Regulation of Salicylic Acid and N-Hydroxy-Pipecolic Acid in Systemic Acquired Resistance
Article information
Abstract
In plants, salicylic acid (SA) is a central immune signal that is involved in both local and systemic acquired resistance (SAR). In addition to SA, several other chemical signals are also involved in SAR and these include N-hydroxy-pipecolic acid (NHP), a newly discovered plant metabolite that plays a crucial role in SAR. Recent discoveries have led to a better understanding of the biosynthesis of SA and NHP and their signaling during plant defense responses. Here, I review the recent progress in role of SA and NHP in SAR. In addition, I discuss how these signals cooperate with other SAR-inducing chemicals to regulate SAR.
The innate immune system of plants detects and responds to pathogens in their environment (Chassot et al., 2008; Osbourn, 1996; Underwood, 2012). Owing to the evolutionary arms race between pathogens and plants, plants have developed layers of immune defense. The immune response typically begins with membrane-localized pattern recognition receptors that detect molecular patterns associated with microbes. This triggers pattern-triggered immunity (PTI). Parallelly, R proteins, mainly nucleotide-binding leucine-rich repeat receptors, perceive effectors secreted by pathogens to suppress plant defenses. Direct and idirect recognition of effector proteins by R proteins activate effector-triggered immunity (ETI) (Jones and Dangl, 2006). PTI and ETI not only induce local defenses but can also induce resistance in the plant’s distal parts by delivering signals, thereby establishing long-lasting and broad-spectrum resistance. This phenomenon is called systemic acquired resistance (SAR). The SAR-induced plants are primed for induction of defense genes thereby enabling plants to defend themselves more efficiently against subsequent pathogen infection (Fu and Dong, 2013; Klessig et al., 2018). Therefore, understanding the regulatory mechanisms of SAR is crucial for enhancing plant disease resistance and reducing yield losses. In recent years, researchers have begun dissecting the key components that regulate the SAR pathway to gain a better understanding of its regulation. SAR mediated long-distance signaling involves several SAR-inducing chemicals, including salicylic acid (SA) (Shah et al., 2014), methyl salicylic acid (MeSA) (Park et al., 2007, 2009), azelaic acid (AzA) (Jung et al., 2009), glycerol-3-phosphate (G3P) (Chanda et al., 2011), dehydroabietinal (Chaturvedi et al., 2012), pipecolic acid (Pip) (Návarová et al., 2012; Zeier, 2013), N-hydroxy-pipecolic acid (NHP) (Hartmann et al., 2018; Návarová et al., 2012), the free radicals nitric oxide (NO) and reactive oxygen species (El-Shetehy et al., 2015; Wang et al., 2014), and galactolipids (Gao et al., 2015). SAR is also associated with factors contributing to cuticle formation (Lim et al., 2020; Xia et al., 2009, 2010), and the lipid transfer proteins defective in induced resistance 1 (DIR1) (Maldonado et al., 2002; Yu et al., 2013), AzA insensitive 1 (AZI1) (Jung et al., 2009), and trans-acting small interfering RNA3a RNAs (TAS3a) (Shine et al., 2022).
Mutants with impaired SA biosynthesis, perception, or signal transduction have weakened disease resistance, whereas those with high SA levels have improved disease resistance (Fu and Dong, 2013; Nawrath and Métraux, 1999; Torrens-Spence et al., 2019; Wu et al., 2012). Additionally, SA and its analogs can enhance disease resistance both locally and systemically (Gao et al., 2014; Vlot et al., 2009). In contrast to the extensive research conducted on the mechanism and regulatory network of SA in SAR, little is known about the actions of many mobile signals. In a recent study, NHP was identified as a crucial molecule in Arabidopsis, which accumulated within 24 h after local infection and triggered SAR. Here, I review the role of SA and NHP in SAR, with an aim to provide an overview of the current advancements and future perspectives in SA and NHP biology.
