 |
 |
| Plant Pathol J > Volume 39(5); 2023 > Article |
|
Abstract
Cowpea mild mottle virus (CPMMV) is a global plant virus that poses a threat to the production and quality of legume crops. Early and accurate diagnosis is essential for effective managing CPMMV outbreaks. With the advancement in isothermal recombinase polymerase amplification and lateral flow strips technologies, more rapid and sensitive methods have become available for detecting this pathogen. In this study, we have developed a reverse transcription recombinase polymerase amplification combined with lateral flow strips (RT-RPA-LFS) method for the detection of CPMMV, specifically targeting the CPMMV coat protein (CP) gene. The RT-RPA-LFS assay only requires 20 min at 40°C and demonstrates high specificity. Its detection limit was 10 copies/μl, which is approximately up to 100 times more sensitive than RT-PCR on agarose gel electrophoresis. The developed RT-RPA-LFS method offers a rapid, convenient, and sensitive approach for field detection of CPMMV, which contribute to controlling the spread of the virus.
Cowpea mild mottle virus (CPMMV), a member of the genus Carlavirus in the family Betaflexiviridae, is a pathogen causing emerging agricultural diseases worldwide (Oliver and Whitfield, 2016). Initially the virus was discovered infect cowpea in Ghana, CPMMV has been found to infect legume plants and various other crops (Brunt and Kenten, 1973). Plants infected with CPMMV often show symptoms including mild mottle, chlorosis, deformity, and necrosis (Zanardo et al., 2014). The virus is naturally transmitted by Bemisia tabaci or by seeds and has spread to Asia, Europe, and South and North America, posing a significant threat to legume production (Wei et al., 2021).
Effective disease management strategies for viruses rely on identifying infected plants in time to prevent virus spread. Therefore, a rapid, sensitive, and specific diagnostic method development is essential for controlling diseases. Over the past few decades, several diagnostic methods have been developed, including reverse transcription-polymerase chain reaction (RT-PCR), multiple RT-PCR, or real-time RT-PCR (Asano et al., 2015; Boonham et al., 2002; Liu et al., 2019; Mortimer-Jones et al., 2009; Nemes and Salánki, 2020; Roberts et al., 2000; Tantiwanich et al., 2018), reverse transcription thermostable helicase-dependent DNA amplification (RT-HDA) (Wu et al., 2016), immunocapture reverse transcription loop-mediated isothermal amplification (IC/RT-LAMP) (Fukuta et al., 2004), enzyme-linked immunosorbent assays (ELISA) (Griep et al., 2000; Huguenot et al., 1990), nondestructive Raman spectroscopy (RS) (Mandrile et al., 2019) and small RNA sequencings (Hagen et al., 2011). However, immune-related diagnostic methods and small RNA were too expensive for field detection. Although nondestructive RS is a rapid and inexpensive method, its accuracy is relatively low. Traditional RT-PCR faces limitations in its ability to rapidly detect viruses due to its reliance on intricate laboratory conditions and equipment. In addition, RT-HDA and RT-LAMP assays require a high temperature of 65°C, which largely diminishes their practicality for on-field detection, hence there is an urgent need to develop a rapid, accurate, convenient, and cost-effective method for detecting CPMMV in order to effectively mitigate the spread of the virus.
