as N. benthamiana.
The development of recombinant DNA technologies and Agrobacterium-mediated transformation enabled expression of foreign genes in plants. Two main expression strategies have been utilized for expression of foreign gene products in plants: i) transient expression through either agroinfiltration or plant virus-based vector and ii) stable transgenic expression by insertion of foreign genes into the plant genome. Plant virus-based vector systems have been revealed to be more beneficial for relatively high level accumulation and rapid production of foreign proteins than regeneration of transgenic plant (Dawson and Folimonova, 2013; Gleba et al., 2007; Hefferon, 2012; Pogue et al., 2002; Shamekova et al., 2014). Moreover, these plant virus vectors can be used for application on other susceptible plants without additional genetic transformation of each host plants (Scholthof et al., 2002; Seaberg et al., 2012).
Plant virus-based vector systems can also be utilized to knock down expression of endogenous genes in plants. This virus-induced gene silencing (VIGS) technique has been widely used to elucidate gene function(s) in plants (Burch-Smith et al., 2004; Kurth et al., 2012; Muruganantham et al., 2009). Even though many VIGS vectors have been developed in recent years, host range of these vectors were mainly restricted in annual or herbaceous plants (Brigneti et al., 2004; Fofana et al., 2004; Holzberg et al., 2002; Kang et al., 2016; Kumagai et al., 1995; Liu et al., 2002). Here, we used grapevine Algerian latent virus (GALV) which can infects both perennial grape varieties and herbaceous Nicotiana benthamiana plants and tested its ability for VIGS vector.
GALV is a member of the genus Tombusvirus, family Tombusviridae and was first isolated in Italy from an Algerian vine (Gallitelli et al., 1989). GALV has subsequently been detected from water samples in agricultural areas, Gypsophila paniculata, nipplefruit, and Limonium sinuatum (Cannizzaro et al., 1990; Fuchs et al., 1994; Fujinaga et al., 2009; Kim et al., 2012; Koenig et al., 2004; Ohki et al., 2006; Yi et al., 1992). Grapevine plants infected by GALV show different symptoms such as chlorotic spots around the veins, mild vein clearing or mottling, malformations or curling of the leaf, and dark green blistering on the leaves in different genotypes (Lovato et al., 2014).
The grape is one of the earliest domesticated fruit crops and the world’s most valuable horticultural crop for the production of wine and table grapes (Myles et al., 2011). With the importance of grapevines, virus-based vectors could provide diverse opportunities and novel possibilities as biotechnological tools for plant improvement or disease protection. There have been only two virus-based vector systems for grapevine plants including grapevine virus A (GVA) and grapevine leafroll-associated virus-2 (GLRaV-2) (Haviv et al., 2006; Kurth et al., 2012; Muruganantham et al., 2009). Diverse virus-derived vector system is required for grape varieties because stability and effectiveness of virus-based vectors depends on both plant traits and virus vectors. In this study, we constructed a GAVL coat protein (CP)-replacement vector system and used to express the foreign gene and knock down the endogenous gene in the model plant N. benthamiana.
