 |
 |
| Plant Pathol J > Volume 36(4); 2020 > Article |
|
Abstract
Host shifting and host expansion of fungal plant pathogens increases the rate of emergence of new pathogens and the incidence of disease in various crops, which threaten global food security. Magnaporthe species cause serious disease in rice, namely rice blast disease, as well as in many alternative hosts, including wheat, barley, and millet. A severe outbreak of wheat blast due to Magnaporthe oryzae occurred recently in Bangladesh, after the fungus was introduced from South America, causing great loss of yield. This outbreak of wheat blast is of growing concern, because it might spread to adjacent wheat-producing areas. Therefore, it is important to understand the host range and population structure of M. oryzae and related species for determining the evolutionary relationships among Magnaporthe species and for managing blast disease in the field. Here, we collected isolates of M. oryzae and related species from various Poaceae species, including crops and weeds surrounding rice fields, in Korea and determined their phylogenetic relationships and host species specificity. Internal transcribed spacer-mediated phylogenetic analysis revealed that M. oryzae and related species are classified into four groups primarily including isolates from rice, crabgrass, millet and tall fescue. Based on pathogenicity assays, M. oryzae and related species can infect different Poaceae hosts and move among hosts, suggesting the potential for host shifting and host expansion in nature. These results provide important information on the diversification of M. oryzae and related species with a broad range of Poaceae as hosts in crop fields.
Rice (Oryza sativa) is a staple food crop for more than half of the global population and 90% of all rice is produced in Asia (Fairhurst and Dobermann, 2002; Gnanamanickam, 2009). Rice blast caused by Magnaporthe oryzae, which belongs to the M. grisea species complex, is one of the most destructive fungal diseases of rice, reducing annual yields by approximately 10-30% (Fernandez and Orth, 2018; Talbot, 2003). In addition to rice, M. oryzae can infect more than 50 species of Poaceae, including economically and agriculturally important crops such as wheat (Triticum spp.), barley (Hordeum vulgare), millet (Setaria and Panicum spp.), and maize (Zea mays), as well as wild grasses surrounding rice fields (Ou, 1985). Rice is a domesticated Poaceae among more than 10,000 species of Poaceae, which may allow pathogens to shift to rice and neighboring plants (Couch et al., 2005; Gaut, 2002; Kellogg, 2001; Zhong et al., 2016). Host shifting and the expansion of M. oryzae have been reported widely and cause the most devastating disease outbreaks in new crops (Woolhouse et al., 2005; Zhong et al., 2016). For example, wheat blast, also caused by M. oryzae, was first reported in Brazil in 1985 and spread to wheat-growing areas in South America (Inoue et al., 2017; Urashima et al., 1993). Wheat blast was also observed in North America in 2011, and in Bangladesh in 2016. The North American isolate was inferred to have evolved from ryegrass (Lolium spp.) through a host jump (Farman et al., 2017; Inoue et al., 2017; Islam et al., 2016; Malaker et al., 2016). The cross-infectivity of isolates from rice and other Poaceae species implies that host shifts in M. oryzae and related species may occur in rice and other plants surrounding rice fields (Choi et al., 1996; Choi et al., 2013).
M. oryzae has a broad host range, but individual isolates have limited host ranges and are divided into different pathotypes based on their compatibility (Choi et al., 2013; Kim et al., 2019; Valent, 1990). The host specificity of the M. grisea species complex, which includes M. oryzae, has been characterized in numerous studies through genomic and genetic comparisons of isolates and their pathogenicity to different Poaceae species (Chiapello et al., 2015; Choi et al., 1996; Choi et al., 2013; Couch and Kohn, 2002; Couch et al., 2005; Hirata et al., 2007; Kato et al., 2000; Yoshida et al., 2016). The representative species M. oryzae and M. grisea are morphologically indistinguishable, but M. oryzae, which is associated with rice and other grasses, is taxonomically distinct from M. grisea, which infects Digitaria spp. (Couch and Kohn, 2002). M. oryzae is also categorized into several host-specific subgroups infecting Oryza spp., Setaria spp., Triticum spp., Panicum spp., and Eleusine spp. (Couch et al., 2005; Hirata et al., 2007; Kato et al., 2000; Yoshida et al., 2016). A major mechanism underlying host species specificity and adaptation of the M. grisea species complex is the gain and loss of effector genes, such as avirulence genes, through chromosomal rearrangements and seems to rely on a few gene families with genetic polymorphisms (Chiapello et al., 2015; Chuma et al., 2011; de Jonge et al., 2013; Seidl and Thomma, 2014; Yoshida et al., 2016).
The gene-for-gene interaction between avirulence and resistance genes controls the host specificity of plant pathogens including M. oryzae by deriving compatible (susceptible) and incompatible (resistant) interactions (Chuma et al., 2011; Couch et al., 2005; Silu├® et al., 1992; Takabayashi et al., 2002; Tosa et al., 2006; Valent and Khang, 2010; Wang et al., 2017). To date, more than 100 blast resistance genes and about 24 avirulence genes have been mapped in the rice and M. oryzae genomes, respectively, of which 35 resistance genes and 12 avirulence genes have been cloned (Wang et al., 2017). Four avirulence genes are involved in host species specificity, PWL1 (Kang et al., 1995), PWL2 (Kang et al., 1995; Sweigard et al., 1995), Avr1-CO39 (Farman et al., 2002), and Avr-Pita (Bryan et al., 2000; Chuma et al., 2011; Khang et al., 2008), and five genes are involved in rice specificity, ACE1 (Tosa et al., 2006), Avr-Piz-t (Li et al., 2009), Avr-Pia (Yoshida et al., 