Recombinase polymerase amplification (RPA) is a novel nucleic acid amplification technology that requires only simple sample preparation and no specialized equipment. RPA assay can be conducted within 30 min at 37-42°C, with high sensitivity and specificity (Piepenburg et al., 2006). Results can be presented by agarose gel electrophoresis (AGE), TwistAmp exo probes (TwistDx, Cambridge, UK) in real time (Boyle et al., 2013) or simply with lateral flow strips (LFS) (Cao et al., 2020). The LFS assay boasts the benefits of obviating the need for specialized equipment and facilitating direct visualization within a mere 5 min, thereby bestowing upon a notably high level of convenience. The combination of RPA and LFS holds tremendous potential for further advancements. To date, RPA has been successfully employed for the detection of plant viruses, including rose rosette virus (Babu et al., 2017), apple stem pitting virus (Kim et al., 2019), chilli veinal mottle virus (Jiao et al., 2020), apple stem grooving virus (Kim et al., 2018), milk vetch dwarf virus (Cao et al., 2020), maize chlorotic mottle virus (Jiao et al., 2019a), and cucumber green mottle mosaic virus (Jiao et al., 2019b).
In this study, we present the development of a reverse transcription recombinase polymerase amplification combined with a lateral flow strips (RT-RPA-LFS) method that specifically targets the coat protein (CP) gene of CPMMV, facilitating and ultra-sensitive detection. The RT-RPA-LFS assay can be completed within a short time-frame of 30 min, and the results can be directly visualized. Through methodical optimization of the RT-RPA-LFS procedure, coupled with a meticulous comparative analysis of its precision and selectivity against conventional RT-PCR, our findings underscore the effectiveness of RT-RPA-LFS as an innovative and potent approach, holding remarkable promise for the precise identification of CPMMV.
A total of 21 different isolate sequences of the CPMMV CP genes were obtained from NCBI GenBank, and a multiple sequence alignment was performed by DNAMAN v8.0 software. In the context of RPA and PCR assay, primers were designed from the conserved region of CP genes of CPMMV. The primer pairs for RPA were formulated to meet specific criteria: (1) The primers should have fewer than six consecutive bases. (2) The structure of stem loop was to encompass no more no more than five bases. (3) The primers should only match the CPMMV CP genes, according to NCBI-BLAST. The RPA primers were designed to be longer than 29 bp, and the length of the amplicons was restrained to a maximum of 500 bp (Supplementary Table 1). Biotin was added in the 5 termini of reverse primers. Probes for RPA-LFS assays carry a FAM antigenic marker and a C3 spacer at the 5′ and 3′ terminal, respectively. The 30th base was replaced with tetrahydrofuran. Additionally, primer pairs for PCR were designed at the same position as RPA primer pairs but with a length of 18-22 base pairs.
Common beans were collected in Zhejiang province and Jiangsu province, China, in the summer of 2021-2022 and identified with RT-PCR agarose gel electrophoresis (RT-PCR-AGE).
Total RNA was extracted from leaves using the EASYspin RNA Plant Mini Kit (Aidlab, Beijing, China) using a random primer and the First Strand cDNA Synthesis Kit (Toyobo, Tokyo, Japan).
Crude leaf extracts were prepared by grinding about 300 mg fresh leaf samples in a mesh bag with 500 μl phosphate buffered saline containing 2% Tween-20. Grinding fluid was then transferred into a 1.5 ml centrifugal tube for centrifugation at 13,000 rpm for 2 min, and the supernatant fluid was used for further assays.
For the PCR assay, a 50 μl reaction system was used, which contained 25 μl buffer, 2 μl of 10 μM forward and reverse primers, 1 μl of template, and 20 μl of ddH2O. The PCR reaction was performed as follows: 95°C for 3 min, followed by 40 cycles of 95°C for 30 s, 56°C for 30 s, 72°C for 30 s, culminating in a final extension at 72°C for 10 min.
RPA-LFS assay was also conducted in a 50 μl supplemented in the kit according to manufacter’s instructions (Zhongce, Hangzhou, China). The reaction composition contained 25 μl of rehydration buffer, 2 μl of 10 μM forward primers, 2 μl of 10 μM reverse primers, 0.6 μl of 10 μM probe, 1 μl of template and 16.9 μl of ddH2O. These components were mixed and added to PCR micro-tubes with a dried enzyme pellet. To initiate the reaction, 2.5 μl 280 mM MgAc was added to tube caps, which were subsequently subjected to vortexing and simultaneous spinning.
