 |
 |
| Plant Pathol J > Volume 37(2); 2021 > Article |
|
Abstract
The transcription factor SHE1 was identified as an interacting partner with the cucumber mosaic virus (CMV) 1a protein in the yeast two-hybrid system, by a pull-down assay, and via bimolecular fluorescent complementation. Using fluorescent-tagged proteins and confocal microscopy, the CMV 1a protein itself was found distributed predominantly between the nucleus and the tonoplast membrane, although it was also found in speckles in the cytoplasm. The SHE1 protein was localized in the nucleus, but in the presence of the CMV 1a protein was partitioned between the nucleus and the tonoplast membrane. SHE1 expression was induced by infection of tobacco with four tested viruses: CMV, tobacco mosaic virus, potato virus X and potato virus Y. Transgenic tobacco expressing the CMV 1a protein showed constitutive expression of SHE1, indicating that the CMV 1a protein may be responsible for its induction. However, previously, such plants also were shown to have less resistance to local and systemic movement of tobacco mosaic virus (TMV) expressing the green fluorescent protein, suggesting that the CMV 1a protein may act to prevent the function of the SHE1 protein. SHE1 is a member of the AP2/ERF class of transcription factors and is conserved in sequence in several Nicotiana species, although two clades of SHE1 could be discerned, including both different Nicotiana species and cultivars of tobacco, varying by the presence of particular insertions or deletions.
The major defense mechanisms against infection by plant viruses are basal immunity, Resistance (R) gene-mediated defense and RNA silencing (Carr et al., 2018b; Guo et al., 2019; Jin et al., 2021; Lu and Tsuda, 2021; Nabori and Tsuda, 2019; Palukaitis and Yoon, 2020; Zvereva and Pooggin, 2012). Plant viruses use various methods to overcome these defense responses, including inhibition of either gene expression or action of some component of the defense response (Carr et al., 2018b; Jin et al., 2021; Palukaitis and Yoon, 2020; Zvereva and Pooggin, 2012). With regard to the latter, this can include a viral-encoded component either binding directly to a host factor involved in various defense pathways or causing the destruction of such host factors (Csorba et al., 2015; Jin et al., 2021; Nakahara and Masuta, 2014).
In the case of cucumber mosaic virus (CMV), various proteins have been identified that affect the host defense mechanisms or the aphid-vector behavior (Carr et al., 2018a; Tungadi et al., 2020; Westwood et al., 2013, 2014; Ziebell et al., 2011). The CMV genome consists of three RNAs expressing five proteins: RNA 1 directly expresses the 1a replication-related protein; RNA 2 directly expresses the RNA-dependent RNA polymerase subunit of the viral replicase, and also expresses the 2b RNA silencing suppressor protein from the subgenomic RNA 4A; RNA 3 directly expresses the 3a movement protein, and also expresses the 3b coat protein (CP) of CMV from the subgenomic RNA 4 (Palukaitis, 2019). The 2b has been shown to bind to and inhibit factors involved in RNA silencing, as well as disrupt several phytohormone-mediated resistance responses (Carr and Murphy, 2019). The 2a protein also causes an increase the biosynthesis of a glucosinolate that discourages aphids from long-term feeding and thus enhancing transmission of the virus in Arabidopsis thaliana (Westwood et al., 2013), By contrast, in tobacco (Nicotiana tabacum), it is the CMV 1a that triggers a strong anti-aphid resistance, which is inhibited by the 2a protein (Tunga et al., 2020; Ziebell et al., 2011). The 1a protein of one CMV strain also conferred breakage of the resistance gene Cmr1 in pepper (Capsicum annuum) (Kang et al., 2012). The 3a movement protein is able to suppress basal immunity to infection by CMV in both A. thaliana and tobacco (Kong et al., 2018). The CP of some strains of CMV can induce a R gene-mediated hypersensitive response (HR) in a few eco-types of A. thaliana (Takahashi et al., 1994, 2001), which is not induced by the CP of most CMV strains (Takahashi et al., 1994), and the 2a protein of CMV legume strains is able to overcome an HR-mediated resistance in cowpea against other CMV strains (Hu et al., 2012; Karasawa et al., 1999; Kim and Palukaitis, 1997). Thus, each protein of CMV is involved in some host interaction that affects either a direct resistance response or the behavior of the aphid vector.
Another response induced by the CMV 1a protein occurred when this protein was expressed transgenically in tobacco and facilitated the local movement and systemic infection of tobacco mosaic virus (TMV) expressing a green fluorescent protein (GFP) reporter gene (TMV-GFP), which was debilitated from such movement in non-transformed tobacco, in contrast to wild-type TMV (Canto and Palukaitis, 2002). The inhibition response in tobacco that was specific to the TMV-GFP was associated with the N gene-mediated resistance to TMV, which normally blocks TMV systemic infection, but this resistance did not occur above 28┬░C allowing systemic infection by wild-type TMV; however, the resistance was still active against TMV-GFP at the higher temperature (Canto and Palukaitis, 2002). The resistance to local movement of TMV-GFP was not dependent on the salicylic acid (SA)-mediated defense pathway (Canto and Palukaitis, 2002). As an initial step in further characterizing the nature of the unknown resistance pathway, we sought to identify other proteins that interact with the CMV 1a protein that may be factors in such a novel resistance pathway. Thus, we used the yeast two-hybrid (Y2H) system to screen a tobacco cDNA library to identify tobacco cDNA clones encoding proteins that interact with the CMV 1a protein and here we describe the analysis of one such tobacco protein, which we have designated SHE1.
N. tabacum cv. Samsun NN plants were germinated and cultivated in the growth room maintained with a 16 h light/8 h dark cycle, a daytime temperature of 24┬░C, and a nighttime temperature of 22┬░C. The plants at the 5 to 6 leaf stage were inoculated mechanically with sap inoculum of TMV-U1, Fny-CMV, potato virus X (PVX-Kr), or potato virus Y (PVY-O) (viruses as described in Baek et al., 2017), after dusting with Carborundum (600 mesh, Thermo Fisher Scientific, Waltham, MA, USA) using 10 mM phosphate buffer, pH 7.2.
A full-length cDNA of the CMV 1a gene was amplified by the polymerase chain reaction (PCR) from the plasmid pF109, using the following primer sets containing attB1 and attB2 site for Gateway system (Invitrogen, Carlsbad, CA, USA): attBF-1 5ŌĆ▓-GGGGACAAGTTTGTACAAAAAAGCAGGCTCCATGGCGACGTCCTCGTTCAA-3ŌĆ▓; attBR-1T 5ŌĆ▓-GGGGACCACTTTGTACAAGAAAGCTGGGTCTAAGCACGAGCAACACATT-3ŌĆ▓. (The underlined sequences are recombination sites for using the Gateway system; the italicized sequences are start and stop codons.) Following the BP reaction, attB-PCR products were introduced into expression clones via the Gateway LR reaction (Invitrogen) using pAS-attR(BD) and pACT2-attR(AD).