2009), Avr-Pii (Yoshida et al., 2009), and Avr-Pik/km/kp (Yoshida et al., 2009). These avirulence genes are mostly a rice-specific gene family. Nevertheless, rice-specific avirulence genes have also been found in isolated from other Poaceae hosts (Khang et al., 2008; Tosa et al., 2005; Valent et al., 1991; Yaegashi and Asaga, 1981). For example, Avr1-CO39, Avr1-M201, and Avr1-YAMO, which confer avirulence specifically toward rice cultivar CO39, M201, and Yashiromochi, respectively, were identified in a weeping lovegrass isolate but is absent in most rice isolates (Farman et al., 2002; Valent et al., 1991). Avr-Pita2 and Avr-Pita3 were also identified in isolates infecting diverse Poaceae species, in contrast, Avr-Pita1 was identified in a rice isolate, which interacts directly with the corresponding resistance gene Pita in the rice cultivar Yashiro-Mochi (Bryan et al., 2000; Jia et al., 2000; Khang et al., 2008; Orbach et al., 2000). On the other hand, the host species specificity determinant, PWL1 and PWL2, which is identified in isolates infecting finger millet and rice, respectively, prevents the infection of weeping lovegrass (Eragrostis curvula) (Kang et al., 1995; Sweigard et al., 1995). Finger millet isolates were divided into two subgroups (EC-I and EC-II), which showed differences in virulence on weeping lovegrass depending on gain and loss of PWL1 (Asuke et al., 2020). In addition, a single non-synonymous substitution in PWL2 results in loss of function with respect to controlling the pathogenicity in weeping lovegrass (Sweigard et al., 1995). It suggests that the presence and absence of a specific avirulence gene and mutation in avirulence genes can lead to a gain of virulence. This study sheds light on the host range and host specificity of M. oryzae and related species by comparing the phylogenetic relationships, cross-infectivity, and distributions of avirulence genes of the collected isolates from different Poaceae species.
A total of 970 isolates were collected from the leaves of rice (O. sativa), crabgrass (Digitaria sanguinalis), green foxtail (Setaria viridis), and other Poaceae infected by blast fungus in eight provinces, Korea, from 2011 to 2013 (Table 1, Supplementary Table 1). All isolates were purified on water agar by single spore isolation and maintained on rice bran agar medium (2% rice bran [w/v], 2% sucrose [w/v], 2% agar powder [w/v]) at 26┬░C.
Pathogenicity was assessed by spraying a conidial suspension on the rice and other Poaceae species. Conidia were harvested from 10-day-old rice bran agar plates further incubated for three days after removing the aerial mycelia. The conidia suspension was adjusted to 105 conidia/ml containing Tween 20 (final concentration, 250 ppm) and 25 ml of the suspension was sprayed on rice and others seedlings at the 4-5 leaf stage. Inoculated plants were maintained in a dew chamber at 26┬░C for 24 h in the dark, and then moved to a greenhouse and incubated for 7 days. Disease severity was scored as follows: 0-3, resistance, 4-5, susceptible. Pathogenicity test for the population assay was performed in one repetition by spraying a mixture of a conidial suspension on more than 30 seedlings each of rice and crabgrass.
Genomic DNA was extracted from mycelia cultured on liquid complete medium (LCM; 0.6% yeast extract [w/v], 1% sucrose [w/v], 0.6% casamino acid [w/v]) or potato dextrose agar (PDA) medium. Mycelia harvested from LCM were ground to a fine powder after lyophilizing. Mycelia cultured on PDA were ground with a sterilized grinder after removing the excess agar. Genomic DNA was extracted using the NuceloSpin Plant II kit (Macherey-Nagel, Duren, Germany) according to the manufacturerŌĆÖs manuals. The internal transcribed spacer (ITS) region and avirulence genes were amplified from genomic DNA (see Supplementary Table 2 for the primers used).
Rep-PCR using the repetitive element Pot2 was performed on single-spore isolates from rice and crabgrass as previously described (George et al., 1998). The PCR reactions were performed in 20 ╬╝l of Maxime PCR PreMix Kit (i-StarMAX II, Intron Biotechnology, Seongnam, Korea) containing 100 ng of genomic DNA and 10 ╬╝M of each outwardly directed primer (Supplementary Table 2). The PCR was run for 2.5 min at 95┬░C as an initial denaturation, followed by four cycles of 1 min at 94┬░C, 1 min at 62┬░C, and 10 min at 65┬░C, and then by 26 cycles of 30 s at 94┬░C, 1 min at 62┬░C, and 10 min at 65┬░C, with a final 15 min extension at 65┬░C. Amplified PCR products containing 1 ╬╝l of Dyne LoadingSTAR loading dye (DyneBio, Seongnam, Korea) were separated by gel electrophoresis on 1.2% agarose gels in 0.5├Ś Tris-borate-EDTA buffer for 3 h at 50 V.
The ITS sequences of 224 isolates and nine reference genomes (M. oryzae Guy11 and KJ201, M. poae ATCC64411, M. salvinii M21, M69, and M71, and M. rhizophila ATCC96043, Mr4757, and CSB F198) were aligned using ClusterW algorithm, and a phylogenetic tree was constructed using the maximum likelihood method in MEGA 7.0 software (1,000 bootstrap replicates). Cluster analysis was conducted based on the presence or absence of the avirulence genes (1 for presence and 0 for absence) using NTSYS-pc ver. 2.2. Genetic similarity was calculated using the JaccardŌĆÖs coefficient. Sequential hierarchical agglomerative non-overlapping clustering was applied for this similarity matrix using the unweighted pair group method with arithmetic averages. The coefficient of similarity was 0.68 (Goh et al., 2013; Park et al., 2008; Prabhu et al., 2007).