The 5 μl RPA products were diluted with 45 μl ddH2O, the strips were immersed in the dilute amplified products in the corresponding well. Samples migrated by capillary action, and the stable incubation time of test strip is 3-5 min at room temperature. Result was detected from the test line (T line) and the control line (C line) on the strips. Solely the presence of the C line denoted a negative outcome, while an invisible C line indicated an inconclusive result.
RPA-LFS assay was optimized by changing reaction incubation time (0-30 min) and incubation temperature (30-48°C), with RNA extracted from collected CPMMV-infected common bean leaves serving as a template.
To evaluate primer pair sensitivity, RNA extracted from infected common bean leaves was used as the template. With an initial RNA concentration of 800 ng/μl, a serial dilution was initiated, spanning from 8 × 106 to 1 fg. This dilution series was applied for both RT-PCR-AGE and RT-RPA-LFS assays, facilitating a thorough sensitivity comparison. In addition, the complete sequence of the CPMMV CP gene isolated from Zhejiang province, China, was cloned into pEASY-T5 Zero (TransGen, Beijing, China) and sequenced (ykang, Hangzhou, China). The pEASY-T5-CPMMV-CP (4,822 bp) was used as a template for sensitivity assay. The plasmids, whose initial copy number was 6 × 109 copies/μl, were diluted with healthy common bean crude extract, and 6 × 106-6 × 1 copies/μl plasmids were also used for the comparison of sensitivity.
To evaluate the specificity of the primer pair, sample leaves infected with cucumber mosaic virus (CMV), bean common mosaic virus (BCMV), bean broad wilt virus 2 (BBWV2), soybean mosaic virus (SMV), tomato spotted wilt virus (TSWV), turnip mosaic virus (TuMV), and pepper mild mottle virus (PMMoV) were collected in Zhejiang province and Shanghai city, China in 2016-2020. It’s noteworthy that each of the aforementioned viruses has been duly documented for their propensity to infect legume plants. These samples were identified by RT-PCR and stored at −80°C for further assays.
A collection of 14 symptomatic common beans from Zhejiang province, accompanied by a non-symptomatic common bean, were used. The evaluation encompassed the application of both RT-RPA-LFS and RT-PCR-AGE techniques. Plasmids with 104 copies/μl were set as a positive control, and healthy crude extracts were established as a negative control.
Primers for RPA were designed from the conserved region of CP genes of CPMMV. Four distinct pairs of primers were designed in this experiment.
The initial assessment of these primer pairs took place within a PCR reaction employing a recombinant plasmid (104 copies/μl). The PCR products were visualized on agarose gel, and all pairs of candidate primers showed positive amplification. Primer pair 3 exhibited a conspicuously robust amplicon brightness, followed by primer pair 2. PCR products amplified by primer pairs 1 and 4 showed fainter amplification signals (Fig. 1A). RPA assay was then conducted, and the products were detected on an agarose gel. Fig. 1B showed the RPA products amplified by all primer pairs, manifesting as nonspecific bands of varying degrees, albeit of a weaker intensity than the target band. Based on the results of PCR-AGE and RPA-AGE, primer pairs 2 and 3 were selected, and their probes were designed for further RPA-LFS assay (Fig. 1C). We first tested their sensitivity and accuracy using recombinant plasmids at low copies (102, 10, and 1 copies/μl). Both candidate primers and probe pairs showed positive amplification, but primers and probe pair 2 showed false positives (Supplementary Fig. 1). This discrepancy could potentially be attributed to complexities within the primer-probe interplay; thus, primers and probe pair 3 were selected for further study.