All assays were carried out according to the protocols described in the YEASTMAKER Yeast Transformation System 2 User Manual (PT1172-1, Clontech, Mountain View, CA, USA). In brief, the pASGW-1a (bait) and pACTGW-SHE1, plasmids were co-transformed into the AH109 yeast strains using a modified lithium acetate protocol, and the transformants were selected on SD/-Trp, SD/-Leu, SD/-Trp/-Leu, SD/-Trp/-Leu/-His, and SD/-Trp/-Leu/-His/-Ade plates without X-gal to detect the initiation of reporter gene (HIS, LEU, TRP, ADE, and MEL1) transcription. Colonies were grown at 30┬░C for about 5-7 days. A pool of cDNA libraries from N. tabacum cv. Xanthi-nc cloned into pAD-GAL4 (Stratagene, La Jolla, CA, USA) (provided by S.H. Kim) was recovered from transformants (1.0 ├Ś 107 cells) on amplicilin-containing Luria-Bertani plates. The cDNA plasmids were co-transformed with pASGW-1a bait plasmid into AH109 yeast cells. Transformants were grown on selective media, SD/-Trp/-Leu and SD/-Trp/-Leu/-His/50 mM 3-AT (3-amino-1,2,4-triazole, Sigma, St. Louis, MO, USA) without X-gal at 30┬░C. Positive colonies were cultured in liquid SD/-Trp/-Leu/-His/-Ade media at 30┬░C for 48 h. Each plasmid was used as a template for PCR to check its inserts with primers, Gal4UP (5ŌĆ▓-ctcgagagtattagtcgactctag-3ŌĆ▓) and Gal4DN (5ŌĆ▓-cctcgtgccgaattctctgc-3ŌĆ▓). The PCR product was cloned into the pGEM-T easy vector (Promega, Madison, WI, USA) and sequenced. DNA sequences were analyzed through the GenBank database.
To confirm the positive reactions, colony-lift filter assay or X-╬▒-gal assay were done. Briefly, fresh grown colonies were transferred to a sterile filter and submerged in a pool of liquid nitrogen for 10 s and then thawed at room temperature for 5 min. The filter was then placed on a pre-soaked filter in Z buffer/X-gal solution (100 ml Z buffer, 0.27 ml 2-mercaptoethanol, 1.67 ml X-╬▓-gal stock solution [20 g/l in dimethylformamide]; Z buffer: Na2HPO4┬Ę7H2O, 16.1 g/l; NaH2PO4┬ĘH2O, 5.50 g/l; KCl, 0.75 g/l; MgSO4┬ĘH2O, 0.246 g/l; pH 7.0), following which the filters were incubated at 30┬░C overnight and the colors of colonies were checked. Alternatively, growth of blue yeast cells representing the positive interactions was analyzed on SD/-Trp/-Leu/-His/-Ade agar containing 40 g/l of X-╬▒-gal (Clontech).
An Escherichia coli expression vector was used to produce maltose binding protein (MBP)-fusion proteins with SHE1. First, the SHE1 gene was cloned in pDONR207 according to the manufacturerŌĆÖs instructions and was then subcloned in pMal-c2 Gateway (Invitrogen), a conversion vector for expressing the N-terminal fusion MBP, and transformed into BL21 codon plus cells (Agilent, Santa Clara, CA, USA). A glutathione S-transferase (GST)-fused CMV 1a clone from AS-CMV RNA 1, and a clone harboring fibrillarin 2 (Fb2)-GST (a nucleolar protein) (both provided by S.H. Kim, Myongji University, Korea) (Kim et al., 2002, 2007), were transformed into BL21 codon plus cells. The Fb2-GST clone and an empty vector were used as negative controls.
GST-fused CMV 1a and MBP-fused SHE1 proteins were induced by 0.2 mM and 0.1 mM IPTG, respectively. The induced cells were sonicated in a buffer containing 50 mM Tris-Cl, pH 8.0, 1 mM EDTA, 50 mM NaCl, 0.1% (v/v) NP40, 10% (v/v) glycerol, 10 mM 2-mercaptoethanol, and one tablet of Protease inhibitor mix (Roche Diagnostics, Laval, QC, Canada). For further purification, amylose resin (NEB, Ipswhich, MA, USA) was mixed with the clarified lysate and incubated with shaking gently at 4┬░C for 1 h, followed by centrifugation at 400 ├Śg for 10 s. The amylose beads bound to MBP-fused proteins were washed in a buffer (20 mM Tris-Cl, pH 8.0, 1 mM EDTA, 150 mM NaCl, 10 mM 2-mercaptoethanol, and 0.1% (v/v) NP40). MBP fusion proteins subsequently were eluted in buffer containing 25 mM Tris-HCl, pH 7.5, 150 mM NaCl and 0.01% (v/v) Triton X-100, supplemented with 20 mM maltose. To assess purification, the eluted proteins were separated by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and were either visualized by Coomassie Brilliant Blue staining or transferred to nitrocellulose membranes (Bio-Rad, Hercules, CA, USA) for western blot analyses with polyclonal anti-GST (Sigma-Aldrich). To assess the protein expression, the clarified lysate of GST-fused CMV 1a and MBP-fused SHE1 proteins were incubated with GST beads (Glutathione Sepharose 4B, GE Healthcare, Little Chalfont, UK) and eluted in elution buffer supplemented with 25 mM glutathione, as previously described. The purified proteins were detected by western blot analyses with polyclonal anti-GST (Sigma-Aldrich).
The membranes were incubated for 30 min at room temperature in blocking solution containing phosphate-buffered saline (PBS) and 0.5% (w/v) skim milk. The blocked membranes then were probed with polyclonal anti-1a (1:1,500 dilution), or anti-GST (1:5,000) antisera, overnight at 4┬░C. The membranes were washed three times with 1├Ś PBST (containing 1├Ś PBS and 0.05% Tween 20). The ECL Plus kit (GE Healthcare) was used for development and exposure to X-ray film (Kodak, Rochester, NY, USA), following incubation with HRP-conjugated anti-rabbit IgG (Sigma-Aldrich) as a secondary antibody.
All plasmids were constructed using the Gateway-compatible bimolecular fluorescent complementation (BiFC) binary system (Gehl et al., 2009; Waadt et al., 2008). Construction of plasmids containing the CMV 1a and SHE1 genes was done using the full-length Entry clones as templates, with the following forward and reverse primers: attBF-1 5ŌĆ▓-GGGGACAAGTTTGTACAAAAAAGCAGGCTCCATGGCGACGTCCTCGTTCAA-3ŌĆ▓, attBR-1 5ŌĆ▓-GGGGACCACTTTGTACAAGAAAGCTGGGTCAGCACGAGCAACACATT-3ŌĆ▓, attBSHE1 - F 5ŌĆ▓-GGGGACAAGTTTGTACAAAAAAGCAGGCTCCATGTCAGTACTCAAGCCCA-3ŌĆ▓, attBSHE1-R 5ŌĆ▓-GGGGACCACTTTGTACAAGAAAGCTGGGTCCAATGAAGTCATTAAAAG-3ŌĆ▓ (Gateway sequences are underlined). The reverse primers contained no stop codon to enable C-terminal fusions. The Entry clones were obtained via BP reaction in pDONR207 according to the manufacturerŌĆÖs instructions (Invitrogen) and all clones were confirmed by DNA sequencing. The SHE1 and CMV 1a genes were transferred into pDEST-GWVYNE (Venus-Nter, amino acids 1-173) and p(MAS)DEST-GWSCYCE(R) (SCFP3A-Cter, amino acids 156-239) by the LR reaction. The BiFC gene-fusion constructs, SCFP3A-C-CMV1a and SHE1-Venus-N, were transformed into Agrobacterium tumefaciens GV3101 and co-agroinfiltrated into leaf cells of Nicotiana benthamiana with a plasmid expressing the P19 protein of tomato bushy stunt virus as a silencing suppressor (Canto et al., 2006; Yoon et al., 2012). As a visualization marker, the concurrent interaction of the protein kinase CIPK24 and the calcium sensors CBL1 and CBL10 (Waadt et al., 2008), localizing at the plasma membrane and tonoplast were used, respectively. Confocal microscopy was done as described previously (Yoon et al., 2012).