The M. oryzae and related species examined in this study included isolates from crabgrass (n=353), rice (n=287), green foxtail (n=153), and other Poaceae (n=177) (Table 1, Supplementary Table 1). To analyze their phylogenetic relationships, the ITS region was sequenced in 224 isolates originating from crabgrass (D. sanguinalis), rice (O. sativa), green foxtail (S. viridis), foxtail millet (S. italica), common millet (Panicum miliaceum), tall fescue (Festuca arundinacea), maize (Zea mays), and barnyard grass (Echinochloa crus-galli). In the phylogenetic tree based on ITS, M. oryzae and related species were divided into four distinct groups mainly composed of isolates from foxtail millet (group M), rice (group R), tall fescue (group F), and crabgrass (group C) (Fig. 1). Isolates from diverse Poaceae species belonged to group C and group M, while group R and group F had only isolates from rice and crabgrass, and rice and tall fescue, respectively. Group M comprised iso lates from foxtail millet (86.9%), common millet (7.1%), crabgrass (4.8%), and barnyard grass (1.2%), and group C comprised isolates from crabgrass (80.0%), green foxtail (15.8%), tall fescue (2.1%), maize (1.1%), and rice (1.1%). Group R contained 91.9% rice isolates, including the reference genomes, and 5.4% crabgrass isolates. Most of the rice isolates were closer to the millet isolates than to the crabgrass isolates, but some of the isolates infecting rice and crabgrass were included in phylogenetically distinct groups. Group F was 77.8% tall fescue isolates and 22.2% rice isolates. The results imply that M. oryzae and related species potentially jump into different hosts and thus expand their host ranges in the field.
M. oryzae and related species from different Poaceae have the potential for host jump and host expansion in the field. To examine this possibility, we performed pathogenicity assays on 100 isolates from rice, crabgrass, green foxtail, common millet, and barnyard grass on rice, barley, wheat, crabgrass, green foxtail, and common millet. The compatibility with each Poaceae species was determined based on the proportion of susceptible isolates to all isolates tested (Table 2, Supplementary Table 3). The isolates from rice and crabgrass could infect all of the Poaceae species tested, and more than half of the tested isolates from both were pathogenic on common millet and barley. Although M. oryzae and M. grisea infect rice and crabgrass, respectively, and are taxonomically distinct (Couch and Kohn, 2002), some of the isolates from rice and crabgrass in this study were pathogenic in the opposite host. Green foxtail isolates were able to infect crabgrass (50%), common millet (100%), and barley (50%), but none of them affected rice and wheat. The isolates from common millet caused disease only in barley (100%). All isolates from barnyard grass developed lesions on common millet (100%) and some infected barley (75%) and wheat (25%). Among the Poaceae species tested, common millet and barley were susceptible to isolates from all of the different hosts.
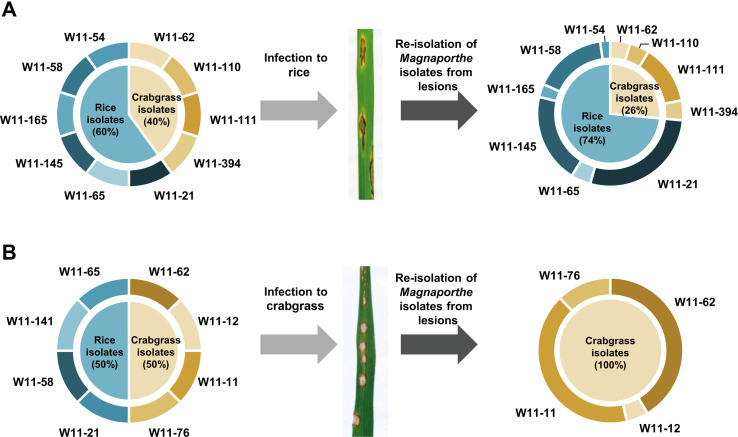
We conducted a population analysis of the rice isolates and crabgrass isolates each on rice and crabgrass to determine whether mixes of M. oryzae and related species on different hosts adjacent to each other led to disease in the opposite hosts in the field (Fig. 2, Supplementary Table 4). A mixture of the rice isolates and crabgrass isolates was inoculated onto healthy rice and crabgrass, respectively. After the lesions had developed, 144 and 41 single spores were isolated from lesions developed on rice and crabgrass, respectively. Their origin was identified by pattern analysis using Pot2 rep-PCR. Six different rice isolates and four different crabgrass isolates were co-inoculated on rice at the same concentrations. Ten different isolates from rice and crabgrass developed lesions on rice leaves, but the re-isolates comprised more isolates originating from rice (74%) than from crabgrass (26%). The dominant isolates originating from rice were W11-21 (27.94%), W11-145 (19.85%), and W11-58 (16.18%), and that from crabgrass was W11-111 (13.24%). When crabgrass was co-inoculated with four different isolates each from rice and crabgrass at the same concentrations, only the isolates originating from crabgrass were identified from among the re-isolates of infected crabgrass. The dominant isolates causing disease in crabgrass were W11-11 (41.46%) and W11-62 (41.46%), both originally from crabgrass.
The distribution of avirulence genes was analyzed in 198 isolates from rice and five other Poaceae species (crabgrass, green foxtail, barnyard grass, common millet, and foxtail millet) by PCR, using specific molecular markers for nine known avirulence genes: Avr-Pita, Avr-Pita3, Avr-Pia, Avr-Pii, Avr-Pikm, AvrPiz-t, Avr1-CO39, PWL2, and ACE1 (Table 3, Supplementary Table 2). The avirulence genes showed different distributions in the isolates depending on their hosts. The isolates from barnyard grass and common millet had the most avirulence genes among the isolates from the six Poaceae species. All of the barnyard grass isolates had Avr-Pita3, Avr-Pia, Avr-Pikm, AvrPiz-t, Avr1-CO39, PWL2, and ACE1, and more than 50% of the isolates also had Avr-Pita and Avr-Pii. Most of the common millet isolates had all of the avirulence genes except Avr-Pita (6.7%). The isolates from crabgrass and green foxtail showed similar tendencies in terms of the distribution of each avirulence gene, and less than 10% of the isolates had Avr-Pii and AvrPiz-t. Of the rice isolates, 82.4%-100% had Avr-Pita, Avr-Pita3, Avr-Pikm, AvrPiz-t, PWL2, and ACE1, while Avr-Pia, Avr-Pii, and Avr1-CO39 were less common. Avr1-CO39 was detected as 300 bp and 150 bp bands, which were differentially distributed among the isolates depending on the Poaceae hosts. The 150 bp fragment was detected more often in the isolates from crabgrass and green foxtail, while the 300 bp fragment was more common in the isolates from barnyard grass, common millet, and foxtail millet. None of the isolates from barnyard grass or common millet had the 150bp Avr1-CO39. Similar to the distribution of the 300 bp Avr1-CO39, Avr-Pii was detected more often in the isolates from barnyard grass, common millet, and foxtail millet than in those from rice, crabgrass, and green foxtail. PWL2 was present in most of the isolates from all Poaceae species except foxtail millet.