To establish and optimize the RT-RPA-LFS assay for detecting CPMMV, a crude extraction from an infected common bean was used as a template. The reaction temperature was varied over eight different gradients (30, 33, 36, 38, 40, 42, 45, and 48°C) for 20 min. The results showed that amplification at 33, 36, 38, 40, 42, and 45°C yielded a clear test line, while assays performed at 30 and 48°C failed to generate amplicons. The optimal temperature of RT-RPA-LFS was found to be 40°C (Fig. 2A). A subsequent experiment to ascertain the ideal reaction time involved the use of infected plant extracts subjected to diverse reaction durations (5-30 min). The RT-RPA-LFS assay produced positive results of CPMMV in 10 min, with a more clear visible test line on the LFS at 20 min. Intriguingly, the intensity of the test line showed minimal fluctuations beyond the 20-min threshold, as observed through visual inspection (Fig. 2B). Therefore, the results suggested that 40°C and 20 min are the optimum reaction conditions for CPMMV detection, using for subsequent assays.
To test the sensitivity of RT-RPA-LFS and RT-PCR-AGE, ten-fold dilutions of infected viral RNAs (106-1 fg/μl) and the recombinant plasmid pEASY-T5-CPMMV-CP (106-1 copies/μl) were used, respectively. As a negative control, total RNA from healthy leaves or from healthy leaf extracts was employed; no amplification was found. At dilutions of 8 × 103 fg/μl for viral RNAs and 6 × 103 copies/μl for recombinant plasmid, PCR using primer pairs CPMMV-PCR3F/R produced a positive amplification result (Fig. 3A and B). However, the RT-RPA-LFS exhibited detection limits of up to 80 fg/μl (for viral RNAs) or 60 copies/μl (for recombinant plasmid, weakly positive) (Fig. 3C and D). The results pointed the significant sensitivity of the RT-RPA-LFS assay for CPMMV, showing a superiority spanning 10-100 times more sensitive than RT-PCR-AGE.
To test primer specificity, both RT-PCR-AGE and RT-RPA-LFS assays were performed using RNA from plants infected with CMV, BCMV, BBWV2, SMV, TSWV, TuMV, PMMoV, and CPMMV, all of which are known to naturally infect legume plants. Total RNA from healthy common beans was used as a negative control. No positive amplification was observed, except for a sample infected with CPMMV in both RT-PCR-AGE and RT-RPA-LFS assays, indicating that the primer pair is specific for CPMMV detection (Fig. 4A and B).
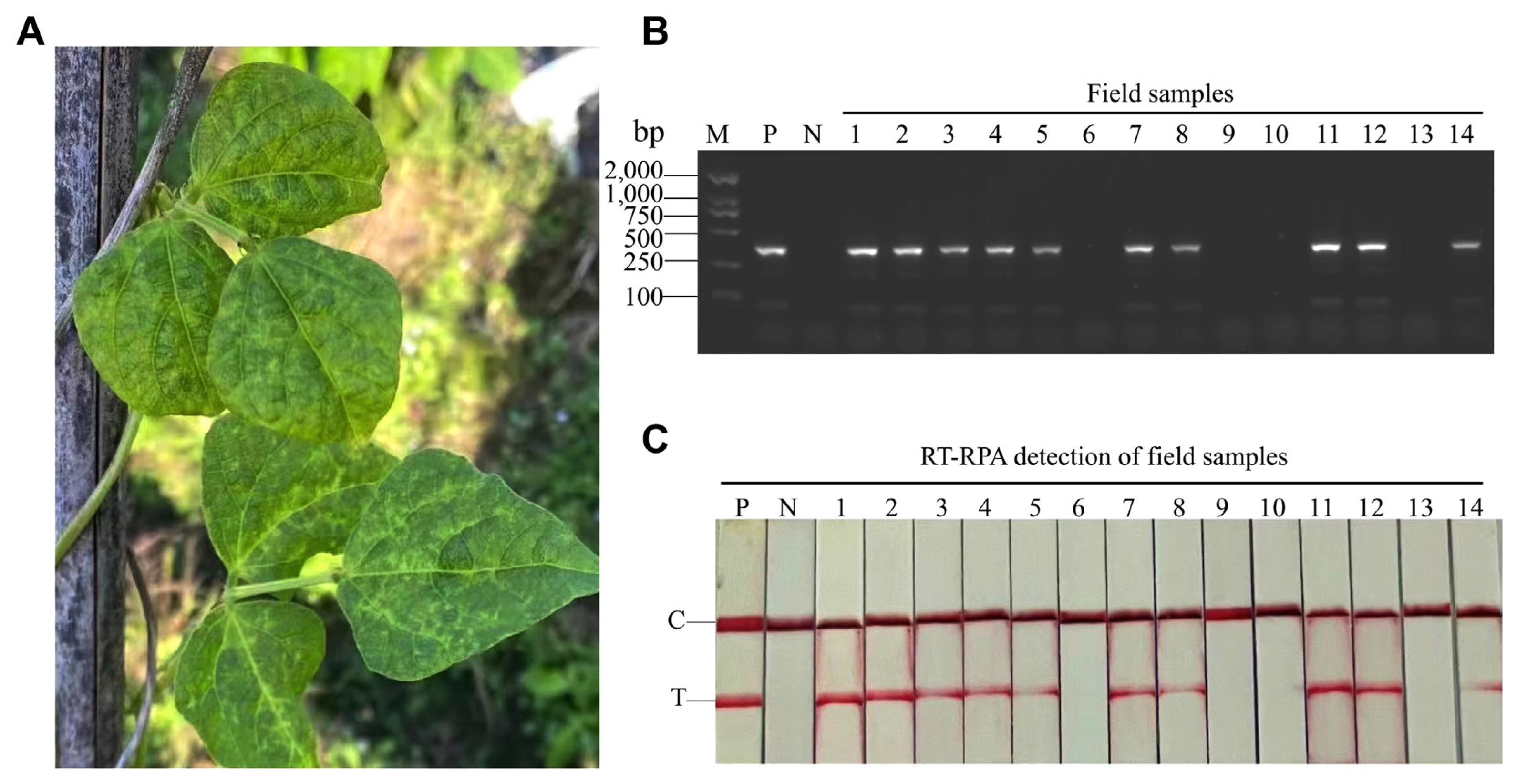
To detect CPMMV in field, 14 diverse common bean samples with typical CPMMV symptoms were collected in Zhejiang Province, China (Fig. 5A). These samples were used for RT-PCR-AGE and RT-RPA-LFS. Ten of 14 samples were infected with CPMMV (Fig. 5B and C). The results showed that RT-RPA-LFS and RT-PCR-AGE had comparable detection accuracies.
CPMMV is a plant virus with a global presence, causing significant economic losses in legume crops. Current disease management strategies hinge on the timely identification and segregation of infected plants to prevent virus dissemination. Consequently, the development of early diagnosis technology is crucial for effective disease management. Previous studies have focused on the development of detection methods such as RT-PCR, ELISA, and RT-LAMP (Fukuta et al., 2004; Huguenot et al., 1990; Liu et al., 2019). In this study, we have developed RT-RPA-LFS for CPMMV detection, which is particularly suitable for field conditions.