Regulation of SA Biosynthesis
In higher plants, the biosynthesis of SA occurs via the isochorismate synthase (ICS)- and/or phenylalanine ammonia-lyase (PAL)-dependent pathways (Huang et al., 2010; Rekhter et al., 2019; Wildermuth et al., 2001). While contribution of these two branches differs among plants, in Arabidopsis a majority (~90%) of pathogen-induced SA is derived from the ICS-catalyzed branch. The Arabidopsis plants express two ICS genes and off these ICS1 contributes to a majority of pathogen-induced SA (Garcion et al., 2008; Nawrath and Métraux, 1999). Interestingly, a mutation in either PAL isoforms or ICS1 impairs SAR, indicating that both SA biosynthesis via both ICS and PAL branches is important for SAR (Huang et al., 2010). The pathogen induced expression of ICS1 is dependent on calmodulin binding protein 60g (CBP60g) and SAR-deficient 1 (SARD1) transcription factors (Truman and Glazebrook, 2012). The cbp60g sard1 double mutant shows impaired ICS1 induction and SA biosynthesis, resulting in a compromised SAR (Zhang and Zhou, 2010). Biosynthesis and transport of SA precursor isochorimate from chloroplast to cytosol is dependent on cytosolic amidotransferase avrPphB susceptible 3 (PBS3) and chloroplastic enhanced disease susceptibility 5 protein (EDS5), respectively (Rekhter et al., 2019). The mutant defective in either PBS3 or EDS5 shows impaired SA accumulation and compromised SAR (Nawrath and Métraux, 1999).
Upon biosynthesis, SA is can be converted to SA 2-O-β-D-glucoside (SAG) and MeSA through glycosylation or methylation, respectively. At least three Arabidopsis UDP-glucosyltransferases are involved in the conversion of SA to SAG (Dean and Delaney, 2008; Song, 2006). In Arabidopsis, mutants of UGT74F1, UGT74F2, and UGT76B1 exhibit reduced SAG levels, increased SA accumulation, and enhanced disease resistance (Noutoshi et al., 2012; von Saint Paul et al., 2011) (Fig. 1).