We previously constructed the CP deletion infectious clone of GALV, pGMG (Fig. 1) and confirmed systemic infection in N. benthamiana (Kim et al., 2012). To evaluate the GALV clone as an expression vector, a green fluorescence protein (GFP) sequence was used as a reporter gene which is expressed from the subgenomic promoter, and Agrobacterium-mediated gene delivery system was utilized for inoculation of N. benthamiana plants. A PCR product (GFP; about 700 bp) was amplified from pPZP-GFP (Powers et al., 2008) by using specific primers, GFP-5-MluI and GFP-3-MluI (Table 1). The PCR product was digested by MluI and inserted into pGMG resulting in pGFG (Fig. 1). Typical GALV systemic symptoms were observed from both pGMG and pGFG-inoculated plants as shown previously (Kim et al., 2012). However, severe necrosis was induced on the inoculated leaves of pGMG and pGFG at 7 days post-infiltration (dpi) and the green fluorescence was not detected on the infiltrated leaves (Fig. 2). In other tombusviruses, the p19 protein of tomato bushy stunt virus (TBSV) and cymbidium ringspot virus is a symptom determinant and responsible for eliciting the severe systemic necrosis on N. benthamiana (Dalmay et al., 1993; Scholthof et al., 1995). To avoid the necrotic cell death of infiltrated leaves, mutation of the p19 coding region, which results in the premature stop of p19, was introduced into pGFG, pGFG19m, as described previously (Fig. 1) (Kim et al., 2012). The amino acid, Ile (isoleucine), at position 157 from the start codon of p24 was changed to a stop-codon by changing ATC to ATT resulting in inactivation of p19 without disruption of the amino acid sequence of p24. Briefly, the region of p19/p24 was amplified by PCR using GALV-5-KpnI and GALV-3-KpnI primers (Table 1), and the PCR product was cloned into pGEM-T Easy vector (Promega, Madison, WI, USA). This plasmid was used for site-directed mutagenesis and PCR was performed with the primers, GALV-sense-157 and GALV-antisense-157 (Table 1). The amplified product was treated with DpnI restriction enzyme (Roche Applied Science, Mannheim, Germany) to remove the template plasmid before the bacterial transformation of the DpnI-treated PCR mixture. The mutated sequence was confirmed by sequencing. As we expected, the green fluorescence was observed when pGFG19m was introduced to leaves (Fig. 2). However, the expression level of GFP was substantially low in the pGFG19m-infiltrated leaves and the fluorescence was abolished at 10 dpi (Fig. 3). This result suggests that wild type p19 inhibits the VIGS and cause severe necrosis, and therefore the GFP transgene was silenced by VIGS in the absence of P19 expression. To determine whether the significant attenuation of GFP expression is associated