The swiftness of RT-RPA-LFS is emphasized by its ability to complete the amplification process within a mere 15-20 min, complemented by the chromogenic process on the LFS, visible to the naked eye in just 3-5 min. For convenience, RT-RPA-LFS does not require any complex reaction conditions and equipment, making it convenient for field use. For sensitivity, the detection limit of the RT-RPA-LFS assay of CPMMV reached 80 fg/μl or 60 copies/μl, 10-100 times higher than that of RT-PCR-AGE, consistent with RT-RPA assays for the detection of other pathogens (Cao et al., 2020). Total RNA was used to evaluated sensitivity in most studies (Jiao et al., 2020; Kim et al., 2018). But it is difficult to obtain an absolute quantification of total RNA. Therefore, we generalized the sensitivity evaluation by using both viral RNAs in total RNA and the cDNA clone containing CPMMV CP gene. It is acknowledged, however, that utilizing in vitro transcripts for sensitivity assessment may enhance precision and should be considered for future investigations.
In comparison, RT-PCR requires complex laboratory equipment and conditions and costs at least 4-5 h to complete which is unsuitable for practical application. Another detection method RT-LAMP, another isothermal amplification technology, is conducted at 60-65°C with six primer pairs, which also has some disadvantages in field detection. For diagnosis accuracy, we have compared the accuracy of both RT-PCR-AGE and RT-RPA-LFS for detecting field samples, and found that the detection rate of the two methods was well matched, indicating that RT-RPA-LFS has a similar detection accuracy to that of RT-PCR-AGE, indicating that it is consistent with previous studies (Cao et al., 2020; Jiao et al., 2019b).
Primer design is crucial for the establishment of RT-RPA-LFS technology. Primers should be designed in the conservative sequence region of CPMMV to ensure that they match with most isolates of CPMMV. Additionally, they must steer clear of forming primer-primer and primer-probe complexes that can lead to false positives. Due to the long length of primers and probes (>29 bp), the number of candidate primers and probes is limited. Despite taking precautionary measures, only one pair has no false-positive. This experience resonates with prior studies that have reported similar challenges (Wu et al., 2020; Yang et al., 2020). Introducing mismatches into the reverse primer and probe has been suggested as a countermeasure against false positives (Wu et al., 2020). Thus, primer design is important not only for sensitivity but also for detection accuracy of the detection technology. In this study, the accuracy and sensitivity requirements were met by primer and probe pairs 3, ensuring the successful application of RT-RPA-LFS in detecting CPMMV.
To the best of our knowledge, this study represents the first report on the development of the RT-RPA-LFS method for CPMMV detection. The entire detection process can be accomplished within a concise timeframe of 40 min, without the need for specialized equipment. This straightforward, highly sensitive, and specific technology can be readily deployed for rapid and visual detection of CPMMV, thereby aiding in disease control efforts in agricultural fields. The RT-PCR-LFS method applied to CPMMV detection can be useful not only for the virus detection but also for the detection of other plant viruses containing genomic RNAs in field.
Acknowledgments
This work is supported by National Nature Science Foundation of China (32202470), Key Research Program of Zhejiang Province (2021C02041), and State Key Laboratory for Managing Biotic and Chemical Threats to the Quality and Safety of Agro-products (2021DC700024-KF202217).
Electronic Supplementary Material
Supplementary materials are available at The Plant Pathology Journal website (http://www.ppjonline.org/).
Fig. 1