To determine the genetic relationships among the isolates from the six different Poaceae species (rice, crabgrass, green foxtail, barnyard grass, common millet, and foxtail millet), cluster analysis of the presence and absence of avirulence genes was conducted (Supplementary Fig. 1). Applying a similarity coefficient of 0.68, the isolates were divided into five clades according to the different distribution of known avirulence genes (Avr-Pita, Avr-Pita3, Avr-Pia, Avr-Pii, Avr-Pikm, AvrPiz-t, Avr1-CO39, PWL2, and ACE1). Isolates in clades G1 and G2 originated from common millet, foxtail millet, barnyard grass, and green foxtail, while isolates in G3 to G5 were from rice, crabgrass, and green foxtail. The dominant isolates in each clade were from common millet (72.2% in G1), foxtail millet (97.6% in G2), rice (83.3% in G3), and crabgrass (81.3% and 93.9% in G4 and G5, respectively). It suggests that the classification of isolates using the distribution of the avirulence genes also reflects their hosts as in the ITS-based phylogenetic analysis. However, the rice isolates (G3) were close to the crabgrass isolates (G4 and G5) than the millet isolates (G1 and G2) unlike the result of the ITS-based phylogenetic analysis.
Plant pathogens can shift from one host to another or acquire new hosts for survival (Zhong et al., 2016). The host ranges and host species specificity of M. oryzae and related species were determined by analyzing the phylogenetic relationships and cross-infectivity, and the distribution of avirulence genes was identified for isolates collected from different Poaceae species. ITS-based phylogenetic analysis divided M. oryzae and related species infecting different Poaceae species into four groups according to their hosts, which were rice (groups R), crabgrass (group C), millets (group M), and tall fescue (group F). Group R was phylogenetically closer to group M than to group C or group F, although some of the rice and crabgrass isolates belonged to phylogenetically distant groups. Previous studies have reported similar phylogenetic relationships between M. oryzae and related species infecting different hosts, albeit with some differences (Choi et al., 2013; Yoshida et al., 2016). M. oryzae isolates were divergent from M. grisea isolates and M. oryzae isolates from Oryza and Setaria closely grouped within a single clade (Choi et al., 2013; Yoshida et al., 2016). As shown by the phylogenetic relationships of M. oryzae and related species, the isolates from rice and crabgrass infected all of the Poaceae species and isolates from green foxtail and barnyard grass also could infect broad ranges of Poaceae. Of the Poaceae, common millet and barley were vulnerable to isolates from all of the different hosts, suggesting that common millet and barley can be an intermediate host for blast fungus. Therefore, M. oryzae and related species can jump into different Poaceae hosts and expand their host range in crop fields, although individual isolates may have host specificity.
Population analysis of mixed isolates from rice and crabgrass revealed that crabgrass isolates are capable of causing disease in both rice and crabgrass, while rice isolates have difficulty infecting crabgrass. The result accords with the result of the pathogenicity assay on different Poaceae species, which showed that the compatibility of crabgrass isolates to rice was higher than that of rice isolates to crabgrass. Among isolates, the crabgrass isolate W11-62 could infect both rice and crabgrass, but the proportion of the reisolates for W11-62 was higher in crabgrass than in rice. It suggests that W11-62 prefers crabgrass to rice and that the individual isolates may have host preferences, which is potentially affected by the genetic variation of pathogenicityrelated genes.
Similar to the result of the ITS-based phylogenetic analysis, the avirulence gene-based classification showed that the isolates from six different Poaceae (rice, crabgrass, green foxtail, barnyard grass, common millet, and foxtail millet) were also divided into five clades (G1 to G5) depending on hosts. The isolates belonging to different clades and different phylogenetic groups showed the different distributions of avirulence genes, especially, Avr-Pii and AvrPiz-t. The clade G1 and the group M had both Avr-Pii and AvrPizt, the clade G3 and the group R had Avr-Pii alone, and the clades G4 to G5 and the group C had none of them. These differences in the genetic composition of avirulence genes seem to be derived by the selection pressure of Poaceae hosts. The clade G1 and the group M had all of the nine avirulence genes we tested, which may contribute to the avirulence of the isolates from common millet and barnyard grass toward rice and crabgrass. However, it is difficult to assume that the infectivity of the isolates for different Poaceae hosts is directly correlated to the distribution of the avirulence genes. Therefore, these results showed that M. oryzae and related species have a broad spectrum of Poaceae species as hosts by easily moving from rice to other Poaceae including weeds and adapting to new hosts. Our data provide important information on the host ranges of M. oryzae and related species, and on latent inoculum of blast disease in crops and weeds surrounding rice fields.
Supplementary materials are available at The Plant Pathology Journal website (http://www.ppjonline.org/).
Acknowledgments
This study was supported by the project PJ01357301 of National Institute of Crop Science, Rural Development Administration, Republic of Korea.
References
Asuke, S., Tanaka, M., Hyon, G.-S., Inoue, Y., Vy, T. T. P., Niwamoto, D., Nakayashiki, H. and Tosa, Y. 2020. Evolution of an Eleusine-specific subgroup of Pyricularia oryzae through a gain of an avirulence gene. Mol. Plant-Microbe Interact. 33:153-165.


