 |
 |
| Plant Pathol J > Volume 36(3); 2020 > Article |
|
Abstract
Type VI secretion system (T6SS) is a contact-dependent secretion system, employed by most gram-negative bacteria for translocating effector proteins to target cells. The present study was conducted to investigate T6SS in Xanthomonas oryzae pv. oryzae (Xoo), which causes bacterial blight in rice, and to unveil its functions. Two T6SS clusters were found in the genome of Xoo PXO99A. The deletion mutants, Δhcp1, Δhcp2, and Δhcp12, targeting the hcp gene in each cluster, and a double-deletion mutant targeting both genes were constructed and tested for growth rate, pathogenicity to rice, and inter-bacterial competition ability. The results indicated that hcp in T6SS-2, but not T6SS-1, was involved in bacterial virulence to rice plants. However, neither T6SS-1 nor T6SS-2 had any effect on the ability to compete with Escherichia coli or other bacterial cells. In conclusion, T6SS gene clusters in Xoo have been characterized, and its role in virulence to rice was confirmed.
Bacterial blight caused by Xanthomonas oryzae pv. oryzae (Xoo) is a widespread and destructive disease of rice. The yield loss can reach 70% in heavily infected fields, and infection of plants at the seedling stage may result in Kresek syndrome, possibly leading to complete destruction of the crop (Mew et al., 1993; Salzberg et al., 2008). Generally, Xoo enters rice leaves through wounds or hydathodes, and systemically spread through the xylem to other plant parts. The pathogenesis of Xoo is a complex process involving several virulence factors such as exopolysaccharides, extracellular enzymes, adhesins, type 3 secretion system, and motility (Das et al., 2009; Rajeshwari et al., 1997; Ray et al., 2002; Song and Yang, 2010). Although several virulence factors involved in bacterial blight have been discovered, the pathogenesis is not yet perfectly understood.
Bacterial secretion systems are classified into seven groups (Types I-VII) according to their structure, function, and the specific effectors involved (Green and Mecsas, 2016). The type 6 secretion system (T6SS) is a contact-dependent injection system that delivers effector proteins into bacteria or eukaryotic targets (Schwarz et al., 2010). It has been first identified in Vibrio cholerae in 2006 and was then found in most gram-negative bacteria, which use it to translocate effector proteins to target cells (Jani and Cotter, 2010; Leiman et al., 2009; Pukatzki et al., 2006).
T6SS has been reported as a virulence factor in various pathogenic bacteria, such as Pseudomonas aeruginosa, V. cholerae, and Burkholderia pseudomallei (Burtnick et al., 2011; Hood et al., 2010; Joshi et al., 2017). P. aeruginosa exhibited decreased virulence to Arabidopsis thaliana when the expression of hcp, which is a main component of T6SS, was restricted (Lesic et al., 2009). However, not all T6SSs are involved in virulence to eukaryotic hosts, and further research indicated that T6SS plays a key role in the competition between bacteria. The pathogenicity of P. syringae pv. tomato to tomato plants was not affected by defective T6SS, yet, its ability to compete with other Escherichia coli and yeast was highly dependent on T6SS (Haapalainen et al., 2012). Although T6SS has been reported as a virulence factor in other plant pathogens, its role in Xoo is still unknown.
T6SS is structurally homologous to T4 bacteriophage tail proteins (Pell et al., 2009). The essential core component consists of 13 proteins named TssA to TssM (Joshi et al., 2017). The Tss complex forms a basal plate anchored to the inner membrane, and the membrane anchoring complex is comprised of TssJ, TssL, and TssM (Aschtgen et al., 2010). TssB and TssC form the outer sheath, which accelerates the delivery of the inner tube to the target cell (Basler et al., 2012). The inner tube is composed of hexamers of Hcp (hemolysin co-regulated protein). VgrG (valine-glycine repeat protein G), which is located at the tip of the Hcp complex, punctures the outer membrane of the target cell. The key components of T6SS are Hcp and VgrG proteins, which resemble the phage tail and needle complex, respectively (Yang et al., 2018). All T6SS clusters essentially contain Hcp and VgrG proteins, and when these proteins are disrupted, the entire T6SS is dysfunctional.