The CMV 1a and SHE1 genes also were introduced into a binary vector encoding the monomeric red fluorescence protein (mRFP), in the pROK2-mRFP plasmid, or the yellow fluorescence protein (YFP) in B7WG2, using the Gateway system. The plasmids containing CMV 1a and SHE1 genes fused to mRFP or YFP, respectively, were transformed into A. tumefaciens, which was then used to co-agroinfiltrate leaves of N. benthamiana as described (Canto et al., 2002), and the fluorescence was observed by confocal microscopy as described, previously (Yoon et al., 2012).
Fresh leaves (1 g) from non-transformed tobacco cv. Samsun NN plants, 1a-transgenic tobacco cv. Samsun NN plants (Canto and Palukaitis, 2002), and virus-inoculated tobacco cv. Samsun NN plants were frozen in liquid nitrogen and ground to a powder in a mortar. Total RNAs were extracted in 1 ml of 50 mM Tris-HCl, pH 8.0, 10 mM EDTA, 2% sodium dodecyl sulfate and 0.5% 2-mercaptoethanol, with an equal volume of phenol and chloroform (1:1). The extracted RNAs were precipitated with LiCl. To isolate the mRNA from total RNA, a PolyATrack mRNA isolation kit (Promega) was used. The isolated mRNAs were fractionated in denaturing 1% agarose gels, transferred to positively-charged nylon membranes, and hybridized to digoxigenin-labeled RNA probes. An SHE1 probe was obtained by linearization of p1aX1-2 with SpeI and transcription using T7 RNA polymerase (Thermo Fisher Scientific). A ubiquitin probe was obtained by linearization of the ubiquitin plasmid clone (provided by T. Canto) with SacI and generation using T7 RNA polymerase. The probed membranes were washed and incubated with anti-digoxigenin antibody (Roche Diagnostics), and were exposed to X-ray film, as the manufacturerŌĆÖs instruction.
For reverse transcription (RT)-PCR, total RNA was extracted from the leaf samples collected in two different plants at 3 dpi, using RNeasy Plant mini kit (Qiagen, Hilden, Germany), according to the manufacturerŌĆÖs instruction. The first strand cDNA was prepared using Superscript II reverse transcriptase (Thermo Fisher Scientific) with random hexamer primers. PCR amplification was done using Ex Taq DNA polymerase (Takara Bio, Tokyo, Japan) with the following primers: SHE1-UP 5ŌĆ▓-atgtcagtactcagca-3ŌĆ▓, SHE1-DN 5ŌĆ▓-tcagtctcgacacgatg-3ŌĆ▓, EF1╬▒-UP 5ŌĆ▓-CGTCAAGAACGTTGCAGTTAAG-3ŌĆ▓, and EF1╬▒-DN 5ŌĆ▓-CAATGGTGGGTACTCAGAGAAG-3ŌĆ▓. Following an initial cDNA denaturation at 95┬░C for 3 min, the PCR conditions consisted of 15, 25 or 35 cycles of 30 s at 95┬░C, 30 s at 53┬░C, 1 min at 72┬░C and a final elongation of 10 min at 72┬░C. The amplified PCR products of the expected size were verified on a 1% agarose gel in 1├Ś TAE buffer using ethidium bromide staining (Thermo Fisher Scientific).
A Y2H screen of a cDNA library made from mRNAs isolated from tobacco cv. Xanthi nc revealed a number of interacting partners (data not shown). The characterization of one of these interacting partners (1aX1-2) is described here. This cDNA clone was sequenced and showed high similarity to the sequence of the tobacco transcription factor (TF) ERF5 (see Supplementary Table 1), the transgenic overexpression of which was shown to enhance resistance to TMV (Fischer and Dr├Čge-Laser, 2004). For reasons described in the Discussion, we have chosen to rename this gene NtSHE1.
The entire open reading frame (ORF) of the SHE1 from clone 1aX1-2 was recloned to confirm the interaction of SHE1 with the CMV 1a protein in the Y2H system, in both orientations, using two separate Y2H assays (Fig. 1). This result showed that the interaction between these two proteins was strong. Additionally, four DNA fragments covering the entire 1a gene were generated by PCR mutagenesis, such that each fragment had an initiation codon, and these fragments (designated 1a-I, 1a-II, 1a-III, and 1a-IV) were cloned into the appropriate vectors of the Y2H system to delimit what region(s) of the 1a protein interacted with the SHE1 protein. These Y2H results indicated that fragments 1a-I, 1a-II, and 1a-III, expressing the 1a protein regions encoded by nucleotide sequences 95-838, 839-1681, and 1670-2533, respectively, interacted with SHE1, while the sequences encoding the C-terminal 19% of the 1a protein (2505-3078) did not interact with SHE1 (Supplementary Fig. 1).
To validate the interaction between 1a and SHE1, an in vitro pull-down assay was used, in which the 1a gene and the NtSHE1 gene were cloned into expression vectors and the corresponding proteins, tagged at their C-termini with either GST or MBP, respectively, were expressed in E. coli, extracted, and incubated with amylose beads, to bind to the MBP moiety. The amylose resin-bound materials were isolated, eluted from the resin, and analyzed by SDS-PAGE and western blotting, using antisera to either GST or the 1a protein to evaluate the specificity of the pull-down assay. The 1a-GST protein was shown to be pulled down by the SHE1-MBP, while the control protein fibrillarin (Fb2-GST) was not pulled down by SHE1-MBP (Fig. 2), but could be pulled down by anti-GST antiserum (Supplementary Fig. 2). Thus, the 1a protein was shown to interact in two different assay systems with the SHE1 TF.
The BiFC system was used to further confirm the interaction of the 1a protein and SHE1, and to examine the subcellular distribution of the complex in planta, with SHE1 and 1a each fused to different parts of modified, enhanced YFP vectors (Waadt et al., 2008). The 1a and SHE1 proteins interacted during BiFC, as shown by confocal microscopic examination of infiltrated leaf epidermal cells, but the complexes found were partitioned largely between the nucleus and membranes (Fig. 3A). The tonoplast membrane could be differentiated from the plasma membrane by the use of membrane-specific fluorescent tags (Fig. 3B), showing that the membrane site of interaction was the tonoplast membrane rather than the plasma membrane (Fig. 3A).
To examine the subcellular distribution further, the 1a protein tagged with mRFP and the SHE1 protein tagged with YFP were expressed in leaf epidermal cells and assessed for their localization. The controls, 1a-mRFP alone, or SHE1-YFP alone, showed the presence of 1a-mRFP both in the nucleus and on tonoplast membranes (Fig. 4A), while the SHE1-YFP was present only in the nucleus (Fig. 4B). By contrast, in the co-infiltrated cells, the images revealed that while some cells only showed 1a-mRFP associated with the tonoplast membrane (Fig. 4C), others showed the 1a-mRFP present on both the tonoplast membrane and in the nucleus (Fig. 4D and E). The SHE1-YFP also was found partitioned between both the nucleus and the tonoplast membrane, but was never found absent from the nucleus (Fig. 4C-E). The 1a-mRFP also was absent from the nucleolus (Fig. 4E).