with RNA silencing and unrelated silencing suppressor can restore the GFP expression, HC-Pro from soybean mosaic virus, was co-infiltrated with pGFG19m. The result showed that the GFP expression of pGFG19m in the presence of HC-Pro was significantly increased when compared to that of pGFG19m at both 6 and 10 dpi (Fig. 3), demonstrating that the attenuation of GFP expression in pGFG19m infection is caused by RNA silencing. To test the stability of the foreign gene, viruses in plant sap were passaged three times from plant to plant by mechanical sap-inoculation. Total RNAs were prepared from each individual plant and analyzed for stable GFP gene insertion in the viral genome by RT-PCR using primers spanning the p92 and p24 cistrons. A unique band with the predicted size was observed for viral GALV-GFP from plants inoculated with progeny viruses through serial passages (data not shown). Taken together, we developed two kinds of the GALV-based virus vectors, pGMG and pGMGm19, which could apply onto herbaceous or grapevine plants and pGMGm19 can be used for expression of a transgene in N. benthamiana expressing HC-Pro in trans.
As the GALV-based vector effectively elicits RNA silencing in N. benthamiana, we decided to investigate the ability of GALV-based vector as a gene silencing vector. In other tombusvirus, CP replacement TBSV vectors were developed to knock down two endogenous genes, magnesium chelatase (ChlH) and phytoene desaturase (PDS) even though no silencing phenotype for PDS was observed (Pignatta et al., 2007). Total RNAs were extracted from N. benthamiana and cDNA was synthesized by using ChlH-3-MluI (Table 1). About 230 bp PCR fragment of ChlH was amplified with primers, ChlH-5-MluI and ChlH-3-MluI (Table 1), and the PCR product was inserted into the pGMG vector in the sense orientation resulting in pGChlH(+). The pGChlH(+) was inoculated in N. benthamiana and examined the silencing phenotype. At 12 to 14 dpi, chlorosis on newly leaves was observed (Fig. 4B). As expected, mRNA accumulation for ChlH gene was significantly lower in the leaves inoculated with pGChlH(+) than in that inoculated with mock (Fig. 4C, D). This result indicates that the ChlH gene was successfully silenced in N. benthamiana plants.
The development of a useful virus-based vector for expressing foreign genes and silencing endogenous genes is considered to be a valuable tool on the production of either useful genes or resistance genes and the study of gene function in the important crop. Until not so long ago, virus-based vectors are mainly applicable to annual plants. In grapevine as one of long-lived perennial crops, however, only two virus-derived vectors including GVA and GLRaV-2 were developed as mentioned above. GVA is a member of the genus Vitivirus, family Flexiviridae and was engineered to express the reporter gene beta-glucuronidase (GUS) and the CP gene of citrus tristeza virus in inoculated N. benthamiana plants (Haviv et al., 2006). The GVA vector also was used to silence the endogenous gene