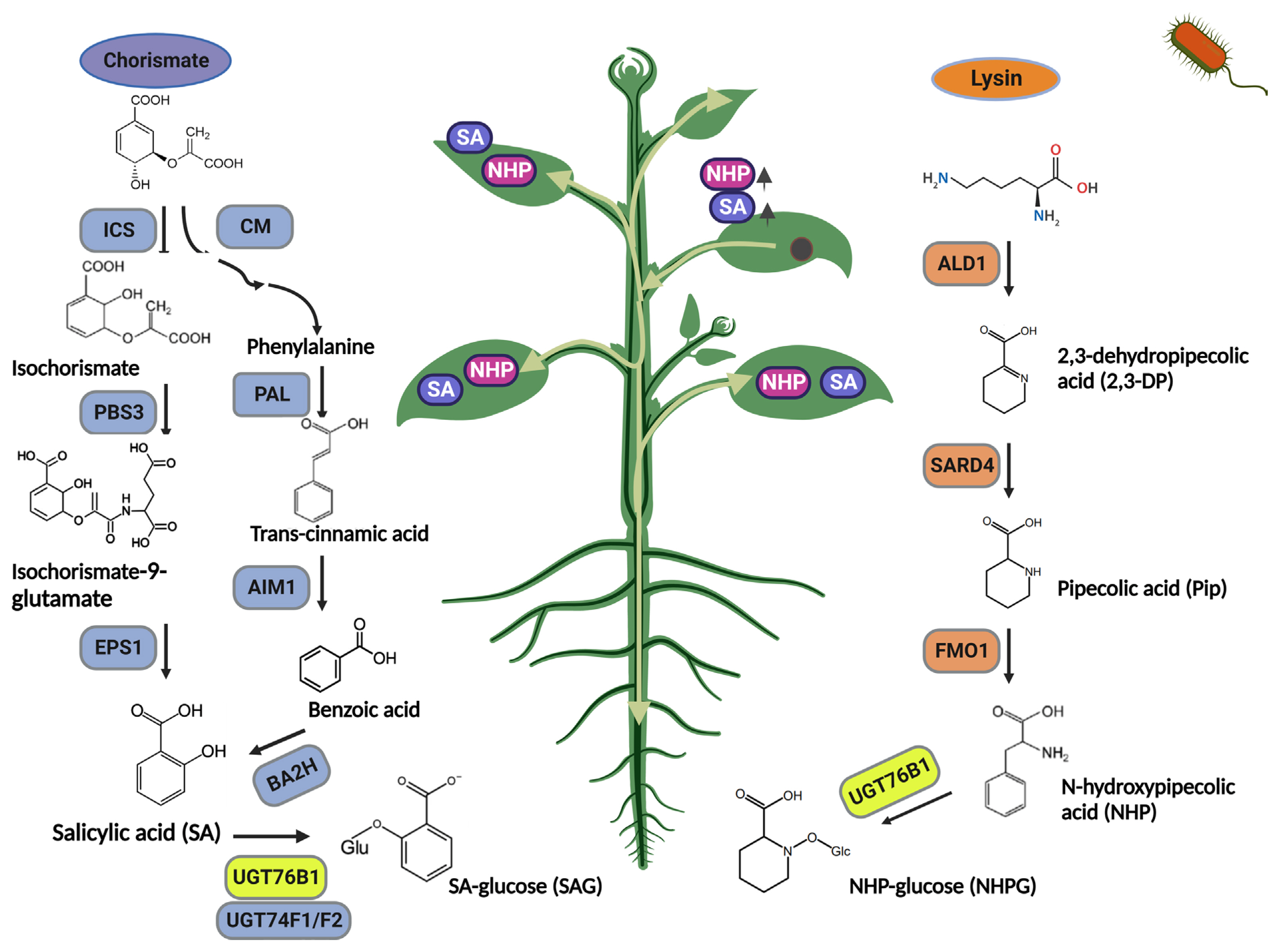
Salicylic acid and N-hydroxypipecolic acid are required for systemic acquired resistance. Abbreviations used are isochosrismate synthase (ICS), avrPphB susceptible 3 (PBS3), EPS1 chorismate mutase (CM), phenylalanine ammonia lyase (PAL), abnormal inflorescence meristem1 (AIM1) benzoic acid 2-hydroxylase (BA2H), AGD2-like defense response protein 1 (ALD1), SAR-deficient 4 (SARD4), flavin-dependent monooxygenase 1 (FMO1), and UDP-glycosyltransferase 76B1 (UGT76B1).
Role of SA in Local and Systemic Defenses
During SAR, SA accumulates both locally and systemically, and early experiments have shown that the degradation of SA by the SA hydroxylase (NahG) compromises both local resistance and SAR (Vernooij et al., 1994). Interestingly, plants lacking the R protein RPS2 accumulate normal levels of SA in their distal tissues after infection with P. syringae pv. tomato (Pst) DC3000 carrying avrRpt2, but still remain compromised in SAR (Cameron et al., 1999). This suggests factors other than SA may contribute to SAR in rps2 plants. G3P or AzA, which induce SAR in wild-type plants, do not induce SA accumulation in plants. However, G3P or AzA are unable to confer SAR in ics1/sid2 plants, which accumulate significantly reduced basal and pathogen-induced SA levels (Chanda et al., 2011). Together, these results suggest that, while SA is clearly important for SAR, this alone is insufficient. Recent work has shown that distal transport of SA is crucial for SAR and is regulated by water potential (Lim et al., 2020).