Using a separate construct expressing 1a-YFP instead of 1a-mRFP for the agroinfiltration, again showed the 1a-YFP was present in the nucleus and on the tonoplast membrane, but also showed the fluorescent signal present as speckles in the cytoplasm (Fig. 4F). These speckles were observed by Watt et al. (2020), who described the speckles as P-bodies. However, those authors did not observe the 1a protein present in the nucleus, although this was not always present here (Fig. 4C and F).
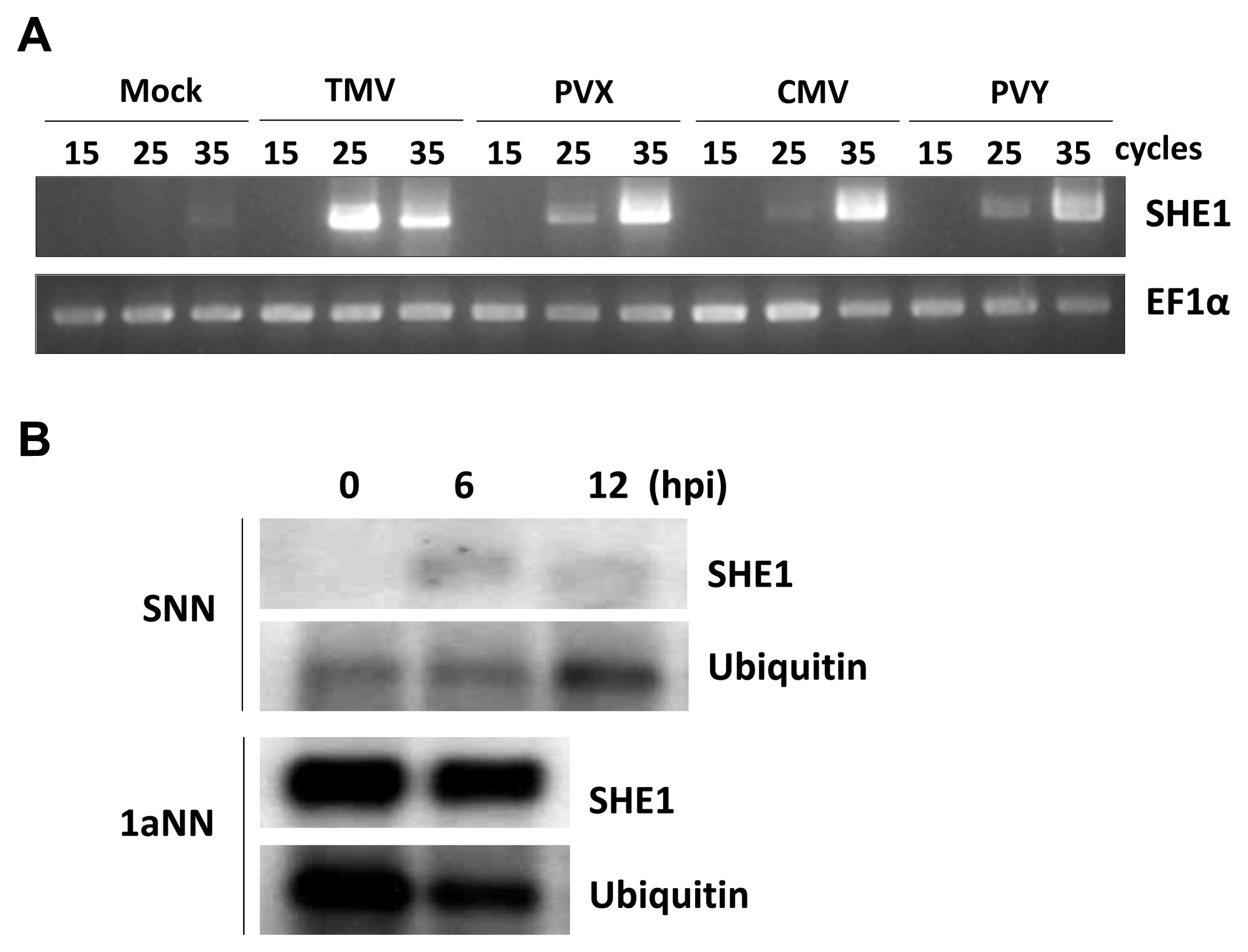
Since TMV infection was known to induce the expression of the tobacco SHE1 gene (Fischer and Dr├Čge-Laser, 2004), we assessed whether infection by CMV or other viruses affected the expression of the tobacco SHE1 gene. Thus, tobacco cv. Samsun NN plants were inoculated with CMV, TMV, PVX, or PVY and expression of SHE1 mRNA in the inoculated leaves was assessed by RT-PCR at 3 dpi. The results showed that all four viruses induced the expression of SHE1 mRNA (Fig. 5A). Thus, the expression of SHE1 is not specific to the N gene-mediated defense response against TMV. The expression of SHE1 mRNA after inoculation with TMV also was assessed at early times (Fig. 5B), which showed that the SHE1 mRNA accumulation could be induced by 6 h after inoculation, consistent with activation of a TF at early times post-inoculation. The expression of SHE1 mRNA in CMV 1a-trangenic tobacco cv. Samsun NN also was examined and was found to be constitutive (Fig. 5B), indicating that the CMV 1a protein induced the expression of the SHE1 gene.
The identification of a new interacting partner for the CMV 1a protein as an AP2/ERF class of TF, which previously was identified as elicited by TMV infection and inhibiting the local and systemic movement of TMV in overexpressing transgenic plant (Fischer and Dr├Čge-Laser, 2004), add a new dimension to the role of this TF, which we have designated SHE1. SHE1 was not induced by SA, jasmonic acid or ethylene (ET) (Fischer and Dr├Čge-Laser, 2004), but was induced during infection by several viruses (Fig. 5A), by an as yet unknown pathway. These results also showed that the induction of SHE1 is not only due to the N gene-mediated resistance response to infection by TMV, which is largely SA-mediated. While SHE1 is similar in structure to AP2/ERF TFs induced by ET, the lack of such an induction by ET indicates this TF should not be referred to as an ethylene response factor. In fact, we have found other defense-related proteins that interact with SHE1 (J.-Y. Yoon and P. Palukaitis, unpublished data), and thus we feel that the re-naming of this TF as ŌĆśsignalling hub effector 1ŌĆÖ (SHE1) is justified.
SHE1 was initially found in a Y2H screen for 1a-interacting partners, but was verified using the same technique (Fig. 1), as well as by a pull-down assay (Fig. 2) and BiFC (Fig. 3). In addition, Y2H data also indicated that SHE1 interacted with several regions of the 1a protein, but not to the C-terminal region of the 1a protein (Supplementary Fig. 1). The Y2H approach has been used previously to identify other 1a-interacting partners; viz., the thaumatin-like protein 1 (designated TLP1) (Kim et al., 2005), two tonoplast intrinsic proteins (designated TIP1 and TIP2) (Kim et al., 2006b); a novel kinase (designated Tcoi2, for tobacco CMV 1a interacting protein 2) (Kim et al., 2006a); a novel methylase transferase (designated Tcoi1) (Kim et al., 2008), and a zinc-finger protein (designated Tsip1) (Huh et al., 2011). In addition, a pull-down assay followed by a proteomic approach was used to identify a number of CMV 1a-interacting proteins, two of which were further characterized; viz., the bromodomain containing RNA binding protein that also interacted with glyceraldehyde 3-phosphate dehydrogenase, the presence of which was required for CMV replicase formation (Chaturvedi and Rao, 2016; Chaturvedi et al., 2016). Thus, a number of proteins are involved in either the intracellular movement of the 1a protein, for its localization to the nucleus or the tonoplast membrane, or for modifying the activity of the 1a protein. This does not seem to be the case in the SHE1:1a interaction.
The SHE1 TF appears to activate a defense response, since the results of overexpression of SHE1 in tobacco showed a negative effect on TMV movement and accumulation (Fischer and Dr├Čge-Laser, 2004). By contrast, constitutive expression of SHE1 in 1a-transgenic tobacco (Fig. 5B), which enhanced the movement of TMV-GFP (Canto and Palukaitis, 2002), indicated that the interaction of 1a with SHE1 inhibited the resistance against TMV-GFP mediated by SHE1. It appears that TMV itself is less inhibited by the defense signaling pathway involving SHE1, since TMV but not TMV-GFP could move further locally during an HR and could also move systemically at higher temperature, in the absence of the 1a protein (Canto and Palukaitis, 2002). Further work will be needed to establish the nature of this defense response and the effect, if any, on CMV infection in tobacco.
In the case of CMV resistance in A. thaliana manifested by the R-gene RCY1, the resistance to CMV was conferred by three signal transduction mechanisms: the SA-mediated pathway, the ET-mediated pathway, and a third novel pathway (Takahashi et al., 2002). In addition, in N-gene tobacco, an early WRKY TF was found to be induced by TMV infection as a non-SA-, non-JA-, and non-ET-mediated response (Yoda et al., 2002). Whether these are related responses or simply manifestations of the actions of other phytohormones needs to be determined.
The CMV replicase was first shown to be localized to the tonoplast membrane, by virtue of the co-localization of the 1a and 2a proteins and (ŌłÆ)-sense CMV RNAs, by immunogold detection and in situ hybridization, respectively (Cillo et al., 2002). However, this does not exclude the ability of the individual 1a and 2a proteins from interacting with other proteins and co-localizing to other subcellular structures. In fact, work by Kim et al. (2002) showed that phosphorylation of the 2a protein occurred, preventing the interaction of the 1a and 2a proteins, providing a pool of 1a (and 2a) proteins that could interact with the various host proteins, such as those mentioned above, as well as other CMV-encodes proteins, such as the 2b protein (Watt et al., 2020). The presence of the 1a protein in speckles in the cytoplasm (Fig. 4D) also is consistent with a recent report (Watt et al., 2020), although the presence of the 1a protein in the nucleus (Fig. 4A, C, and D) was not observed in that study. The co-localization of the 1a protein and SHE1 both in the nucleus and on the tonoplast membrane (Fig. 4C) suggests that the 1a protein is able to relocalize some of the SHE1 from the nucleus to the tonoplast membrane. This is reminiscent to the 1a protein being able to relocalize the 2b from both the nucleus and diffused throughout the cytoplasm to the speckles designated P-bodies (Watt et al., 2020). It is not known by what process these relocations take place.