As T6SS has not yet been examined in Xoo, this study aimed to characterize T6SS in Xoo and to explore its possible involvement in the virulence to host plants and/or in inter-bacterial competition.
The genome of Xoo PXO99A, the Philippine race 6 strain, which is composed of a single chromosome of 5,240,075 bp, has been previously analyzed (Keshri et al., 2014). The analysis revealed two T6SS gene clusters, T6SS-1 (PXO_02040-PXO_02050, PXO_04696, PXO_04699), containing all the 13 essential proteins of T6SS, and T6SS 2 (PXO_00245-PXO_00266), lacking TssJ, one of the membrane anchoring complexes (Fig. 1A).
In order to study the functions of T6SS-1 and T6SS-2, markerless deletion mutants targeting the hcp gene in each cluster, as a crucial component of T6SS, were constructed. The Δhcp1 mutant is deleted for the 492 bp of the hcp1 gene (PXO_02047) of T6SS-1, Δhcp2 is deleted for the 427 bp of the hcp2 gene (PXO_00264) in T6SS-2, and Δhcp12 is a double mutant deleted for both genes (PXO_02047 and PXO_00264).
The wild-type strain, Xoo PXO99A, and each mutant strain were cultured at 28┬░C with shaking at 200 rpm in nutrient broth and nutrient agar (MBcell, Seoul, Korea), Escherichia coli strains were cultured in Luria-Bertani (LB; Difco, Detroit, MI, USA) agar. For the growth curve, the bacteria were cultured in broth medium at 37┬░C with shaking at 200 rpm, while tryptone yeast extract glucose with 0.6% agar (tryptone, 1 g/l; yeast extract, 0.5 g/l; glucose, 10 g/l; NaCl, 1 g/l; agar 0.6%) was used for the swarming motility assay.
Deletion mutants in Xoo PXO99A were designed using the pK18mobsacB plasmid, which can induce deletion by double-crossover homologous recombination (Sch├żfera et al., 1994). The primers for amplification of fragments upstream and downstream of each deletion mutant strains were designed according to Gibson assembly reaction (Gibson et al., 2010). All primers used in this study are shown in Supplementary Table 1. The upstream and downstream DNA fragment were amplified by PCR, mixed at the same concentration, and re-amplified by secondary PCR using the LF and RR primers of each strain. LF primers contained EcoRI restriction enzyme digestion sites, while RR primers contained HindIII sites. The product of secondary PCR was digested with EcoRI and HindIII (NEB, Ipswich, MA, USA). To generate Xoo with the recombinant plasmids, the wild-type Xoo (recipient strain) and E. coli S17-1 (donor strain), harboring identical recombinant plasmids, were conjugated by the bi-parental mating method (Shen et al., 2002). Properly transformed cells were incubated in NB medium for 2 days, and smeared on peptone sucrose agar (PSA) containing 20 ╬╝g/ml of cephalexin and 10% sucrose (w/v). The obtained deletion mutants were confirmed by PCR assays and sequencing (Fig. 1B). All bacterial strains and plasmids used are listed in Supplementary Table 2. The growth rates and bacterial swarming motility of the obtained mutants were compared to those of the wild-type Xoo PXO99A to evaluate the role of T6SS.