Bryan, G. T., Wu, K.-S., Farrall, L., Jia, Y., Hershey, H. P., McAdams, S. A., Faulk, K. N., Donaldson, G. K., Tarchini, R. and Valent, B. 2000. A single amino acid difference distinguishes resistant and susceptible alleles of the rice blast resistance gene Pi-ta. Plant Cell. 12:2033-2046.



Chiapello, H., Mallet, L., Gu├®rin, C., Aguileta, G., Amselem, J., Kroj, T., Ortega-Abboud, E., Lebrun, M.-H., Henrissat, B., Gendrault, A., Rodolphe, F., Tharreau, D. and Fournier, E. 2015. Deciphering genome content and evolutionary relationships of isolates from the fungus Magnaporthe oryzae attacking different host plants. Genome Biol. Evol. 7:2896-2912.



Choi, J., Park, S.-Y., Kim, B.-R., Roh, J.-H., Oh, I.-S., Han, S.-S. and Lee, Y.-H. 2013. Comparative analysis of pathogenicity and phylogenetic relationship in Magnaporthe grisea species complex. PLoS ONE. 8:e57196.



Choi, W.-B., Chun, S.-J. and Lee, Y.-H. 1996. Host range of Korean isolates of Magnaporthe grisea. Korean J. Plant Pathol. 12:453-454.
Chuma, I., Isobe, C., Hotta, Y., Ibaragi, K., Futamata, N., Kusaba, M., Yoshida, K., Terauchi, R., Fujita, Y., Nakayashiki, H., Valent, B. and Tosa, Y. 2011. Multiple translocation of the AVRPita effector gene among chromosomes of the rice blast fungus Magnaporthe oryzae and related species. PLoS Pathog. 7:e1002147.



Couch, B. C., Fudal, I., Lebrun, M.-H., Tharreau, D., Valent, B., Van Kim, P., Nott├®ghem, J.-L. and Kohn, L. M. 2005. Origins of host-specific populations of the blast pathogen Magnaporthe oryzae in crop domestication with subsequent expansion of pandemic clones on rice and weeds of rice. Genetics. 170:613-630.



Couch, B. C. and Kohn, L. M. 2002. A multilocus gene genealogy concordant with host preference indicates segregation of a new species, Magnaporthe oryzae, from M. grisea. Mycologia. 94:683-693.


de Jonge, R., Bolton, M. D., Kombrink, A., den Berg, G. C. M., Yadeta, K. A. and Thomma, B. P. H. J. 2013. Extensive chromosomal reshuffling drives evolution of virulence in an asexual pathogen. Genome Res. 23:1271-1282.