The major differences in sequence between the NtSHE1 ORF of tobacco cv. Samsun NN (GenBank no. AY655738) vs. NtSHE1 of tobacco cv. Xanthi nc (this study) were found to be in four regions (Supplementary Fig. 3): (1) a deletion/insertion at nucleotides 147-149, corresponding to a single codon; (2) a deletion/insertion of nucleotides 228-230 (Samsun NN SHE1 sequence numbers; one codon); (3) a deletion/insertion of nine nucleotides in an 11 nucleotide stretch between nucleotides 289-299 (affecting three codons); and (4) a deletion or readthrough of the end of the ORF, by 21 nucleotides (seven codons). Interestingly, these sequence differences were specific to the SHE1 sequences both of the tobacco cv. Samsun NN and the species N. tomentosiformis, while the alternative sequence variations were observed in tobacco cvs. Xanthi nc and TN90, as well as in the species N. sylvestris, and N. attenuata, although N. attenuata also contained a three-nucleotide deletion between nucleotides 289-293 (Supplementary Fig. 3). The SHE1 sequence of the species N. umbratica was more similar to the sequences of the Xanthi nc group, although it also contained two unique changes: a deletion of nucleotides 59-61 and an insertion of 27 nucleotides after nucleotide 285 (Supplementary Fig. 3). The later insertion encodes an increased array of glutamate (E) and lysine (K) sequences (KEKEEEEEE), inside a similar array found in both the Xanthi nc group (flanked by EEKEKEKE and EEEEK in N. umbratica) and the Samsun NN group (EEKEKEEŌĆ” EK) (Supplementary Fig. 4), although this array is shorter in the right border of the Samsun NN group flanking the insertion in N. umbratica. The deletion of nine nucleotides in the Samsun NN group also resulted in the deletion of three amino acids immediately flanking the insertion site in N. umbratica. These sequences are located immediately before the conserved amino acid sequences found in AP2/ERF TFs (amino acids 99-157 of the Samsun NN SHE1 sequence) that represent a DNA-binding domain, as described for SHE1 by Fischer and Dr├Čge-Laser (2004) (Supplementary Fig. 4). In this domain, the equivalent positions to SHE1 amino acids A112 and D117 were shown to be required for GCC-binding activity in other EFR TFs (Hao et al., 2002). The bold sequences within the DNA-binding domain (TAEEAALAYDQAALSM) form an amphipathic alpha helix (amino acids 132-147). A nearby basic region (amino acids 186-203), HKTRRVKHKRSSRKKKNK, could represent a nuclear localization domain for the SHE1 protein. Stretches of acidic amino acids, such as are located in the N-terminal and C-terminal regions, could be associated with transcriptional activation (Lemon and Tjian, 2000) (Supplementary Fig. 4).
The identification of two different SHE1 genes from tobacco, one from cv. Samsun NN (Fischer and Dr├Čge-Laser, 2004) and one from cv. Xanthi nc (this report) (Supplementary Fig. 3), may simply be due to cultivar differences. The genome of N. tabacum is allotetraploid, formed by a hybridization of the ancestors of the genomes of N. sylvestris and N. tomentosiformis (Sierro et al., 2014). Thus, the sequence variation may be due to the silencing of one parental copy in one cultivar and the silencing of the other parental copy in the other cultivar. In fact, the sequences of the SHE1 genes from these two parental genomes correspond closely to the sequences represented by the two N. tabacum genes identified from the two tobacco cultivars, as well as the two groups seen in the sequence comparisons described above with the tobacco cv. TN90 and other Nicotiana spp. shown in Supplementary Fig. 3. In addition, the percentage sequence similarities between and within these two SHE1 gene groups (Supplementary Table 1) supports this evolution relationship. Whether the SHE1 gene is also functional in defense signaling in these other species is unknown; however, from a search of N. benthamiana expressed genes for a homologue of the tobacco cv. Samsum NN SHE1 gene, we were not able to identify an intact SHE1-like gene, but rather only clusters of limited sequences showing similarity to the DNA-binding domain of AP2/ERF TFs (data not shown). Given that some transgenic lines of N. benthamiana containing the tobacco N gene are not resistant to systemic infection by wild-type TMV, or even TMV-GFP (Peart et al., 2002), it would be interesting to determine whether this is related to the absence of a resistance pathway in N. benthamiana involving the SHE1 gene.
Acknowledgments
The authors wish to thank Sang Hyon Kim (Myongji University, Yongin, South Korea) and Seung-Kook Choi (RDA, DRPC, Jeonju, South Korea) for providing DNA constructs and for useful discussions.
Electronic Supplementary Material
Supplementary materials are available at The Plant Pathology Journal website (http://www.ppjonline.org/).
Supplementary┬ĀTable┬Ā1.
Comparison of nucleotide and amino acid sequences between SHE1 and other ERF genes
Supplementary┬ĀFig.┬Ā1.
Y2H interactions between the CMV 1a protein and four fragments the CMV 1a protein (1a-I, 1a-II, 1a-III, and 1a-IV) and the SHE1 TF. The diagram above the Y2H results shows the regions of the 1a protein expressed in the four DNA fragments cloned into the Y2H BD vector. Y2H, yeast two-hybrid; CMV, cucumber mosaic virus; BD, binding domain; AD, activation domain. SD-LT and SD-LTHA represent assays in which transformants were selected on SD/-Trp/-Leu, and SD/-Trp/-Leu/-His/-Ade plates containing X-╬▒-gal to detect the initiation of transcription of amino acid biosynthesis reporter genes (TRP, LEU, HIS, and ADE); Positive interactions represents growth of blue-colored yeast cells on SD/-Trp/-Leu/-His/-Ade/ X-╬▒-gal; +, positive interaction; -, negative interaction.
Supplementary┬ĀFig.┬Ā2.
Construction and analysis of MBP and GST fusion proteins. (A) The diagram shows the nature of the fusion protein constructs for expression of SHE1-MBP, CMV 1a-GST, and Fb2-GST. (B) All GST tagged protein was pulled down on glutathione Sepharose beads and confirmed using western blotting with GST antiserum. MBP, maltose binding protein; GST, glutathione S-transferase.
Supplementary┬ĀFig.┬Ā3.
Comparison of nucleotide sequences between SHE1 and highly related ERF-like open reading frames (ORFs) of similar size among cultivars of Nicotiana tabacum and other Nicotiana spp. The nucleotide sequences of ORFs of the SHE1 gene from three cultivars of N. tabacum (Samson NN, Xanthi nc and TN90), and of highly related ERF-like genes from N. sylvestris, N. tomentosiformis, N. umbratica, and N. attenuata are shown. The GenBank numbers for these ORFs, their nucleotide lengths and their percentage sequence identity to SHE1 from tobacco cv. Xanth nc are given in Supplementary Table 1.