A pathogenicity test was performed to evaluate the involvement of T6SS in Xoo virulence to rice plants. Six-week-old rice (Oryza sativa L. Saeilmi) plants were inoculated by the mutant and the wild-type strains using the scissor clipping method (Mew et al., 1993; Staskawicz et al., 1995). Briefly, each strain was cultured on peptone sucrose broth, and the cells were harvested by centrifugation, washed twice, and re-suspended in sterilized distilled water (SDW). The final OD600 was adjusted to 0.8. At 14 days post-inoculation, the disease severity was evaluated by measuring the lesion lengths. The bacterial population colonizing the rice leaves was assessed by grinding the leaves in SDW, spotting serial dilutions on PSA plates, and expressed as colony forming units (cfu)/leaf.
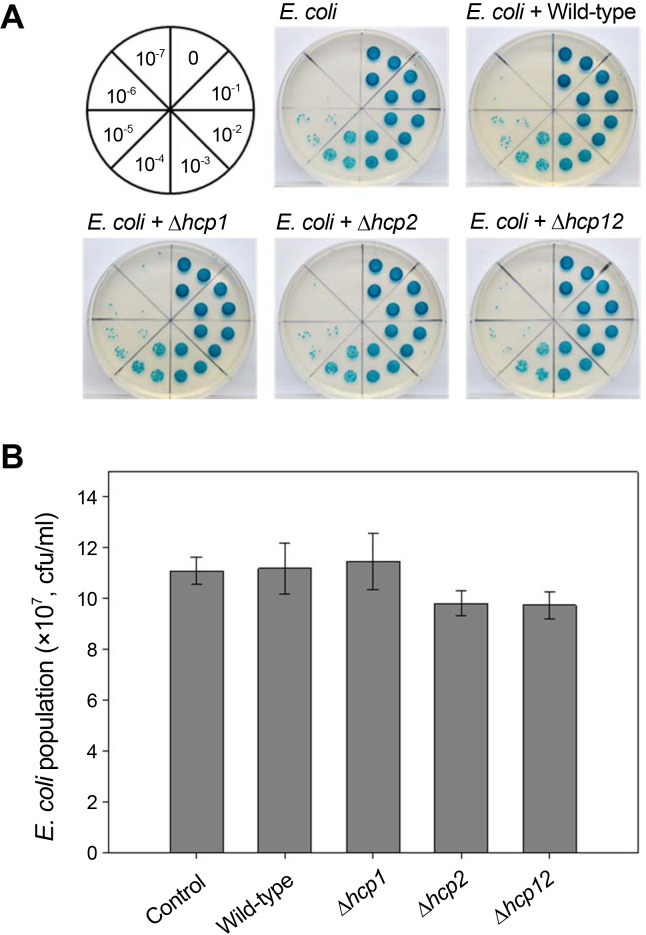
To test whether T6SS was involved in the competition with other bacteria, an inter-bacterial competition assay was conducted using E. coli DH5╬▒ cells as a prey, following a previously described method with some modifications (Ma et al., 2014). The mutants and the wild-type Xoo strains were co-cultured with E. coli DH5╬▒ cells harboring the pCRISPomyces-2 plasmid, allowing for survival in apramycin media and alpha complementation for ╬▓-galactosidase. Bacteria were cultured at a 1:1 ratio in forms of patches on LB plates, and incubated for 12 h. Following re-suspension of the patches, the survival of the E. coli prey cells was assessed after spotting serially diluted patches from 0 to 10-7 on LB plates containing apramycin (100 ╬╝g/ml) and 5-bromo-4-chloro-indolyl-╬▓-D-galactopyranoside (X-gal, 40 ╬╝g/ml). The number of blue colonies on the plate indicated the number of surviving E. coli cells.
The interbacterial competition was also performed using the wild-type and the double mutant Δhcp12 against other bacterial strains as preys (B. glumae BGR1, B. gladioli KACC18962, B. plantarii KACC18965, Ralstonia solanacearum and Burkholderia sp. KJ006). Briefly, Xoo wild-type or Δhcp12 were inoculated into LB gar 0.8% to a final OD600 value of 0.2 that was then poured on LB agar 1.5%. 10 μl from the prey cultures adjusted to OD600 = 0.2 were spotted onto the plates and incubated for 24 h at 30ºC. Bacterial growth was then observed in comparison to the controls.
Statistical analysis was performed using the Statistical Analysis Systems (SAS Institute, Cary, NC, USA). Analysis of variance was performed using the general linear model, and separation of means was determined by Tukey's Honest Significant Difference test, setting P < 0.05 as the statistical significance threshold.
The growth rates and bacterial swarming motility of Δhcp1, Δhcp2, and Δhcp12 mutant strains were not significantly different from those of the wild-type (Fig. 2). These results indicated that neither T6SSs were involved in Xoo growth or motility, which are critical virulence factors in these bacteria (Tian et al., 2015).
When the mutant and wild-type strains were tested for virulence to rice plants, the mutant strains defective in Δhcp2 and Δhcp12 showed a significant reduction in disease severity compared to the wild-type strain (Fig. 3A and B). However, there was no significant difference in disease severity between the Δhcp1 mutant compared to the wildtype strain (Fig. 3A and B). The assessment of bacterial population surviving in rice leaves, expressed as (cfu/leaf), was consistent with the results of disease severity, as the mutant strains, Δhcp2 and Δhcp12, showed significantly reduced survival compared to the control and the Δhcp1 mutant (Fig. 3C). These results implied that Hcp of T6SS-2 was involved in the virulence of Xoo to host rice plants, while no apparent role was associated with T6SS-1. When the complemented mutant Δhcp2C was tested to confirm hcp2 involvement in the virulence to rice, the complemented strain showed recovery of virulence and Xoo population surviving of the leaves comparable to the wild-type (Fig. 3D-F).