in N. benthamiana and Vitis vinifera cv. Prime plantlets (Muruganantham et al., 2009). The vector successfully expressed the reporter genes in N. benthamiana plants and silenced the PDS gene in both N. benthamiana and V. vinifera plantlets (Haviv et al., 2006; Muruganantham et al., 2009). However, this vector was not stable and the inserted sequence was removed from the genome (Haviv et al., 2006). Another GLRaV-2-based vector was utilized to express GFP and knock down the PDS and the magnesium-protoporphyrin IX chelatase (Chl1) genes in V. vinifera plants (Kurth et al., 2012). The vector had the genetic capacity to hold inserts of at least ~2 kb in size and the durability to accommodate the intact inserted cassettes for more than a year (Kurth et al., 2012).
As tombusviruses are well-studied plant viruses in the model plants such as Nicotiana species and have a wide host range, the development of tombusvirus-based vector for grapevine is expected to be a powerful tool for analysis of gene functions and delivery of resistance gene for pathogen control. To our best of knowledge, the GALV-based vector is the only tombusvirus-derived expression and VIGS vector for grapevine.
Acknowledgments
This work was supported in part by grants from the Vegetable Breeding Research Center (No. 710001-07-05) through the Agriculture Research Center program from the Ministry for Food, Agriculture, Forestry and Fisheries and the Next-Generation BioGreen 21 Program (No. PJ01101401), Rural Development Administration, Republic of Korea. SH Park, H Choi, S Kim, and WK Cho were supported by research fellowships from the Ministry of Education, Science, and Technology through the Brain Korea 21 Project.
Fig. 1
Schematic diagrams of the grapevine Algerian latent virus (GALV)-derived vectors used in this study. (A) The GALV CP-deletion clone, pGMG, was adopted from Kim et al. (2012). (B) The reporter gene, green fluorescence protein (GFP), was inserted to pGMG resulting in pGFG. (C) Site-directed mutagenesis was used to create the premature stop of the p19 coding region. The asterisk (*) indicates the stop codon (UAG). (D) The reporter gene, magnesium chelatase (ChlH), was introduced into pGMG in the sense (+) or antisense (−) orientations. LB, a left border of T-DNA; RB, a right border of T-DNA; Rz, a cis-cleaving ribozyme sequence; NOSt, a NOS terminator; sg1, subgenomic RNA1; sg2, subgenomic RNA2; MCS, multicloning site; GFP, green fluorescence protein.
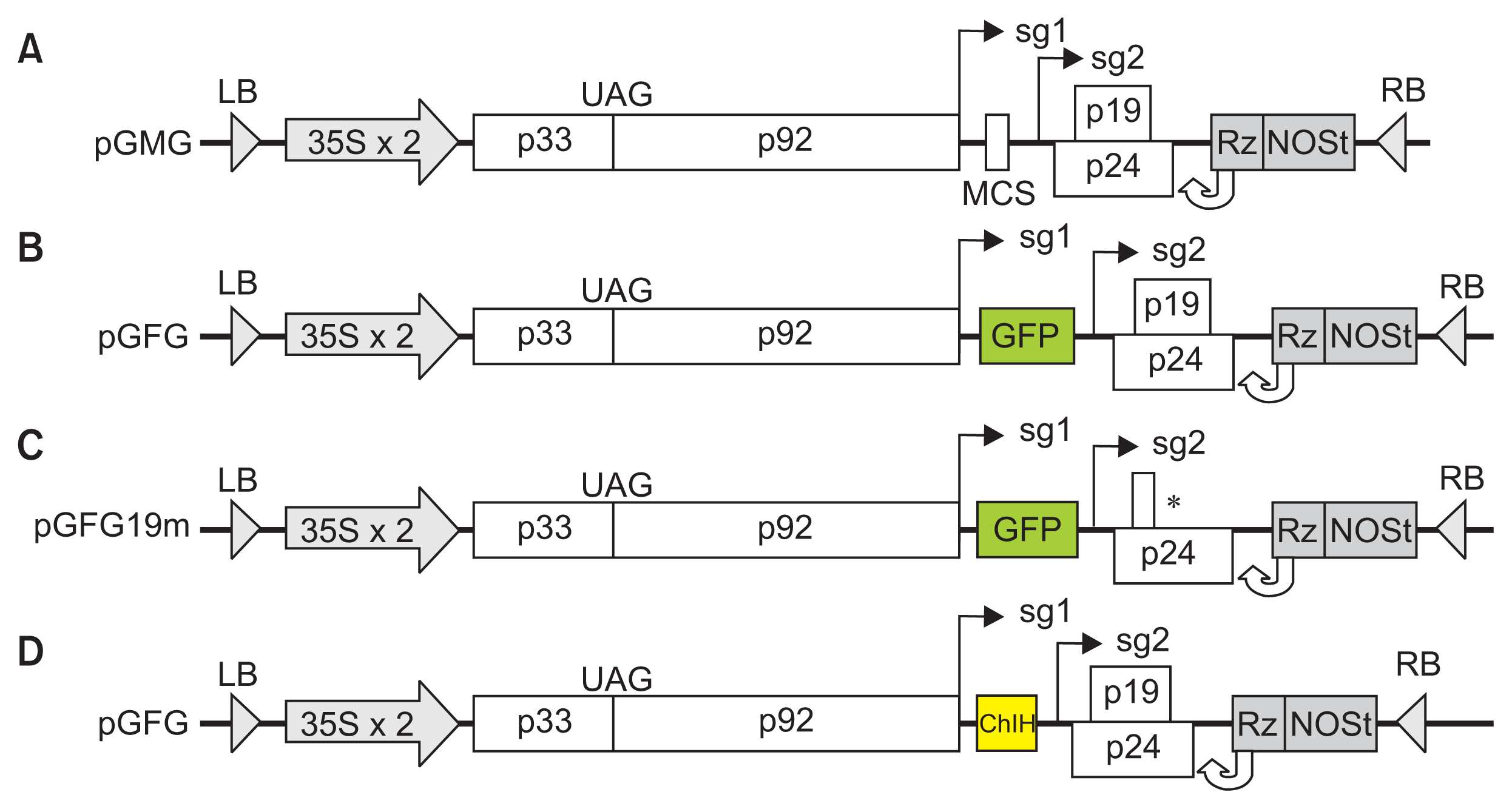
Fig. 2
Observation of expression of green fluorescence protein in Nicotiana benthamiana infected by pGFG at 7 dpi. Green fluorescence was observed in whole plants (A) using an ultraviolet (UV) lamp and the infiltrated leaves (B) by a confocal microscope (× 200).
Fig. 3
Effect of silencing suppressors on expression of a foreign gene in the grapevine Algerian latent virus-based vectors. Green fluorescence protein expression was visualized upon either pGFG19m only (A) or co-infiltration of pGFG19m and HC-Pro (B) at 6 and 10 dpi.