Supplementary┬ĀFig.┬Ā4.
Amino acid sequence of the 234-amino acid tobacco cv. Samsun NN SHE1 protein, and proposed conserved domains for functionality. Identical or highly conserved amino acids between ERFs and related proteins are underlined (amino acids 99-157) corresponding to the AP2/ERF DNA-binding domain found in AP2/ERF transcription factors. The sequences at the lowercase residues (a112 and d117) are required for GCC binding. Bold sequences (amino acids 132-147) form an amphipathic alpha-helix. The italicized basic region (amino acids 186-203) could be a nuclear localization signal. The amino acids at the N and C termini resemble those shown to be involved in transcription activation. The N and C termini are also acidic. Nicotiana umbratica contains a nine amino acid sequence (KEKEEEEEE) insertion within the similar array (EEKEKEEŌĆ”EK) found at amino acids 89-98, immediately before the conserved AP2/ERF DNA-binding domain.
Fig.┬Ā1
Yeast two-hybrid reciprocal interactions between the CMV 1a protein and the SHE1 TF. CMV, cucumber mosaic virus; TF, transcription factor; BD, binding domain; AD, activation domain. SD-LT and SD-LTHA represent assays in which transformants were selected on SD/-Trp/-Leu, and SD/-Trp/-Leu/-His/-Ade plates without X-gal to detect the initiation of transcription of amino acid biosynthesis reporter genes (TRP, LEU, HIS, and ADE). Colony-lift filter assay represents colonies transferred to filters and assayed for reaction with X-╬▓-gal; +, positive interaction; ŌłÆ, negative interaction.
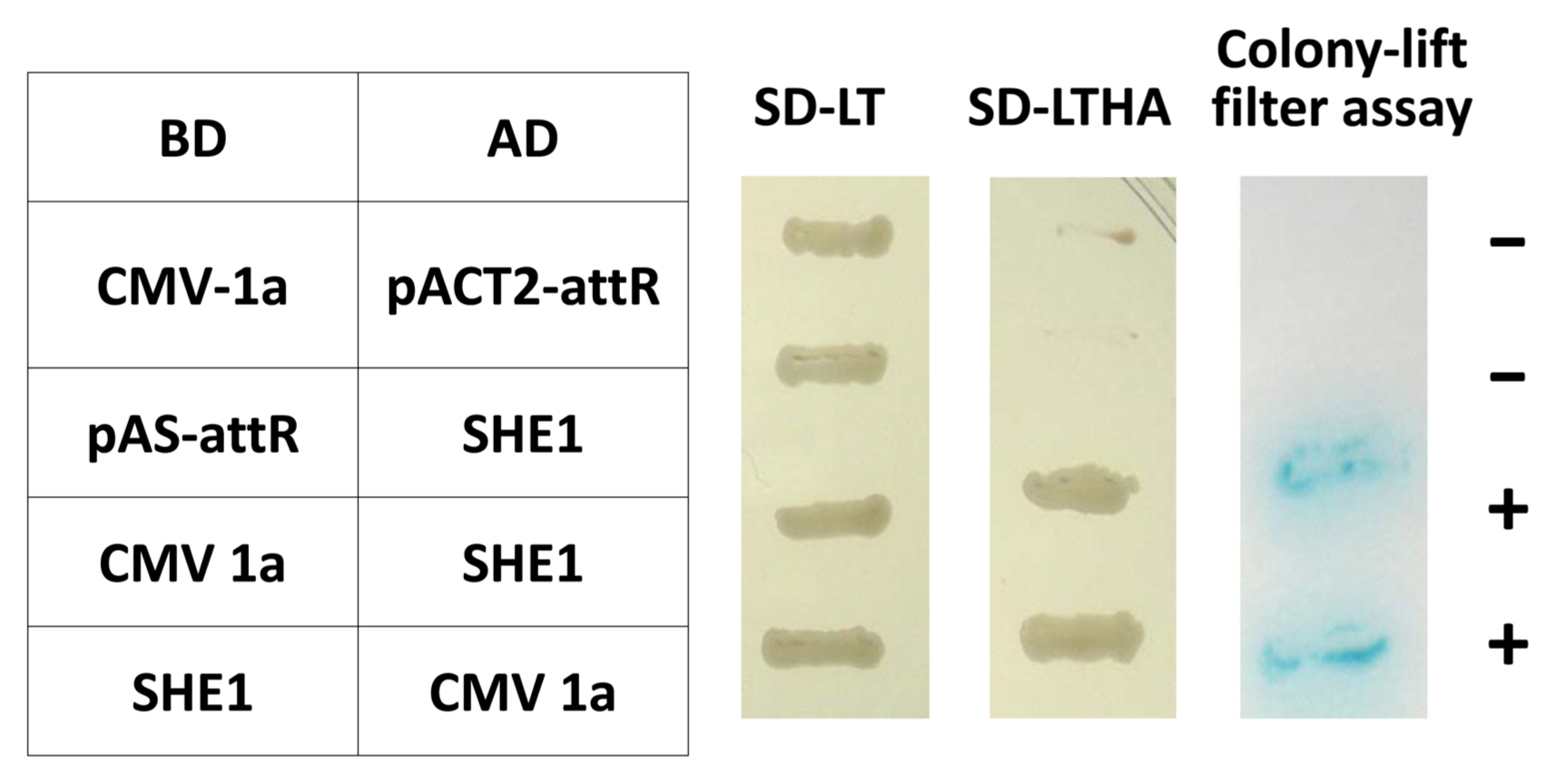
Fig.┬Ā2
Pull-down assay for detection of CMV 1a:SHE1 interaction. Extracted Escherichia coli-expressed fusion proteins (SHE1-MBP, 1a-GST, and Fb2-GST), AP, for SHE1-MBP and associated proteins, using amylose beads, were fractionated by SDS-PAGE and either stained with CBB (lower panel) or subjected to western blotting and probing with antisera to GST (upper panel) or CMV 1a protein (middle panel). The GST-fusion with nucleolar protein fibrillarin (Fb2) and free GST were used as negative controls. The CBB stained proteins served as loading controls. CMV, cucumber mosaic virus; AP, affinity-purified; SDS-PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis; CBB, Coomassie Brilliant Blue; GST, glutathione S-transferase; Fb2, fibrillarin 2. AP expressed protein samples loaded onto the gel were as follows: lane 1, SHE1-MBP/Empty-GST; lane 2, SHE1-MBP/CMV 1a-GST; lane 3, SHE1-MBP/Fb2-GST; and lane 4, SHE1-MBP only. The mol. wt. markers are shown to the left of the upper and middle panels. Protein sizes were as follows: MBP, 42 kDa; GST, 26 kDa; Fb2, 39 kDa; SHE1, 26.5 kDa; and CMV 1a, 111 kDa. The doublet band of ca. 80 kDa present in the anti-CMV 1a probed blot is a non-specific cross-reacting protein.
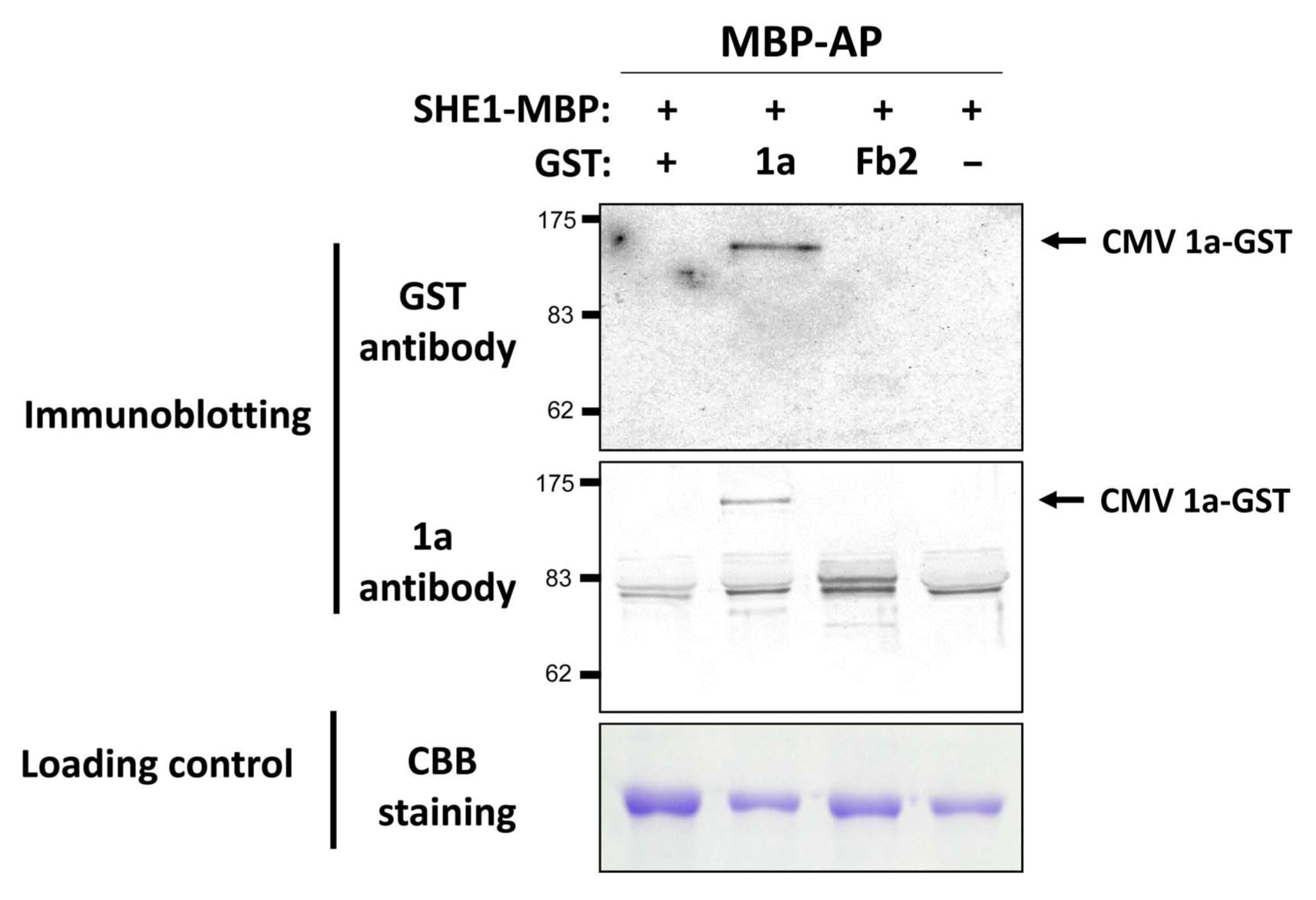
Fig.┬Ā3
BiFC visualization of interaction between SCFP3A-C-CMV1a and SHE1-Venus-N expressed in infiltrated Nicotiana benthamiana epidermal cells. (A) The fusion proteins expressed from the constructs given above the confocal microscopy images showed the interaction of the fusion proteins SCFP3A-C-CIPK24 and CBL1-SCFP3A-N localizing to the plasma membrane (left, upper and lower panels; blue) and the interaction of the fusion proteins SCFP3A-C-CMV1a and SHE1-Venus-N localizing to the nucleus and membranes (middle, upper and lower panels; yellow). The right upper and lower panels show the merging of the left and central panels, demonstrating that the membrane yellow fluorescence of SCFP3A-C-CMV1a interacting with SHE1-Venus-N is not associated with the plasma membrane (magnified in the far right upper panel). The right lower panel also shows localization to the nucleus of the yellow fluorescence. (B) The fusion proteins expressed from the constructs given above the images showed the interaction of the fusion proteins SCFP3A-C-CIPK24 and CBL1-SCFP3A-N localizing to the plasma membrane (left, upper and lower panels; blue) and the interaction of the fusion proteins SCFP3A-C-CIPK24 and CBL10-Venus-N localizing to the tonoplast membrane (middle, upper and lower panels; yellow). The right upper and lower panels show the merging of the left and central panels, demonstrating that these two membranes can de distinguished using these fluorescent markers. BiFC, bimolecular fluorescent complementation; CMV, cucumber mosaic virus.
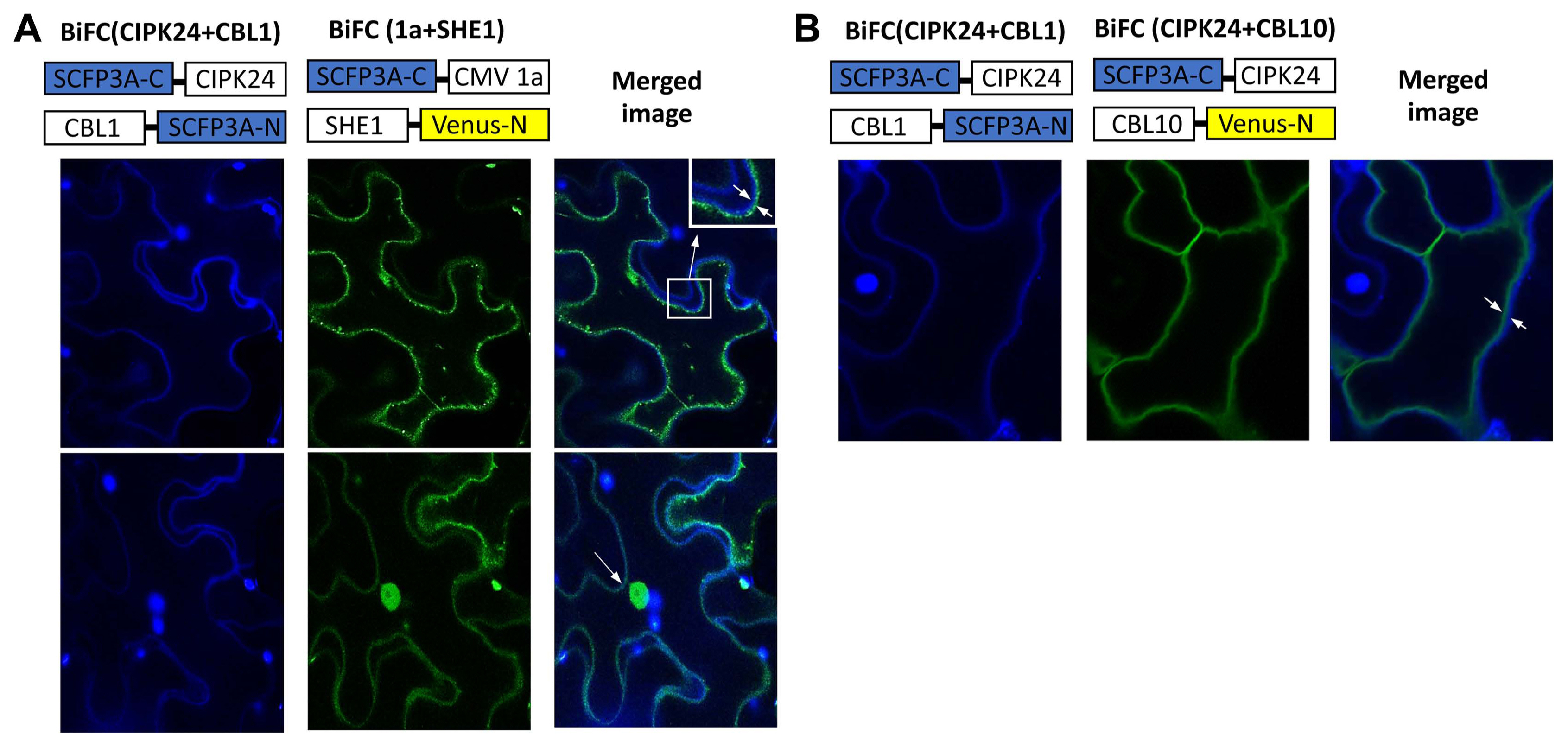
Fig.┬Ā4
Localization of fluorescently tagged CMV 1a, SHE1 and co-localization CMV 1a and SHE1 in infiltrated Nicotiana benthamiana epidermal cells. (A) Diagram showing constructs used for agroinfiltration to express CMV 1a-mRFP in cells (above) and confocal microscopy image of CMV 1a-mRFP localization, with the NU and TO of one cell indicated (below). (B) Diagram showing constructs used for agroinfiltration to express SHE1-YFP in cells (above) and confocal microscopy image of SHE1-YFP localization in the nucleus of various cells, at different magnifications (below). (C) Diagram showing constructs used for agroinfiltration to express CMV 1a-mRFP and SHE1-YFP together in cells (above), and (below) co-localization of CMV 1a-mRFP and SHE1-YFP in cells after co-infiltration, with CMV-1a mRFP images (left), SHE1-YFP images (middle) and merged images (right). (D) Diagram showing constructs used for agroinfiltration to express CMV 1a-YFP in cells (above) and confocal microscopy image of CMV 1a-YFP distribution (below). The yellow dotted circles highlight fluorescence signals in speckles. CMV, cucumber mosaic virus; NU, nucleus; TO, tonoplast membrane.
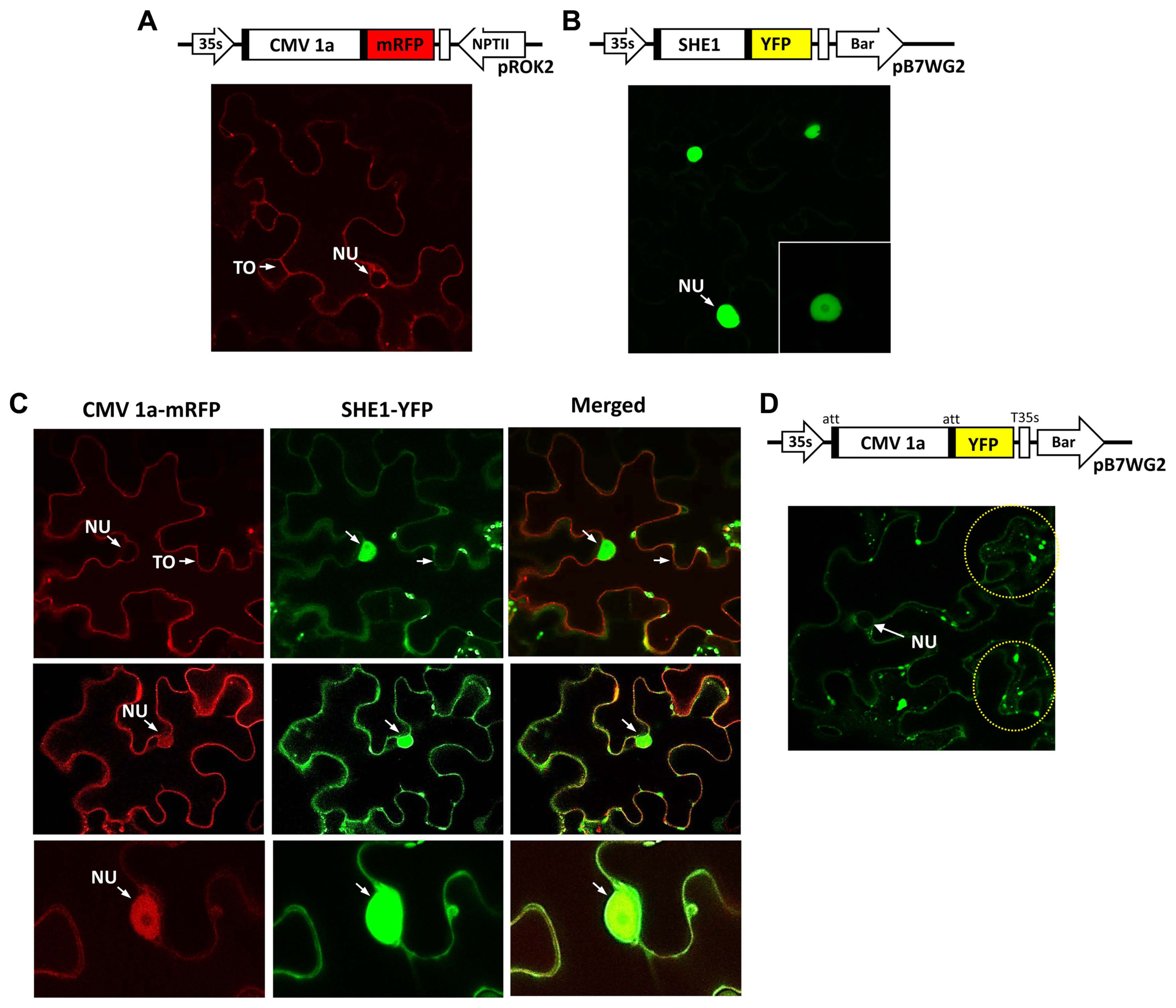
Fig.┬Ā5
Expression analysis of SHE1 mRNA. (A) Expression of SHE1 mRNA as determined by reverse transcription-PCR of total RNAs extracted at 3 dpi from the inoculated leaves of tobacco cv. Samsun NN either mock-inoculated or inoculated with TMV, PVX, CMV, or PVY. The results obtained from different numbers of PCR cycles (15, 25, or 35) are shown. The internal reference gene used was mRNA of the house-keeping gene EF1╬▒. (B) Northern blot of SHE1 mRNA from either inoculated leaves of TMV-inoculated tobacco cv. Samsun NN at various times (h) post-inoculation (upper panel), or non-inoculated leaves of CMV 1a-expressing transgenic tobacco cv. Samsun NN (lower panel). Detection of mRNA to the house-keeping gene Ubiquitin was used as a loading control. TMV, tobacco mosaic virus; PVX, potato virus X; CMV, cucumber mosaic virus; PVY, potato virus Y.
References
Baek, E., Yoon, J.-Y. and Palukaitis, P. 2017. Validation of reference genes for quantifying changes in gene expression in virus-infected tobacco. Virology. 510:29-39.