According to the inter-bacterial competition assay, there was no difference in the number of surviving E. coli prey cells after co-cultivation with the mutants Δhcp1, Δhcp2, and Δhcp12, as compared to the wild-type (Fig. 4). Additionally, there were no interactions with other pathogenic bacterial spp. (B. glumae BGR1, B. gladioli KACC18962, B. plantarii KACC18965, R. solanacearum and Burkholderia sp. KJ006) as shown in Supplementary Fig. 1. Based on these findings, it could be inferred that T6SS in Xoo is not involved in inter-bacterial competition. However, further research is required to confirm that T6SS has no role in the interaction with other surrounding microbes.
In this study, screening of the Xoo PXO99A genome resulted in the discovery of two T6SS clusters. A deletion mutation of the hcp gene in each cluster was constructed and the obtained strains were tested for virulence to rice plants and for inter-bacterial competition compared to the wild-type strain. The results indicated that the hcp deletion in T6SS-2 partially impaired the bacterial virulence to rice plants, whereas its deletion in T6SS-1 did not result in any significant difference in virulence to rice. This finding was in accordance with previous work on the association of T6SS with the virulence to eukaryotic hosts. Regarding phytopathogenic bacteria, T6SS was reported to be important for the virulence of R. solanacearum to host plants (Zhang et al., 2014).
It has been previously reported that, in some bacterial species, T6SS is involved in the translocation of toxic effectors to other bacteria as a mean of survival competitive advantage (Haapalainen et al., 2012; Russell et al., 2011). However, the inter-bacterial competition assay conducted in this study indicated that disruption of both T6SS clusters did not affect Xoo ability to compete with other bacteria.
The NCBI BLAST analysis demonstrated that Hcp2 in Xoo has a highly conserved amino acid sequence, with respect to the Hcp homolog harbored by X. citri (Xac4145), which is involved in the resistance to the amoeba, Dictyostelium (Bayer-Santos et al., 2018). Therefore, Hcp2 in Xoo might be involved in the interaction with other eukaryotic cells, including amoeba and yeast, in addition to rice plant. The amino acid sequence of Hcp1, however, did not correspond to other Hcp sequences with known functions.
Most T6SSs are strictly regulated at the transcriptional level to ensure the activation of the system only under appropriate conditions (Bernard et al., 2010). T6SS regulation in Xoo has not been studied yet. In V. cholerae, P. aeruginosa and Pectobacterium atrosepticum, the regulation of T6SS is mediated by quorum sensing (Ishikawa et al., 2009; Lesic et al., 2009; Liu et al., 2008). In addition, according to the first relevant study in the genus Xanthomonas, the T6SS of X. citri, was regulated by EcfK, an alternative sigma factor of the ECF family and a transmembrane eukaryotic-like serine threonine kinase (Bayer-Santos et al., 2018).
Since the main role of T6SS is the translocation of various effectors to host cells, it is important to study the secreted effectors. Comparative proteomics analysis in wild-type and T6SS deficient strain could be utilized to identify effectors that are delivered by Xoo to host cells via T6SS (Lien and Lai, 2017; Schwarz et al., 2010).
In conclusion, the present study is the first to functionally characterize T6SS in Xoo and to unveil the role of T6SS-2 in bacterial virulence to rice plants. The presented results are a step toward the complete understanding of the pathogenesis of bacterial blight caused by Xoo, and may facilitate the development of novel disease management options.
Supplementary materials are available at The Plant Pathology Journal website (http://www.ppjonline.org/).
Acknowledgments
This work was supported by a 2-year Research Grant of Pusan National University (Y.-S.S.).
References
Aschtgen, M.-S., Gavioli, M., Dessen, A., Lloub├©s, R. and Cascales, E. 2010. The SciZ protein anchors the enteroaggregative Escherichia coli Type VI secretion system to the cell wall. Mol. Microbiol. 75:886-899.