Fig. 4
Silencing the magnesium chelatase (ChlH) gene in Nicotiana benthamiana plants by pGChlH(+). Photographs show mock plants (A) and plants infiltrated by the silencing vector (B). The photobleached phenotype was observed in systemic leaves of N. benthamiana plants infected by both pGChlH(+) at 21 dpi (B). (C, D) Semi-quantitative and real-time PCR validations of the expression level of ChlH mRNA. Significantly down-regulated ChlH gene in pGChlH(+) inoculated plants at 21 dpi was observed.
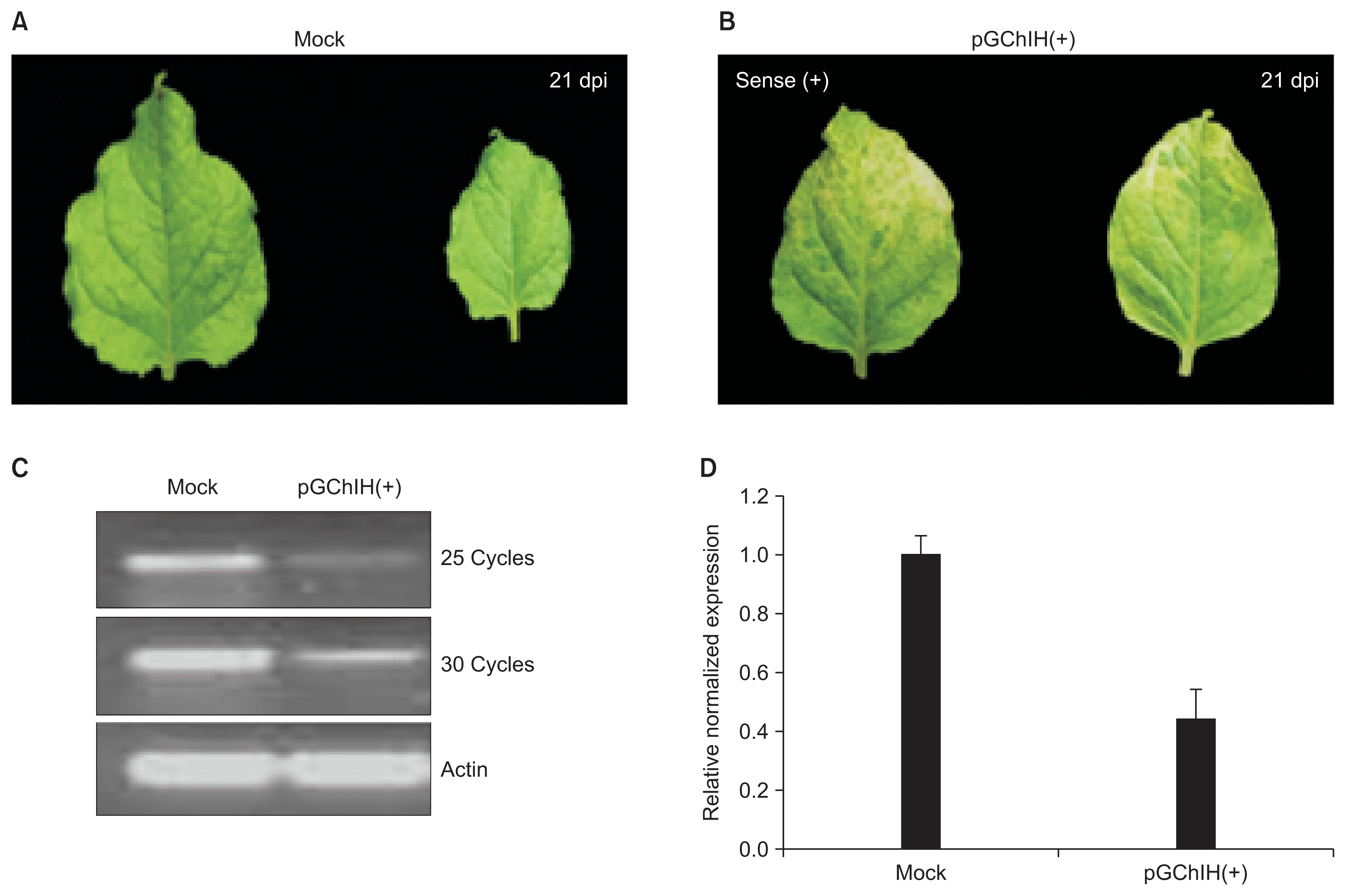
Table 1
Oligonucleotides used in this study
| Oligonucleotides | Sequences (5′ → 3′)* |
|---|---|
| GFP-5-MluI | CGACGCGTCATCATCATCATCATCATGTGAGC |
| GFP-3-MluI | CGACGCGTCTTGTACAGCTCGTCCATGCC |
| GALV-5-KpnI | GGGGTACCCTCGAGAGATCTTAGTGCATGCCATGCAAGAAAGGGG |
| GALV-3-KpnI | GGGGTACCGGGCTGCATTTCTGAAT |
| GALV-sense-157 | GAGTGAGGATTAGGATCATCC |
| GALV-antisense-157 | GGATGATCCTAATCCTCACTC |
| ChlH-5-MluI | CGACGCGTAGCCAGAAGACC |
| ChlH-3-MluI | CGACGCGTTGCCAACACCAG |
References
Brigneti, G, Martín-Hernández, AM, Jin, H, Chen, J, Baulcombe, DC, Baker, B and Jones, JD 2004. Virus-induced gene silencing in Solanum species. Plant J. 39:264-272.