Canto, T., Cillo, F. and Palukaitis, P. 2002. Generation of siRNAs by T-DNA sequences does not require active transcription or homology to sequences in the plant. Mol. Plant-Microbe Interact. 15:1137-1146.


Canto, T. and Palukaitis, P. 2002. Novel N gene-associated, temperature-independent resistance to the movement of tobacco mosaic virus vectors neutralized by a cucumber mosaic virus RNA1 transgene. J. Virol. 76:12908-12916.



Canto, T., Uhrig, J. F., Swanson, M., Wright, K. M. and MacFarlane, S. A. 2006. Translocation of tomato bushy stunt virus p19 protein into the nucleus by ALY proteins compromises its silencing suppressor activity. J. Virol. 80:9064-9072.



Carr, J. P., Donnelly, R., Tungadi, T., Murphy, A. M., Jiang, S., Bravo-Cazar, A., Yoon, J.-Y., Cunniffe, N. J., Glover, B. J. and Gilligan, C. A. 2018a. Viral manipulation of plant stress responses and host interactions with insects. Adv. Virus Res. 102:177-197.


Carr, J. P. and Murphy, A. M. 2019. Chapter 12:Suppression of plant defense. In: Cucumber mosaic virus, eds. by P. Palukaitis and F. Garc├Ła-Arenal, 133-144. APS Press, St. Paul, MN, USA.
Carr, J. P., Murphy, A. M., Tungadi, T. and Yoon, J.-Y. 2018b. Plant defense signals: players and pawns in plant-virus-vector interactions. Plant Sci. 279:87-95.