Basler, M., Pilhofer, M., Henderson, G. P., Jensen, G. J. and Mekalanos, J. J. 2012. Type VI secretion requires a dynamic contractile phage tail-like structure. Nature. 483:182-186.



Bayer-Santos, E., Lima, L. D. P., Ceseti, L. M., Ratagami, C. Y., de Santana, E. S., da Silva, A. M., Farah, C. S. and Alvarez-Martinez, C. E. 2018. Xanthomonas citri T6SS mediates resistance to Dictyostelium predation and is regulated by an ECF Žā factor and cognate Ser/Thr kinase. Environ. Microbiol. 20:1562-1575.


Bernard, C. S., Brunet, Y. R., Gueguen, E. and Cascales, E. 2010. Nooks and crannies in type VI secretion regulation. J. Bacteriol. 192:3850-3860.



Burtnick, M. N., Brett, P. J., Harding, S. V., Ngugi, S. A., Ribot, W. J., Chantratita, N., Scorpio, A., Milne, T. S., Dean, R. E., Fritz, D. L., Peacock, S. J., Prior, J. L., Atkins, T. P. and Deshazer, D. 2011. The cluster 1 type VI secretion system is a major virulence determinant in Burkholderia pseudomallei. Infect. Immun. 79:1512-1525.



Das, A., Rangaraj, N. and Sonti, R. V. 2009. Multiple adhesin-like functions of Xanthomonas oryzae pv. oryzae are involved in promoting leaf attachment, entry, and virulence on rice. Mol. Plant-Microbe Interact. 22:73-85.


Gibson, D. G., Glass, J. I., Lartigue, C., Noskov, V. N., Chuang, R.-Y., Algire, M. A., Benders, G. A., Montague, M. G., Ma, L., Moodie, M. M., Merryman, C., Vashee, S., Krishnakumar, R., Assad-Garcia, N., Andrews-Pfannkoch, C., Denisova, E. A., Young, L., Qi, Z.-Q., Segall-Shapiro, T. H., Calvey, C. H., Parmar, P. P., Hutchison, C. A., Smith, H. O. and Venter, J. C. 2010. Creation of a bacterial cell controlled by a chemically synthesized genome. Science. 329:52-56.


Green, E. R. and Mecsas, J. 2016. Bacterial secretion systems: an overview. Microbiol. Spectr. 4:VMBF-0012-2015.

Haapalainen, M., Mosorin, H., Dorati, F., Wu, R.-F., Roine, E., Taira, S., Nissinen, R., Mattinen, L., Jackson, R., Pirhonen, M. and Lin, N.-C. 2012. Hcp2, a secreted protein of the phytopathogen Pseudomonas syringae pv. tomato DC3000, is required for fitness for competition against bacteria and yeasts. J. Bacteriol. 194:4810-4822.