Burch-Smith, TM, Anderson, JC, Martin, GB and Dinesh-Kumar, SP 2004. Applications and advantages of virus-induced gene silencing for gene function studies in plants. Plant J. 39:734-746.


Cannizzaro, G, Rosciglione, B and Castellano, MA 1990. The presence of phytopathogenic viruses in waterways of western Sicily. Inf Fitopatol. 40:55-56.
Dalmay, T, Rubino, L, Burgyán, J, Kollár, A and Russo, M 1993. Functional analysis of cymbidium ringspot virus genome. Virology. 194:697-704.


Dawson, WO and Folimonova, SY 2013. Virus-based transient expression vectors for woody crops: a new frontier for vector design and use. Annu Rev Phytopathol. 51:321-337.


Fofana, IB, Sangaré, A, Collier, R, Taylor, C and Fauquet, CM 2004. A geminivirus-induced gene silencing system for gene function validation in cassava. Plant Mol Biol. 56:613-624.



Fuchs, E, Grüntzig, M, Auerbach, I, Einecke, I, Müller, C and Krägenow, M 1994. On the occurrence of plant pathogenic viruses in waters in the region of Halle/Saale (German Federal State of Saxony-Anhalt). Arch Phytopathol Plant Prot. 29:133-141.
Fujinaga, M, Ogiso, H, Wakabayashi, H, Morikawa, T and Natsuaki, T 2009. First report of a grapevine Algerian latent virus disease on statice plants (Limonium sinuatum) in Japan. J Gen Plant Pathol. 75:157-159.


Gallitelli, D, Martelli, GP and Di Franco, A 1989. Grapevine Algerian latent virus, a newly recognized tombusvirus. Phytoparasitica. 17:61-62.
Gleba, Y, Klimyuk, V and Marillonnet, S 2007. Viral vectors for the expression of proteins in plants. Curr Opin Biotechnol. 18:134-141.


Haviv, S, Galiakparov, N, Goszczynski, DE, Batuman, O, Czosnek, H and Mawassi, M 2006. Engineering the genome of grapevine virus A into a vector for expression of proteins in herbaceous plants. J Virol Methods. 132:227-231.


Hefferon, KL 2012. Plant virus expression vectors set the stage as production platforms for biopharmaceutical proteins. Virology. 433:1-6.


Holzberg, S, Brosio, P, Gross, C and Pogue, GP 2002. Barley stripe mosaic virus-induced gene silencing in a monocot plant. Plant J. 30:315-327.


Kang, M, Seo, JK, Cho, H, Choi, HS and Kim, KH 2016. Establishment of a simple and rapid gene delivery system for cucurbits by using engineered zucchini yellow mosaic virus. Plant Pathol J. 32:70-76.



Kim, S, Cho, WK, Lee, HG, Park, SH, Sohn, SH and Kim, KH 2012. The p19 protein of grapevine Algerian latent virus is a determinant of systemic infection of Chenopodium quinoa. Virus Res. 165:81-89.


Koenig, R, Verhoeven, JT, Fribourg, CE, Pfeilstetter, E and Lesemann, DE 2004. Evaluation of various species demarcation criteria in attempts to classify ten new tombusvirus isolates. Arch Virol. 149:1733-1744.