Chaturvedi, S. and Rao, A. L. N. 2016. A shift in plant proteome profile for a bromodomain containing RNA binding protein (BRP1) in plants infected with cucumber mosaic virus and its satellite RNA. J. Proteomics. 131:1-7.


Chaturvedi, S., Seo, J.-K. and Rao, A. L. N. 2016. Functionality of host proteins in cucumber mosaic virus replication: GAP-DH is obligatory to promote interactions between replication-associated proteins. Virology. 494:47-55.


Cillo, F., Roberts, I. M. and Palukaitis, P. 2002. In situ localization and tissue distribution of the replication-associated proteins of cucumber mosaic virus in tobacco and cucumber. J. Virol. 76:10654-10664.



Csorba, T., Contra, L. and Burgy├Īn, J. 2015. Viral silencing suppressors: tools forged to fine-tune host-pathogen coexistence. Virology. 479-480:85-103.

Fischer, U. and Dr├Čge-Laser, W. 2004. Overexpression of NtERF5, a new member of the tobacco ethylene response transcription factor family enhances resistance to tobacco mosaic virus. Mol. Plant-Microbe Interact. 17:1162-1171.


Gehl, C., Waadt, R., Kudla, J., Mendel, R.-R. and H├żnsch, R. 2009. New GATEWAY vectors for high throughput analysis of protein-protein interactions by bimolecular fluorescence complementation. Mol. Plant. 2:1051-1058.


Guo, Z., Li, Y. and Ding, S.-W. 2019. Small RNA-based antimicrobial immunity. Nat Rev Immunol. 19:31-44.


Hao, D., Yamasaki, K., Sarai, A. and Ohme-Tagaki, M. 2002. Determinants in the sequence specific binding of two plant transcription factors, CBF1 and NtERF2, to the DRE and GCC motifs. Biochemistry. 41:4202-4208.


Hu, Z., Zhang, T., Yao, M., Feng, Z., Miriam, K., Wu, J., Zhou, X. and Tao, X. 2012. The 2a protein of cucumber mosaic virus induces a hypersensitive response in cowpea independently of its replicase activity. Virus Res. 170:169-173.


Huh, S. U., Kim, M. J., Ham, B.-K. and Paek, K.-H. 2011. A zinc finger protein Tsip1 controls cucumber mosaic virus infection by interacting with the replication complex on vacuolar membranes of the tobacco plant. New Phytol. 191:746-762.


Jin, Y., Zhao, J.-H. and Guo, H.-S. 2021. Recent advances in understanding plant antiviral RNAi and viral suppressors of RNAi. Curr. Opin. Virol. 46:65-72.


Kang, W.-H., Seo, J.-K., Chung, B. N., Kim, K.-H. and Kang, B.- C. 2012. Helicase domain encoded by cucumber mosaic virus RNA1 determines systemic infection of Cmr1 in pepper. PLoS ONE. 7:e43136.



Karasawa, A., Okada, I., Akashi, K., Chida, Y., Hase, S., Nakazawa-Nasu, Y., Ito, A. and Ehara, Y. 1999. One amino acid change in cucumber mosaic virus RNA polymerase determines virulent/avirulent phenotypes on cowpea. Phytopathology. 89:1186-1192.


Kim, C.-H. and Palukaitis, P. 1997. The plant defense response to cucumber mosaic virus in cowpea is elicited by the viral polymerase gene and affects accumulation in single cells. EMBO J. 16:4060-4068.



Kim, M. J., Ham, B.-K., Kim, H. R., Lee, I.-J., Kim, Y. J., Ryu, K. H., Park, Y. I. and Paek, K.-H. 2005. In vitro and in planta interaction evidence between Nicotiana tabacum thaumatin-like protein 1 (TLP1) and cucumber mosaic virus proteins. Plant Mol Biol. 59:981-994.


Kim, M. J., Ham, B.-K. and Paek, K.-H. 2006a. Novel protein kinase interacts with the cucumber mosaic virus 1a methyltransferase domain. Biochem Biophys Res Commun. 340:228-235.


Kim, M. J., Huh, S. U., Ham, B.-K. and Paek, K.-H. 2008. A novel methyltransferase methylates cucumber mosaic virus 1a protein and promotes systemic spread. J. Virol. 82:4823-4833.



Kim, M. J., Kim, H. R. and Paek, K.-H. 2006b. Arabidopsis tonoplast proteins TIP1 and TIP2 interact with the cucumber mosaic virus 1a replication protein. J. Gen Virol. 87:3425-3431.


Kim, S. H., MacFarlane, S., Kalinina, N. O., Ryabov, E. V., Gillespie, T., Haupt, S., Brown, J. W. S. and Taliansky, M. 2007. Interaction of a plant virus-encoded protein with the major nucleolar protein fibrillarin is required for systemic virus infection. Proc Natl Acad Sci U S A. 104:11115-11120.



Kim, S. H., Palukaitis, P. and Park, Y. I. 2002. Phosphorylation of cucumber mosaic virus RNA polymerase 2a protein inhibits formation of replicase complex. EMBO J. 21:2292-2300.



Kong, J., Wei, M., Li, G., Lei, R., Qiu, Y., Wang, C., Li, Z.-H. and Zhu, S. 2018. The cucumber mosaic virus movement protein suppresses PAMP-triggered immune responses in Arabidopsis and tobacco. Biochem Biophys Res Commun. 498:395-401.


Lemon, B. and Tjian, R. 2000. Orchestrated response: a symphony of transcription factors for gene control. Genes Dev. 14:2551-2569.


Lu, Y. and Tsuda, K. 2021. Intimate association of PRR- and NLR-mediated signaling in plant immunity. Mol. Plant-Microbe Interact. 34:3-14.


Nabori, T. and Tsuda, K. 2019. The plant system in heterogeneous environments. Curr Opin Plant Biol. 50:58-66.

Nakahara, K. S. and Masuta, C. 2014. Interaction between viral RNA silencing suppressors and host factors in plant immunity. Curr Opin Plant Biol. 20:88-95.


Palukaitis, P. 2019. Chapter 10:Genome structure and expression. In: Cucumber mosaic virus, eds. by P. Palukaitis and F. Garc├Ła-Arenal, 113-121. APS Press, St. Paul, MN, USA.
Palukaitis, P. and Yoon, J.-Y. 2020. R gene mediated defense against viruses. Curr. Opin. Virol. 45:1-7.


Peart, J. R., Cook, G., Feys, B. J., Parker, J. E. and Baulcombe, D. C. 2002. An EDS1 orthologue is required for N-mediated resistance against tobacco mosaic virus. Plant J. 29:569-579.


Sierro, N., Battey, J. N. D., Ouadi, S., Bakaher, N., Bovet, L., Willig, A., Goepfert, S., Peitsch, M. C. and Ivanov, N. V. 2014. The tobacco genome sequence and its comparison with those of tomato and potato. Nat Commun. 5:3833.



Takahashi, H., Goto, N. and Ehara, Y. 1994. Hypersensitive response in cucumber mosaic virus-inoculated Arabidopsus thaliana. Plant J. 6:369-377.

Takahashi, H., Miller, J., Nozaki, Y., Takeda, M., Shah, Y., Hase, S., Ikegami, M., Ehara, Y. and Dinesh-Kumar, S. P. 2002. RCY1, an Arabidopsis thaliana RPP8/HRT family resistance gene, conferring resistance to cucumber mosaic virus requires salicylic acid, ethylene and a novel signal transduction mechanism. Plant J. 32:655-667.


Takahashi, H., Suzuki, M., Natsuaki, K., Shigyo, T., Hino, K., Teraoka, T., Hosokawa, D. and Ehara, Y. 2001. Mapping the virus and host genes involved in the resistance response in cucumber mosaic virus-infected Arabidopsis thaliana. Plant Cell Physiol. 42:340-347.


Tungadi, T., Donnelly, R., Qing, L., Iqbal, J., Murphy, A. M., Pate, A. E., Cunniffe, N. J. and Carr, J. P. 2020. Cucumber mosaic virus 2b proteins inhibit virus-induced aphid resistance in tobacco. Mol. Plant Pathol. 21:250-257.


Waadt, R., Schmidt, L. K., Lohse, M., Hashimoto, K., Bock, R. and Kudla, J. 2008. Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J. 56:505-516.


Watt, L. G., Crawshaw, S., Rhee, S.-J., Murphy, A. M., Canto, T. and Carr, J. P. 2020. The cucumber mosaic virus 1a protein regulates interactions between the 2b protein and ARGO-NAUTE 1 while maintaining the silencing suppressor activity of the 2b protein. PLoS Pathog. 16:e1009125.



Westwood, J. H., Groen, S. C., Du, Z., Murphy, A. M., Anggoro, D. T., Tungadi, T., Luang-In, V., Lewsey, M. G., Rossiter, J. T., Powell, G., Smith, A. G. and Carr, J. P. 2013. A trio of viral proteins tunes aphid-plant interactions in Arabidopsis thaliana. PLoS ONE. 8:e83066.



Westwood, J. H., Lewsey, M. G., Murphy, A. M., Tungadi, T., Bates, A., Gilligan, C. A. and Carr, J. P. 2014. Interference with jasmonic acid-regulated gene expression is a general property of viral suppressors of RNA silencing but only partly explains virus-induced changes in plant-aphid interactions. J. Gen Virol. 95:733-739.



Yoda, H., Ogawa, M., Yamaguchi, Y., Koizumi, N., Kusano, T. and Sano, H. 2002. Identification of early-response genes associated with the hypersensitive response to tobacco mosaic virus and characterization of a WRKY-type transcription factor in tobacco plants. Mol. Genet Genomics. 267:154-161.


Yoon, J.-Y., Han, K.-S., Park, H.-Y. and Choi, S.-K. 2012. Comparative analysis of RNA silencing suppression activities between viral suppressors and an endogenous plant RNA-dependent RNA polymerase. Virus Genes. 44:495-504.


- TOOLS
-
METRICS

- ORCID iDs
-
Ju-Yeon Yoon

https://orcid.org/0000-0003-1646-7310 - Related articles



 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Supplement1
Supplement1 Print
Print