Design and screening of RPA-LFS primers. (A) Electrophoresis analysis of candidate primers screened by PCR-AGE. PCR-AGE assay was conducted with four pairs of primers using recombinant plasmid (104 copies/μl). (B) Electrophoresis analysis of candidate primers screened by RPA-AGE. RPA-AGE assay was conducted with four pairs of primers using recombinant plasmid (104 copies/μl). White arrows indicate nonspecific bands. (C) Positions of primer and probe pairs 2 and 3 within the CPMMV-CP gene. Multiple sequence alignment was performed using DNAMAN v8.0 software. RPA-LFS, recombinase polymerase amplification combined with lateral flow strips; PCR-AGE, polymerase chain reaction agarose gel electrophoresis; RPA-AGE, recombinase polymerase amplification agarose gel electrophoresis; CPMMV, cowpea mild mottle virus; CP, coat protein.
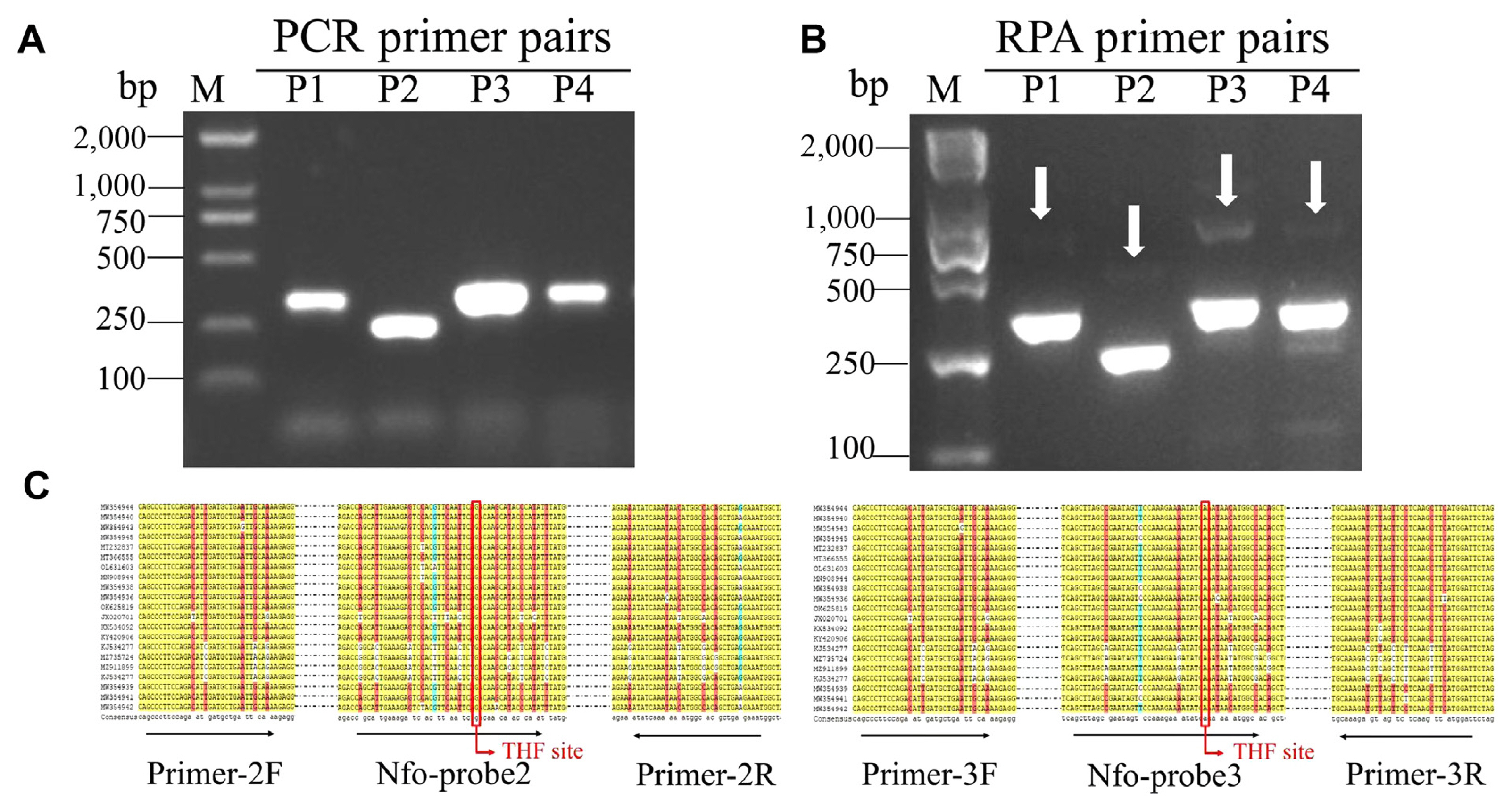
Fig. 2
Effects of temperature and time on RT-RPA-LFS. (A) Evaluation of different temperatures (30-48°C) for RT-RPA-LFS. (B) Evaluation of different times (0-30 min) for RT-RPA-LFS. RT-RPA-LFS, reverse transcription recombinase polymerase amplification combined with a lateral flow strips. C, quality control band; T, test band.
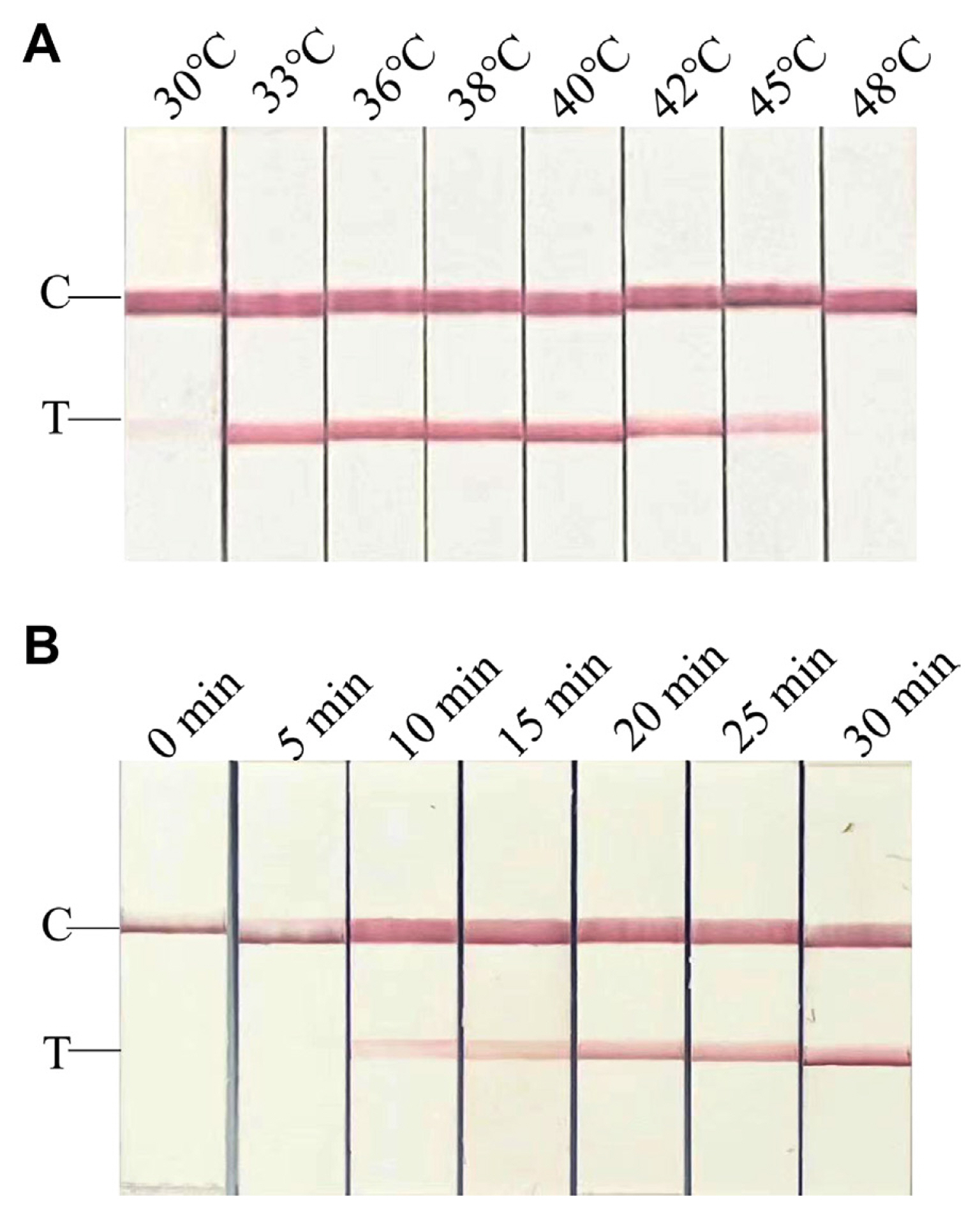
Fig. 3
Sensitivity tests of RT-RPA-LFS. Ten-fold dilutions of RNA of infected common bean leaves (8 × 106-8 × 1 fg/μl) and recombinant plasmid (6 × 106-6 × 1 copies/μl) were used for evaluating the sensitivity of RT-RPA-LFS and RT-PCR-AGE, respectively. RNA of healthy leaves or healthy leaf extracts was used as a negative control, where no amplification was detected. (A) In the PCR-AGE assay, the detection limit was 8 × 103 fg/μl for RNA. M, Trans2K plus DNA marker; N, negative control. (B) In the PCR-AGE assay, the detection limit was 103 copies/μl for recombinant plasmid. M, Trans2K plus DNA marker; N, negative control. (C) In RPA-LFS assay, the detection limit was 8 × 10 fg/μl (RNA). N, negative control; C, quality control band; T, test band. (D) In RPA-LFS assay, the detection limit was 10 copies/μl (recombinant plasmid). N, negative control; C, quality control band; T, test band. RT-RPA-LFS, reverse transcription recombinase polymerase amplification combined with a lateral flow strips; RT-PCR-AGE, reverse transcription-polymerase chain reaction agarose gel electrophoresis.
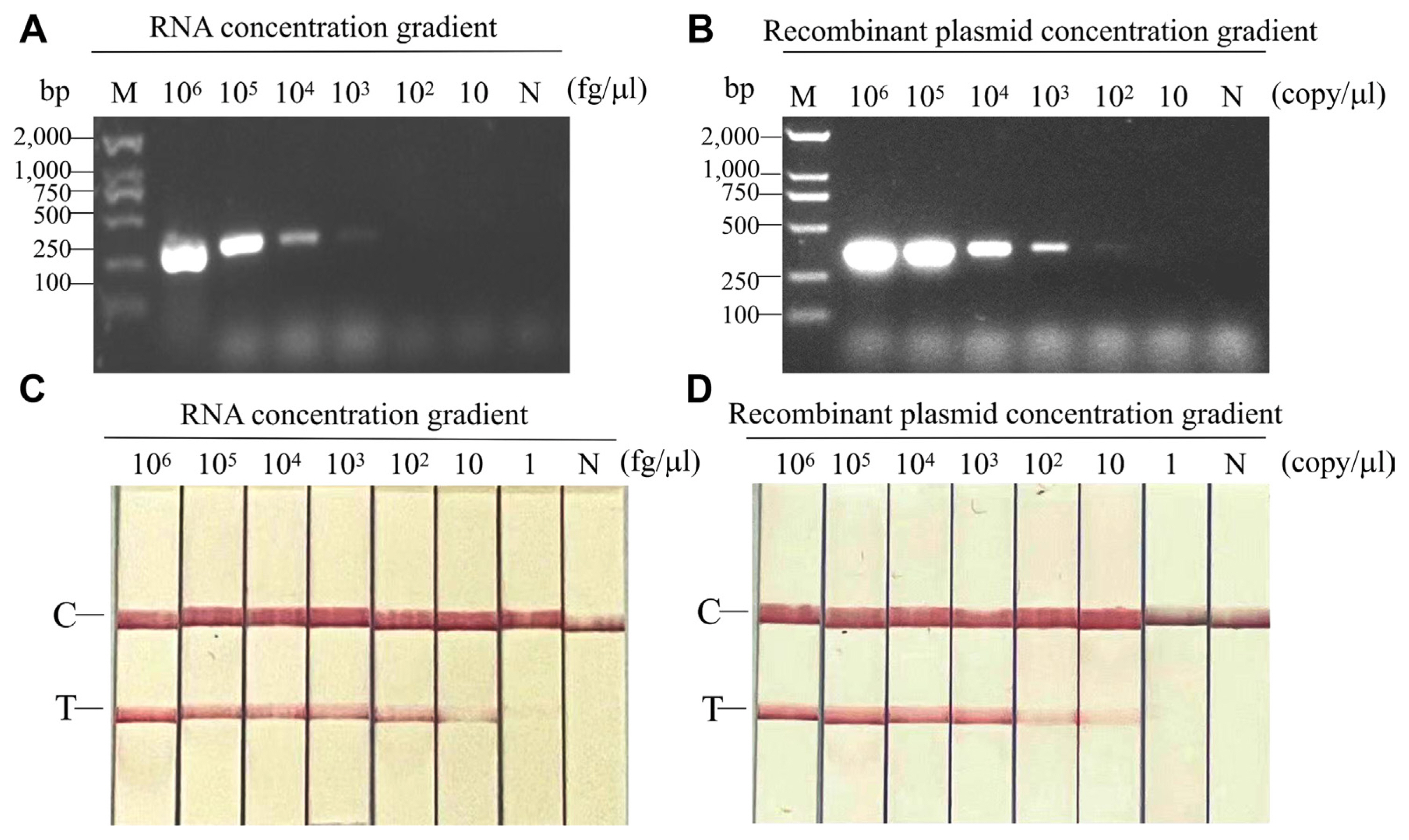
Fig. 4
Detection specificity of RT-RPA-LFS and RT-PCR-AGE. (A) Specificity assay was conducted with RNA samples from plants infected with different viruses by RT-PCR-AGE. (B) Specificity assay was conducted with RNA samples from plants infected with different viruses by RT-RPA-LFS. RT-RPA-LFS, reverse transcription recombinase polymerase amplification combined with a lateral flow strips; RT-PCR-AGE, reverse transcription-polymerase chain reaction agarose gel electrophoresis; M, marker; CPMMV, cowpea mild mottle virus; TSWV, tomato spotted wilt virus; CMV, cucumber mosaic virus; TuMV, turnip mosaic virus; BCMV, bean common mosaic virus; SMV, soybean mosaic virus; BBWV2, bean broad wilt virus 2; PMMoV, pepper mild mottle virus; C, quality control band; T, test band.
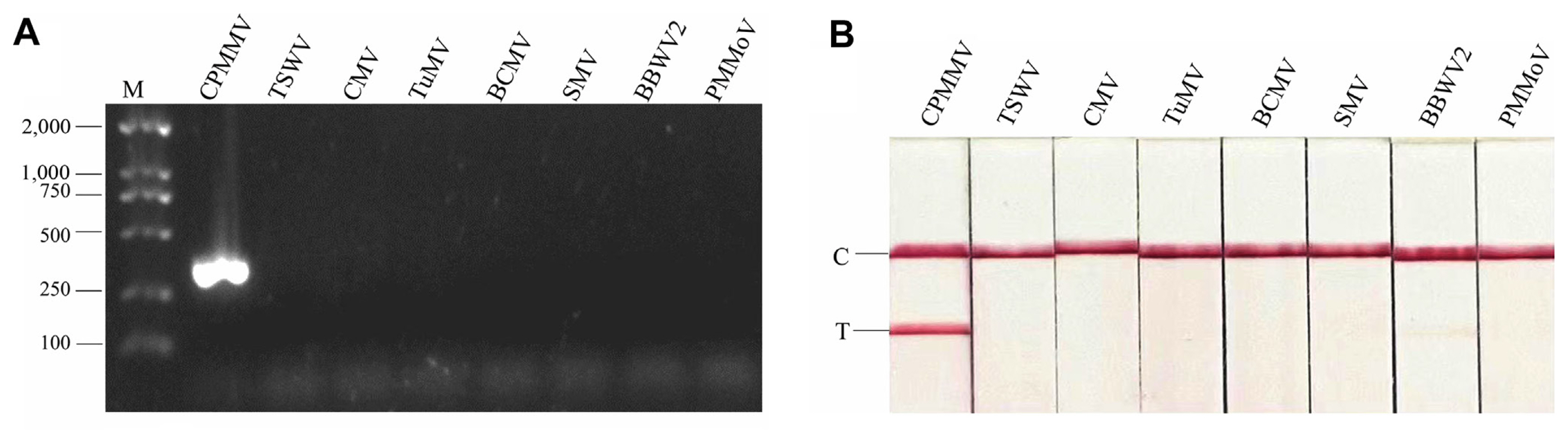
Fig. 5
Clinical trial comparing RT-RPA-LFS and RT-PCR-AGE for the detection of field samples. (A) A symptom of CPMMV-infected common bean collected in Zhejiang province, China. (B) RT-PCR-AGE assay for the detection of field samples. Fourteen field samples were collected from Zhejiang province, China. Recombinant plasmid and ddH2O were set as the positive and negative control, respectively. M, Trans2K plus DNA marker; N, negative control; P, positive control. (C) RT-RPA-LFS assay for the detection of field samples. 1-14: 14 samples were collected. RT-RPA-LFS, reverse transcription recombinase polymerase amplification combined with a lateral flow strips; RT-PCR-AGE, reverse transcription-polymerase chain reaction agarose gel electrophoresis; CPMMV, cowpea mild mottle virus; C, quality control band; T, test band.
References
Asano, S., Matsushita, Y., Hirayama, Y. and Naka, T. 2015. Simultaneous detection of Tomato spotted wilt virus, Dahlia mosaic virus and Chrysanthemum stunt viroid by multiplex RT-PCR in dahlias and their distribution in Japanese dahlias. Lett. Appl. Microbiol 61:113-120.