Hood, R. D., Singh, P., Hsu, F., G├╝vener, T., Carl, M. A., Trinidad, R. R. S., Silverman, J. M., Ohlson, B. B., Hicks, K. G., Plemel, R. L., Li, M., Schwarz, S., Wang, W. Y., Merz, A. J., Goodlett, D. R. and Mougous, J. D. 2010. A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe. 7:25-37.



Ishikawa, T., Rompikuntal, P. K., Lindmark, B., Milton, D. L. and Wai, S. N. 2009. Quorum sensing regulation of the two hcp alleles in Vibrio cholerae O1 strains. PLoS ONE. 4:e6734.



Jani, A. J. and Cotter, P. A. 2010. Type VI secretion: not just for pathogenesis anymore. Cell Host Microbe. 8:2-6.



Joshi, A., Kostiuk, B., Rogers, A., Teschler, J., Pukatzki, S. and Yildiz, F. H. 2017. Rules of engagement: the type VI secretion system in Vibrio cholerae. Trends Microbiol. 25:267-279.


Keshri, V., Singh, D. P., Prabha, R., Rai, A. and Sharma, A. K. 2014. Genome subtraction for the identification of potential antimicrobial targets in Xanthomonas oryzae pv. oryzae PXO99A pathogenic to rice. 3 Biotech. 4:91-95.


Leiman, P. G., Basler, M., Ramagopal, U. A., Bonanno, J. B., Sauder, J. M., Pukatzki, S., Burley, S. K., Almo, S. C. and Mekalanos, J. J. 2009. Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc. Natl. Acad. Sci. U. S. A. 106:4154-4159.



Lesic, B., Starkey, M., He, J., Hazan, R. and Rahme, L. G. 2009. Quorum sensing differentially regulates Pseudomonas aeruginosa type VI secretion locus I and homologous loci II and III, which are required for pathogenesis. Microbiology. 155:2845-2855.



Lien, Y. W. and Lai, E. M. 2017. Type VI secretion effectors: methodologies and biology. Front. Cell. Infect. Microbiol. 7:254



Liu, H., Coulthurst, S. J., Pritchard, L., Hedley, P. E., Ravensdale, M., Humphris, S., Burr, T., Takle, G., Brurberg, M.-B., Birch, P. R. J., Salmond, G. P. C. and Toth, I. K. 2008. Quorum sensing coordinates brute force and stealth modes of infection in the plant pathogen Pectobacterium atrosepticum. PLoS Pathog. 4:e1000093.



Ma, L.-S., Hachani, A., Lin, J.-S., Filloux, A. and Lai, E.-M. 2014. Agrobacterium tumefaciens deploys a superfamily of type VI secretion DNase effectors as weapons for interbacterial competition in planta. Cell Host Microbe. 16:94-104.



Mew, T. W., Alvarez, A. M., Leach, J. E. and Swings, J. 1993. Focus on bacterial blight of rice. Plant Dis. 77:5-12.

Pell, L. G., Kanelis, V., Donaldson, L. W., Howell, P. L. and Davidson, A. R. 2009. The phage lambda major tail protein structure reveals a common evolution for long-tailed phages and the type VI bacterial secretion system. Proc. Natl. Acad. Sci. U. S. A. 106:4160-4165.



Pukatzki, S., Ma, A. T., Sturtevant, D., Krastins, B., Sarracino, D., Nelson, W. C., Heidelberg, J. F. and Mekalanos, J. J. 2006. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc. Natl. Acad. Sci. U. S. A. 103:1528-1533.



Rajeshwari, R., Yashitola, J., Sonti, R. V. and Reddy, A. P. K. 1997. Characteristics of stationary-phase variation affecting virulence in Xanthomonas oryzae pv. oryzae. Can. J. Microbiol. 43:862-867.