Kumagai, MH, Donson, J, della-Cioppa, G, Harvey, D, Hanley, K and Grill, LK 1995. Cytoplasmic inhibition of carotenoid biosynthesis with virus-derived RNA. Proc Natl Acad Sci U S A. 92:1679-1683.



Kurth, EG, Peremyslov, VV, Prokhnevsky, AI, Kasschau, KD, Miller, M, Carrington, JC and Dolja, VV 2012. Virus-derived gene expression and RNA interference vector for grapevine. J Virol. 86:6002-6009.




Liu, Y, Schiff, M and Dinesh-Kumar, SP 2002. Virus-induced gene silencing in tomato. Plant J. 31:777-786.


Lovato, A, Faoro, F, Gambino, G, Maffi, D, Bracale, M, Polverari, A and Santi, L 2014. Construction of a synthetic infectious cDNA clone of grapevine Algerian latent virus (GALV-Nf) and its biological activity in Nicotiana benthamiana and grapevine plants. Virol J. 11:186




Muruganantham, M, Moskovitz, Y, Haviv, S, Horesh, T, Fenigstein, A, du Preez, J, Stephan, D, Burger, JT and Mawassi, M 2009. Grapevine virus A-mediated gene silencing in Nicotiana benthamiana and Vitis vinifera. J Virol Methods. 155:167-174.


Myles, S, Boyko, AR, Owens, CL, Brown, PJ, Grassi, F, Aradhya, MK, Prins, B, Reynolds, A, Chia, JM, Ware, D, Bustamante, CD and Buckler, ES 2011. Genetic structure and domestication history of the grape. Proc Natl Acad Sci U S A. 108:3530-3535.



Ohki, T, Uematsu, S, Nakayama, Y, Lesemann, DE, Honda, Y, Tsuda, S and Fujisawa, I 2006. Characterization of grapevine Algerian latent virus isolated from nipplefruit (Solanum mammosum) in Japan. J Gen Plant Pathol. 72:119-122.


Pignatta, D, Kumar, P, Turina, M, Dandekar, A and Falk, BW 2007. Quantitative analysis of efficient endogenous gene silencing in Nicotiana benthamiana plants using tomato bushy stunt virus vectors that retain the capsid protein gene. Mol Plant-Microbe Interact. 20:609-618.


Pogue, GP, Lindbo, JA, Garger, SJ and Fitzmaurice, WP 2002. Making an ally from an enemy: plant virology and the new agriculture. Annu Rev Phytopathol. 40:45-74.


Powers, JG, Sit, TL, Heinsohn, C, George, CG, Kim, KH and Lommel, SA 2008. The red clover necrotic mosaic virus RNA-2 encoded movement protein is a second suppressor of RNA silencing. Virology. 381:277-286.


Scholthof, HB, Scholthof, KB and Jackson, AO 1995. Identification of tomato bushy stunt virus host-specific symptom determinants by expression of individual genes from a potato virus X vector. Plant Cell. 7:1157-1172.



Scholthof, KB, Mirkov, TE and Scholthof, HB 2002. Plant virus gene vectors: biotechnology applications in agriculture and medicine. Genet Eng (N Y). 24:67-85.


Seaberg, BL, Hsieh, YC, Scholthof, KB and Scholthof, HB 2012. Host impact on the stability of a plant virus gene vector as measured by a new fluorescent local lesion passaging assay. J Virol Methods. 179:289-294.


Shamekova, M, Mendoza, MR, Hsieh, YC, Lindbo, J, Omarov, RT and Scholthof, HB 2014. Tombusvirus-based vector systems to permit over-expression of genes or that serve as sensors of antiviral RNA silencing in plants. Virology. 452-453:159-165.

Yi, L, Lesemann, DE, Koenig, R, Rüdel, M and Pfeilstetter, E 1992. Isometric plant viruses in ditches and streams in agricultural areas: recovery of previously found viruses and identification of hitherto unrecorded carmoand tombusviruses including grapevine Algerian latent virus. J Phytopathol. 134:121-132.

- TOOLS



 PDF Links
PDF Links PubReader
PubReader Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Print
Print