Regulation of NHP Biosynthesis
NHP, another recently discovered plant metabolite, is also essential for SAR (Chen et al., 2018; Hartmann and Zeier, 2018). In Arabidopsis, NHP is synthesized from Pip (Fig. 1). Three distinct enzymes are required for biosynthesis of NHP and are encoded by genes that are highly responsive to biotic stress (Zeier, 2021). Among them, AGD2-like defense response protein 1 (ALD1) encodes a Lys aminotransferase that generates 2,3-dehydro-pipecolic acid (dehydro-Pip; 2,3-DP) from lysine. SAR-deficient 4 (SARD4), which encodes bacterial ornithine cyclodeaminase, converts 2,3-DP to Pip. Lastly, flavin-dependent monooxygenase 1 (FMO1) generates NHP by adding hydroxyl amine to Pip. The ald1 and fmo1 mutants prevent the accumulation of NHP, resulting in reduced pathogen resistance and compromised SAR (Mishina and Zeier, 2006; Song et al., 2004). Additionally, pathogen inoculated fmo1 plants accumulate Pip to higher than wild-type levels. NHP exists in both the free and glycosylated forms. Plants infected with pathogens accumulate NHP and NHP-N-O-glucoside (NHPG) (Chen et al., 2018; Hartmann et al., 2018). UDP-glycosyltransferases (UGTs) play a crucial role in the regulation of signaling molecules via glycosylation (Chen et al., 2020; Dean and Delaney, 2008; Hou et al., 2004; Jin et al., 2013; Song, 2005). A number of UGTs are closely associated with plant disease resistance. For example, UGT73B3 and UGT73B5 are required for resistance to Pst DC3000 in Arabidopsis (Langlois-Meurinne et al., 2005). In addition, UGTs can recognize certain defense-related metabolites as substrates and alter them to an inactive form. NHP are glycosylated by UGT76B1 to produce NHPG, which is inactive (Bauer et al., 2021; Cai et al., 2021; Holmes et al., 2021; Mohnike et al., 2021) (Fig. 1).
Role of Pip and NHP in Local and Systemic Defenses
Plants locally treated with Pip induce SAR (Li et al., 2020; Wang et al., 2018). Exogenous Pip application also enhances local resistance to P. syringae, inducing defense priming and the expression of genes associated with plant defense (Bernsdorff et al., 2016; Hartmann et al., 2018; Návarová et al., 2012). Pip can be detected in vascular exudates after local infection (Návarová et al., 2012; Wang et al., 2018) and localized application of 14C- Pip is detected in distal leaves (Wang et al., 2018). However, petiole exudate from Pip deficient ald1 plants can induce SAR, suggesting that transport of Pip or NHP is not required for SAR (Shine et al., 2022; Wang et al., 2018). Exogenous application of NHP restores SAR in ald1 and fmo1 mutant, suggesting that NHP functions downstream of Pip (Chen et al., 2018; Hartmann et al., 2018; Zeier, 2021). However, no endogenous free NHP was detected at infection sites on wild-type seedlings or adult plants (Chen et al., 2018). Jiang et al. (2021) were also unable to detect NHP in local exudate and distal leaves in ald1 mutants and DEX-induced transgenic ALD1. It remains unclear whether NHP is converted to additional SAR signaling molecules (Shan and He, 2018; Yildiz et al., 2021).
Transport of SA and NHP in SAR
During SAR, SA preferentially transports via the apoplast, while AzA and G3P load via the symplast (Lim et al., 2016). AzA and G3P are transported by symplastic transport through the plasmodesmata (PD). PDLP1 and PDLP5 (plasma localizing protein 1 and 5), two PD localizing proteins, regulate SAR by controlling PD gating and subcellular partitioning (Lim et al., 2016). Recent research further suggests that a portion of the total SA is incorporated into cuticle wax during systemic SA transport (Lim et al., 2020). As a result, mutants with defects in the cuticle show reduced SA transport to distal tissues and compromised SAR (Lim et al., 2020). Cuticle defects prevent SA from moving through the apoplast since increased transpiration in these mutants leads to a reduction in apoplastic hydrostatic pressure (Kachroo et al., 2022; Lim et al., 2020).