Ray, S. K., Rajeshwari, R., Sharma, Y. and Sonti, R. V. 2002. A high-molecular-weight outer membrane protein of Xanthomonas oryzae pv. oryzae exhibits similarity to non-fimbrial adhesins of animal pathogenic bacteria and is required for optimum virulence. Mol. Microbiol. 46:637-647.


Russell, A. B., Hood, R. D., Bui, N. K., LeRoux, M., Vollmer, W. and Mougous, J. D. 2011. Type VI secretion delivers bacteriolytic effectors to target cells. Nature. 475:343-347.



Salzberg, S. L., Sommer, D. D., Schatz, M. C., Phillippy, A. M., Rabinowicz, P. D., Tsuge, S., Furutani, A., Ochiai, H., Delcher, A. L., Kelley, D., Madupu, R., Puiu, D., Radune, D., Shumway, M., Trapnell, C., Aparna, G., Jha, G., Pandey, A., Patil, P. B., Ishihara, H., Meyer, D. F., Szurek, B., Verdier, V., Koebnik, R., Dow, J. M., Ryan, R. P., Hirata, H., Tsuyumu, S., Seo, Y.-S., Sriariyanum, M., Ronald, P. C., Sonti, R. V., Van Sluys, M.-A., Leach, J. E., White, F. F. and Bogdanove, A. J. 2008. Genome sequence and rapid evolution of the rice pathogen Xanthomonas oryzae pv. oryzae PXO99A. BMC Genomics. 9:204



Sch├żfera, A., Tauch, A., J├żger, W., Kalinowski, J., Thierbach, G. and P├╝hler, A. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene. 145:69-73.


Schwarz, S., West, T. E., Boyer, F., Chiang, W.-C., Carl, M. A., Hood, R. D., Rohmer, L., Tolker-Nielsen, T., Skerrett, S. J. and Mougous, J. D. 2010. Burkholderia type VI secretion systems have distinct roles in eukaryotic and bacterial cell interactions. PLoS Pathog. 6:e1001068.



Shen, Y., Sharma, P., da Silva, F. G. and Ronald, P. 2002. The Xanthomonas oryzae pv. oryzae raxP and raxQ genes encode an ATP sulphurylase and adenosine-5'-phosphosulphate kinase that are required for AvrXa21 avirulence activity. Mol. Microbiol. 44:37-48.


Song, C. and Yang, B. 2010. Mutagenesis of 18 type III effectors reveals virulence function of XopZ (PXO99) in Xanthomonas oryzae pv. oryzae. Mol. Plant-Microbe Interact. 23:893-902.


Staskawicz, B. J., Ausubel, F. M., Baker, B. J., Ellis, J. G. and Jones, J. D. 1995. Molecular genetics of plant disease resistance. Science. 268:661-667.


Tian, F., Yu, C., Li, H., Wu, X., Li, B., Chen, H., Wu, M. and He, C. 2015. Alternative sigma factor RpoN2 is required for flagellar motility and full virulence of Xanthomonas oryzae pv. oryzae. Microbiol. Res. 170:177-183.