Babu, B., Washburn, B. K., Ertek, T. S., Miller, S. H., Riddle, C. B., Knox, G. W., Ochoa-Corona, F. M., Olson, J., Katırcıoğlu, Y. Z. and Paret, M. L. 2017. A field based detection method for Rose rosette virus using isothermal probe-based reverse transcription-recombinase polymerase amplification assay. J. Virol. Methods 247:81-90.


Boonham, N., Smith, P., Walsh, K., Tame, J., Morris, J., Spence, N., Bennison, J. and Barker, I. 2002. The detection of Tomato spotted wilt virus (TSWV) in individual thrips using real time fluorescent RT-PCR (TaqMan). J. Virol. Methods 101:37-48.


Boyle, D. S., Lehman, D. A., Lillis, L., Peterson, D., Singhal, M., Armes, N., Parker, M., Piepenburg, O. and Overbaugh, J. 2013. Rapid detection of HIV-1 proviral DNA for early infant diagnosis using recombinase polymerase amplification. mBio 4:e00135-13.




Brunt, A. A. and Kenten, R. H. 1973. Cowpea mild mottle, a newly recognized virus infecting cowpeas (Vigna unguiculata) in Ghana. Ann. Appl. Biol 74:67-74.


Cao, Y., Yan, D., Wu, X., Chen, Z., Lai, Y., Lv, L., Yan, F., Chen, J., Zheng, H. and Song, X. 2020. Rapid and visual detection of milk vetch dwarf virus using recombinase polymerase amplification combined with lateral flow strips. Virol. J 17:102.