Fairhurst, T. and Dobermann, A. 2002. Rice in the global food supply. World. 5:454349-511675.
Farman, M. L., Eto, Y., Nakao, T., Tosa, Y., Nakayashiki, H., Mayama, S. and Leong, S. A. 2002. Analysis of the structure of the AVR1-CO39 avirulence locus in virulent rice-infecting isolates of Magnaporthe grisea. Mol. Plant-Microbe Interact. 15:6-16.


Farman, M., Peterson, G., Chen, L., Starnes, J., Valent, B., Bachi, P., Murdock, L., Hershman, D., Pedley, K., Fernandes, J. M. and Bavaresco, J. 2017. The Lolium pathotype of Magnaporthe oryzae recovered from a single blasted wheat plant in the United States. Plant Dis. 101:684-692.


Fernandez, J. and Orth, K. 2018. Rise of a cereal killer: the biology of Magnaporthe oryzae biotrophic growth. Trends Microbiol. 26:582-597.



George, M. L. C., Nelson, R. J., Zeigler, R. S. and Leung, H. 1998. Rapid population analysis of Magnaporthe grisea by using rep-PCR and endogenous repetitive DNA sequences. Phytopathology. 88:223-229.


Gnanamanickam, S. S. 2009. Biological control of rice diseases. Springer, Dordrecht, Netherlands. 108
Goh, J., Kim, B.-R., Lee, S.-W., Roh, J.-H., Shin, D.-B., Jeung, J.-U., Cho, Y.-C. and Han, S.-S. 2013. Selection of representative Magnaporthe oryzae isolates and rice resistant gene types for screening of blast-resistant rice cultivars. Res. Plant Dis. 19:243-253.

Hirata, K., Kusaba, M., Chuma, I., Osue, J., Nakayashiki, H., Mayama, S. and Tosa, Y. 2007. Speciation in Pyricularia inferred from multilocus phylogenetic analysis. Mycol. Res. 111:799-808.


Inoue, Y., Vy, T. T. P., Yoshida, K., Asano, H., Mitsuoka, C., Asuke, S., Anh, V. L., Cumagun, C. J. R., Chuma, I., Terauchi, R., Kato, K., Mitchell, T., Valent, B., Farman, M. and Tosa, Y. 2017. Evolution of the wheat blast fungus through functional losses in a host specificity determinant. Science. 357:80-83.


Islam, M. T., Croll, D., Gladieux, P., Soanes, D. M., Persoons, A., Bhattacharjee, P., Hossain, M. S., Gupta, D. R., Rahman, M. M., Mahboob, M. G., Cook, N., Salam, M. U., Surovy, M. Z., Sancho, V. B., Maciel, J. L. N., NhaniJ├║nior, A., Castroagud├Łn, V. L., de Assis Reges, J. T., Ceresini, P. C., Ravel, S., Kellner, R., Fournier, E., Tharreau, D., Lebrun, M.-H., McDonald, B. A., Stitt, T., Swan, D., Talbot, N. J., Saunders, D. G. O., Win, J. and Kamoun, S. 2016. Emergence of wheat blast in Bangladesh was caused by a South American lineage of Magnaporthe oryzae. BMC Biol. 14:84



Jia, Y., McAdams, S. A., Bryan, G. T., Hershey, H. P. and Valent, B. 2000. Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J. 19:4004-4014.



Kang, S., Sweigard, J. A. and Valent, B. 1995. The PWL host specificity gene family in the blast fungus Magnaporthe grisea. Mol. Plant-Microbe Interact. 8:939-948.


Kato, H., Yamamoto, M., Yamaguchi-Ozaki, T., Kadouchi, H., Iwamoto, Y., Nakayashiki, H., Tosa, Y., Mayama, S. and Mori, N. 2000. Pathogenicity, mating ability and DNA restriction fragment length polymorphisms of Pyricularia populations isolated from Gramineae, Bambusideae and Zingiberaceae plants. J. Gen. Plant Pathol. 66:30-47.

Khang, C. H., Park, S.-Y., Lee, Y.-H., Valent, B. and Kang, S. 2008. Genome organization and evolution of the AVR-Pita avirulence gene family in the Magnaporthe grisea species complex. Mol. Plant-Microbe Interact. 21:658-670.


Kim, K.-T., Ko, J., Song, H., Choi, G., Kim, H., Jeon, J., Cheong, K., Kang, S. and Lee, Y.-H. 2019. Evolution of the genes encoding effector candidates within multiple pathotypes of Magnaporthe oryzae. Front. Microbiol. 10:2575



Li, W., Wang, B., Wu, J., Lu, G., Hu, Y., Zhang, X., Zhang, Z., Zhao, Q., Feng, Q., Zhang, H., Wang, Z., Wang, G., Han, B., Wang, Z. and Zhou, B. 2009. The Magnaporthe oryzae avirulence gene AVRPiz-t encodes a predicted secreted protein that triggers the immunity in rice mediated by the blast resistance gene Piz-t. Mol. Plant-Microbe Interact. 22:411-420.