Fig.┬Ā1
(A) Schematic diagram showing the genetic organization of the two clusters of type VI secretion system (T6SS) found in the genome of Xanthomonas oryzae pv. oryzae PXO99A. The genes are represented by colored arrows and the directions of arrows indicate the transcription direction. The hcp genes examined in this study are indicated by arrows. (B) Molecular confirmation of hcp1 and hcp2 deletion mutant strains by PCR assay using the 5ŌĆ▓-upstream and 3ŌĆ▓-downstream primers of each gene. PCR products were loaded on 1% agarose gel. M, 100 bp DNA ladder; WT, wild-type X. oryzae pv. oryzae PXO99A genomic DNA.
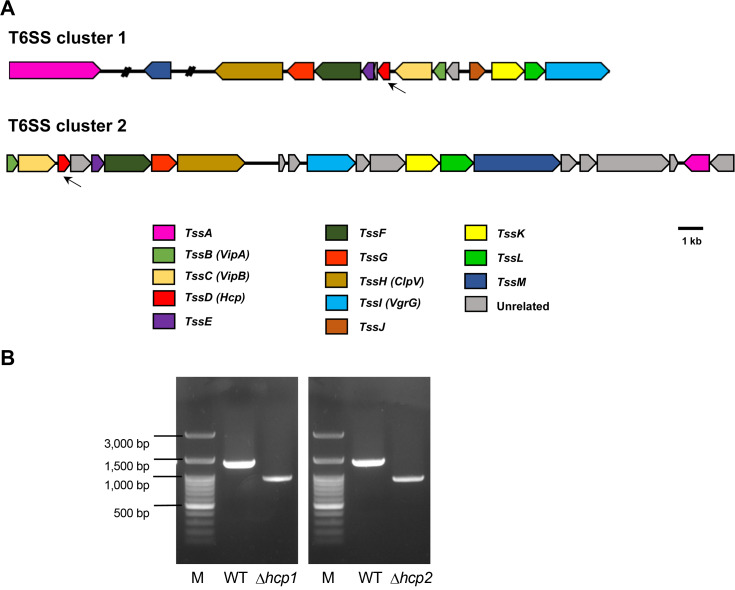
Fig.┬Ā2
Growth rate and bacterial swarming motility are not affected with hcp deletions in both type VI secretion system clusters in Xanthomonas oryzae pv. oryzae (Xoo). (A) Growth rate of Xoo wild-type and hcp mutant strains was monitored on rich peptone sucrose broth by measuring the optical density (O.D) at 600 nm at 2 h intervals. All strains were grown at 28°C with shaking at 200 rpm. (B) Swarming motility assay was performed on tryptone yeast extract glucose with 0.6% agar. Two microliters from cultures of Xoo wild-type and Δhcp1, Δhcp2, and Δhcp12, adjusted to OD600 = 0.8, were spotted on the plates and incubated at 28°C for 48 h. Three independent replicates were maintained for and each experiment was repeated with similar results.

Fig.┬Ā3
Pathogenicity assay of Xanthomonas oryzae pv. oryzae (Xoo), hcp deletion mutant strains and hcp2 complemented strain on rice plants. (A, D) Photographs from the pathogenicity assays indicating the blight lesions on inoculated leaves at 14 days post-inoculation (dpi) with Xoo PXO99A wild-type, Δhcp1 mutant, Δhcp2 mutant, the double mutant, Δhcp12, and the complemented strain Δhcp2C. The end of each lesions is indicated by red arrows. Plants treated with sterile distilled water served as negative control. (B, E) Bar graphs representing the lesion lengths measured at 14 dpi. Bars are the mean values ± standard deviations of 9 independent leaves. (C, F) Bar graphs representing the bacterial populations from inoculated leaves at 14 dpi. Bars are the mean values ± standard deviations of 3 different leaves. Significant differences between the wild-type and each mutant strain are indicated by different letters on the error bar (P < 0.05).

Fig.┬Ā4
Inter-bacterial competition assay between Xanthomonas oryzae pv. oryzae (Xoo) and Escherichia coli DH5α. The prey, E. coli DH5α, carries a lacZ-encoding gene allowing it to survive on Luria-Bertani (LB) agar plates containing 100 μg/ml apramycin (Apm). The prey E. coli was co-cultured with Xoo PXO99A wild-type, Δhcp1 mutant, Δhcp2 mutant, or the double mutant strain, Δhcp12, for 12 h on LB plates. The patch was resuspended, serially diluted, and spotted on LB plates containing X-gal, IPTG, and Apm. (A) Photographs from the inter-bacterial assay showing the serially diluted cultures spotted on LB plates, divided into eight sections; blue colonies indicate the surviving cells. (B) Bar graph representing the surviving E. coli colonies. Bars are the means ± standard deviations of three replicates.



 PDF Links
PDF Links PubReader
PubReader Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Supplement
Supplement Print
Print