During pathogen infection, SA is synthesized in the cytoplast (Rekhter et al., 2019). In contrast, Pip appears to be synthesized in plastids based on the localization of ALD1 and SARD4 (Cecchini et al., 2015; Sharma et al., 2013; Wang et al., 2018). Pip is likely transported to cytosol where it is converted to NHP via cytosol localized FMO1 (Hartmann et al., 2018; Kachroo et al., 2021). Interestingly, it was recently found that UV-induced NHP accumulation is markedly reduced in eds5 mutant plants (Rekhter et al., 2019). Exogenous application of SA could not recover NHP accumulation in eds5 mutant plants, indicating that EDS5 is required for NHP biosynthesis (Rekhter et al., 2019). It is probable that besides SA precursor, EDS5 may also facilitates the transport of Pip from the plastid to the cytosol, where Pip is converted into NHP by FMO1 (Rekhter et al., 2019). It remains unclear whether Pip promotes plant immunity by exerting its function in plastids or through translocation.
SA-NHP Interaction during Plant Immunity
In view of the common overlap between the SA and NHP regulators, it is not surprising that SA and NHP could cooperatively influence each other to induce SAR (Shields et al., 2022). SAR and/or the priming of associated defenses may also involve interaction between various SAR associated chemicals (Bernsdorff et al., 2016; Hartmann et al., 2018; Kachroo et al., 2022; Koo et al., 2020). A recent ChIP analysis revealed that SARD1 and CBP60g target not only genes involved in the biosynthesis of SA but also genes involved in the synthesis of NHPs, such as ALD1, SARD4, and FMO1 (Sun et al., 2015). The expression levels of ALD1, SARD4, and FMO1 are significantly reduced in sard1 cbp60g double mutant inoculated with Pseudomonas syiringae pv. maculicola (Psm) ES4326 (Huang et al., 2020). In contrast, overexpression of SARD1 increases the expression of ALD1 and SARD4 as well as the level of Pip (Sun et al., 2018). The sard1 cbp60g double mutant shows significantly lower Pip and NHP levels after infection with Psm ES4326 than the wild-type plants, suggesting that SARD1 and CBP60g activate Pip and NHP biosynthesis by inducing their biosynthesis genes. Additionally, UGT76B1 accepts both NHP and SA as substrates (Bauer et al., 2021; Cai et al., 2021). These results suggest interaction between SA- and NHP-regulated processes leading to SAR. Notably, D9-NHP of leaf- to leaf movement was also observed in the ics1 mutant, suggesting that SA signaling is not required for transport of NHP (Yildiz et al., 2021). Although, numerous transcription factors regulating SA and NHP biosynthesis have been identified, their connection to upstream defense signaling components remains unclear.
Concluding Remarks and Perspectives
The SAR pathway are conserved between diverse plants including Arabidopsis, soybean, tobacco, cucumber, tomato, and the monocot Brachypodium dystachyon (Holmes et al., 2019; Schnake et al., 2020; Shine et al., 2019). Higher levels of SA and NHP result in dwarfed plants (Cai et al., 2021; Rivas-San Vicente and Plasencia, 2011), suggesting that optimal levels of these chemical govern normal development and defense. The transient expression of Arabidopsis ALD1 and FMO1 in N. benthamiana can increase NHP production 100–1,000 times more than the native plants (Holmes et al., 2019). The transient overexpression of ALD1, SARD4, and FMO1 in tomato plants can also induce disease resistance and activate the SAR pathway (Holmes et al., 2019). Similarly, transient expression of UGT76B1 glycosylates NHP and suppresses defense signals in tomato (Holmes et al., 2021). This suggests that while engineering plants to produce more NHP may confer disease resistance it can also affect their normal growth and development.
Acknowledgments
This work was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF-2022R1C1C1012729) and Pusan National University Research Grant, 2021.
Notes
Conflicts of Interest
No potential conflict of interest relevant to this article was reported.