Malaker, P. K., Barma, N. C. D., Tiwary, T. P., Collis, W. J., Duveiller, E., Singh, P. K., Joshi, A. K., Singh, R. P., Braun, H.-J., Peterson, G. L., Pedley, K. F., Farman, M. L. and Valent, B. 2016. First report of wheat blast caused by Magnaporthe oryzae pathotype triticum in Bangladesh. Plant Dis. 100:2330

Orbach, M. J., Farrall, L., Sweigard, J. A., Chumley, F. G. and Valent, B. 2000. A telomeric avirulence gene determines efficacy for the rice blast resistance gene Pi-ta. Plant Cell. 12:2019-2032.



Ou, S. H. 1985. Rice diseases. 2nd ed. Commonwealth Mycological Institute, Kew, Surrey, UK. 380.
Park, S.-Y., Milgroom, M. G., Han, S. S., Kang, S. and Lee, Y.-H. 2008. Genetic differentiation of Magnaporthe oryzae populations from scouting plots and commercial rice fields in Korea. Phytopathology. 98:436-442.


Prabhu, A. S., Ara├║jo, L. G., Silva, G. B. and Trindade, M. G. 2007. Virulence and rep-PCR analysis of Pyricularia grisea isolates from two Brazilian upland rice cultivars. Fitopatol. Bras. 32:13-20.

Seidl, M. F. and Thomma, B. P. H. J. 2014. Sex or no sex: evolutionary adaptation occurs regardless. BioEssays. 36:335-345.



Silu├®, D., Notteghem, J. L. and Tharreau, D. 1992. Evidence of a gene-for-gene relationship in the Oryza sativa-Magnaporthe grisea pathosystem. Phytopathology. 82:577-580.

Sweigard, J. A., Carroll, A. M., Kang, S., Farrall, L., Chumley, F. G. and Valent, B. 1995. Identification, cloning, and characterization of PWL2, a gene for host species specificity in the rice blast fungus. Plant Cell. 7:1221-1233.



Takabayashi, N., Tosa, Y., Oh, H. S. and Mayama, S. 2002. A gene-for-gene relationship underlying the species-specific parasitism of Avena/Triticum isolates of Magnaporthe grisea on wheat cultivars. Phytopathology. 92:1182-1188.


Talbot, N. J. 2003. On the trail of a cereal killer: exploring the biology of Magnaporthe grisea. Annu. Rev. Microbiol. 57:177-202.


Tosa, Y., Osue, J., Oh, H.-S., Nakayashiki, H., Mayama, S. and Leong, S. A. 2005. Evolution of an avirulence gene, AVR1-CO39, concomitant with the evolution and differentiation of Magnaporthe oryzae. Mol. Plant-. Microbe Interact. 18:1148-1160.

Tosa, Y., Tamba, H., Tanaka, K. and Mayama, S. 2006. Genetic analysis of host species specificity of Magnaporthe oryzae isolates from rice and wheat. Phytopathology. 96:480-484.


Urashima, A. S., Igarashi, S. and Kato, H. 1993. Host range, mating type, and fertility of Pyricularia grisea from wheat in Brazil. Plant Dis. 77:1211-1216.

Valent, B., Farrall, L. and Chumley, F. G. 1991. Magnaporthe grisea genes for pathogenicity and virulence identified through a series of backcrosses. Genetics. 127:87-101.



Valent, B. and Khang, C. H. 2010. Recent advances in rice blast effector research. Curr. Opin. Plant Biol. 13:434-441.


Wang, B.-H., Ebbole, D. J. and Wang, Z.-H. 2017. The arms race between Magnaporthe oryzae and rice: diversity and interaction of Avr and R genes. J. Integr. Agric. 16:2746-2760.

Woolhouse, M. E. J., Haydon, D. T. and Antia, R. 2005. Emerging pathogens: the epidemiology and evolution of species jumps. Trends Ecol. Evol. 20:238-244.



Yaegashi, H. and Asaga, K. 1981. Further studies on the inheritance of pathogenicity in crosses of Pyricularia oryzae with Pyricularia sp. from finger millet. Ann. Phytopathol. Soc. Jpn. 47:677-679.

Yoshida, K., Saitoh, H., Fujisawa, S., Kanzaki, H., Matsumura, H., Yoshida, K., Tosa, Y., Chuma, I., Takano, Y., Win, J., Kamoun, S. and Terauchi, R. 2009. Association genetics reveals three novel avirulence genes from the rice blast fungal pathogen Magnaporthe oryzae. Plant Cell. 21:1573-1591.



Yoshida, K., Saunders, D. G. O., Mitsuoka, C., Natsume, S., Kosugi, S., Saitoh, H., Inoue, Y., Chuma, I., Tosa, Y., Cano, L. M., Kamoun, S. and Terauchi, R. 2016. Host specialization of the blast fungus Magnaporthe oryzae is associated with dynamic gain and loss of genes linked to transposable elements. BMC Genomics. 17:370



Zhong, Z., Norvienyeku, J., Chen, M., Bao, J., Lin, L., Chen, L., Lin, Y., Wu, X., Cai, Z., Zhang, Q., Lin, X., Hong, Y., Huang, J., Xu, L., Zhang, H., Chen, L., Tang, W., Zheng, H., Chen, X., Wang, Y., Lian, B., Zhang, L., Tang, H., Lu, G., Ebbole, D. J., Wang, B. and Wang, Z. 2016. Directional selection from host plants is a major force driving host specificity in Magnaporthe species. Sci. Rep. 6:25591




Fig.┬Ā1
Maximum likelihood tree based on internal transcribed spacer (ITS) sequences of Magnaporthe oryzae and related species. ITS sequences were obtained from 233 isolates of M. oryzae and related species, including the reference genomes Guy11, KJ201, M. poae, M. salvinii, and M. rhizophila. Abbreviations for the Poaceae species are as follows: Os, Oryza sativa; Zm, Zea mays; Fa, Festuca arundinacea; Sv, Setaria viridis; Ds, Digitaria sanguinalis; Si, Setaria italica; Ec, Echinochloa crus-galli; Pm, Panicum miliaceum.
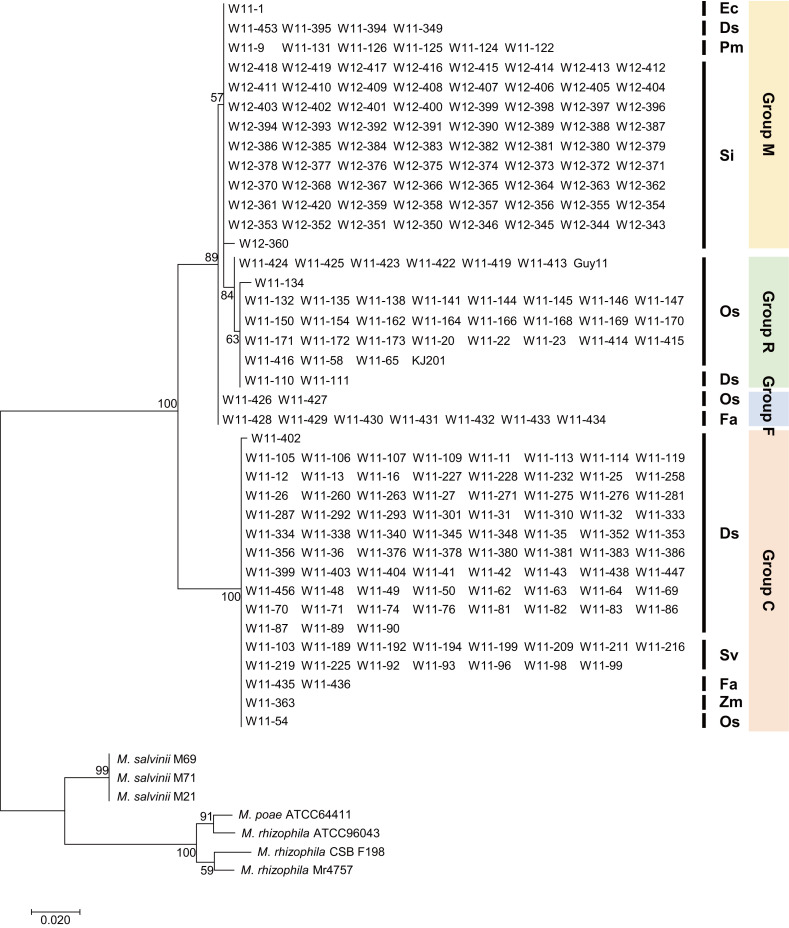
Fig.┬Ā2
Transition of the fungal population among isolates from rice and crabgrass. The colors in the chart indicate the original host of the isolates: blue, rice; brown, crabgrass. Mixed isolates from rice and crabgrass were inoculated on rice (A) and crabgrass (B), respectively.
Table┬Ā1
Number of blast isolates from different host plants
Table┬Ā2
Cross-infection of blast fungus isolated from rice and weeds among different host plants
Table┬Ā3
Distribution of avirulence genes in blast fungus isolated from rice and weeds
| Proportion of avirulence gene (%)a | ||||||
|---|---|---|---|---|---|---|
|
|
||||||
| Oryza sativa | Digitaria sanguinalis | Setaria viridis | Echinochloa crus-galli | Panicum miliaceum | Setaria italica | |
| Avr-Pita | 100.0 | 83.6 | 80.0 | 75.0 | 6.7 | 1.3 |
| Avr-Pita3 | 100.0 | 43.8 | 40.0 | 100.0 | 100.0 | 92.4 |
| Avr-Pia | 17.6 | 95.9 | 90.0 | 100.0 | 93.3 | 7.6 |
| Avr-Pii | 35.3 | 6.8 | 0.0 | 50.0 | 100.0 | 57.0 |
| Avr-Pikm | 100.0 | 75.3 | 60.0 | 100 | 93.3 | 84.8 |
| AvrPiz-t | 82.4 | 2.7 | 10.0 | 100.0 | 100.0 | 93.7 |
| Avr1-CO39b | 23.5/23.5 | 0.0/93.2 | 10.0/90.0 | 100.0/0.0 | 100.0/0.0 | 92.4/1.3 |
| PWL2 | 100.0 | 100.0 | 90.0 | 100.0 | 100.0 | 0.0 |
| ACE1 | 88.2 | 42.5 | 60.0 | 100.0 | 93.3 | 17.7 |



 PDF Links
PDF Links PubReader
PubReader Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Supplement
Supplement Print
Print