Fukuta, S., Ohishi, K., Yoshida, K., Mizukami, Y., Ishida, A. and Kanbe, M. 2004. Development of immunocapture reverse transcription loop-mediated isothermal amplification for the detection of tomato spotted wilt virus from chrysanthemum. J. Virol. Methods 121:49-55.


Griep, R. A., Prins, M., van Twisk, C., Keller, H. J., Kerschbaumer, R. J., Kormelink, R., Goldbach, R. W. and Schots, A. 2000. Application of phage display in selecting tomato spotted wilt virus-specific single-chain antibodies (scFvs) for sensitive diagnosis in ELISA. Phytopathology 90:183-190.


Hagen, C., Frizzi, A., Kao, J., Jia, L., Huang, M., Zhang, Y. and Huang, S. 2011. Using small RNA sequences to diagnose, sequence, and investigate the infectivity characteristics of vegetable-infecting viruses. Arch. Virol 156:1209-1216.



Huguenot, C., van den Dobbelsteen, G., de Haan, P., Wagemakers, C. A., Drost, G. A., Osterhaus, A. D. and Peters, D. 1990. Detection of tomato spotted wilt virus using monoclonal antibodies and riboprobes. Arch. Virol 110:47-62.



Jiao, Y., Jiang, J., An, M., Xia, Z. and Wu, Y. 2019a. Recombinase polymerase amplification assay for rapid detection of maize chlorotic mottle virus in maize. Arch. Virol 164:2581-2584.



Jiao, Y., Jiang, J., Wu, Y. and Xia, Z. 2019b. Rapid detection of Cucumber green mottle mosaic virus in watermelon through a recombinase polymerase amplification assay. J. Virol. Methods 270:146-149.


Jiao, Y., Xu, C., Li, J., Gu, Y., Xia, C., Xie, Q., Xie, Y., An, M., Xia, Z. and Wu, Y. 2020. Characterization and a RT-RPA assay for rapid detection of Chilli Veinal mottle virus (ChiVMV) in tobacco. Virol. J 17:33.




Kim, N.-Y., Lee, H.-J. and Jeong, R.-D. 2019. A portable detection assay for Apple stem pitting virus using reverse transcription-recombinase polymerase amplification. J. Virol. Methods 274:113747.


Kim, N.-Y., Oh, J., Lee, S.-H., Kim, H., Moon, J. S. and Jeong, R.-D. 2018. Rapid and specific detection of Apple stem grooving virus by reverse transcription-recombinase polymerase amplification. Plant Pathol. J 4:575-579.


Liu, H., Wu, K., Wu, W., Mi, W., Hao, X. and Wu, Y. 2019. A multiplex reverse transcription PCR assay for simultaneous detection of six main RNA viruses in tomato plants. J. Virol. Methods 265:53-58.


Mandrile, L., Rotunno, S., Miozzi, L., Vaira, A. M., Giovannozzi, A. M., Rossi, A. M. and Noris, E. 2019. Nondestructive raman spectroscopy as a tool for early detection and discrimination of the infection of tomato plants by two economically important viruses. Anal. Chem 91:9025-9031.


Mortimer-Jones, S. M., Jones, M. G. K., Jones, R. A. C., Thomson, G. and Dwyer, G. I. 2009. A single tube, quantitative real-time RT-PCR assay that detects four potato viruses simultaneously. J. Virol. Methods 161:289-296.


Nemes, K. and Salánki, K. 2020. A multiplex RT-PCR assay for the simultaneous detection of prevalent viruses infecting pepper (Capsicum annuum L.). J. Virol. Methods 278:113838.


Oliver, J. E. and Whitfield, A. E. 2016. The genus tospovirus: emerging bunyaviruses that threaten food security. Annu. Rev. Virol 3:101-124.


Piepenburg, O., Williams, C. H., Stemple, D. L. and Armes, N. A. 2006. DNA detection using recombination proteins. PLoS Biol 4:e204.



Roberts, C. A., Dietzgen, R. G., Heelan, L. A. and Maclean, D. J. 2000. Real-time RT-PCR fluorescent detection of tomato spotted wilt virus. J. Virol. Methods 88:1-8.


Tantiwanich, Y., Chiemsombat, P., Naidu, R. A. and Adkins, S. 2018. Integrating local lesion assays with conventional RT-PCR for detection of interspecies tospovirus reassortants and mixed tospovirus infections. Plant Dis 102:715-719.


Wei, Z., Mao, C., Jiang, C., Zhang, H., Chen, J. and Sun, Z. 2021. Identification of a new genetic clade of Cowpea mild mottle virus and characterization of its interaction with Soybean mosaic virus in co-infected soybean. Front. Microbiol 12:650773.



Wu, H., Zhao, P., Yang, X., Li, J., Zhang, J., Zhang, X., Zeng, Z., Dong, J., Gao, S. and Lu, C. 2020. A recombinase polymerase amplification and lateral flow strip combined method that detects Salmonella enterica serotype Typhimurium with no worry of primer-dependent artifacts. Front. Microbiol 11:1015.



Wu, X., Chen, C., Xiao, X. and Deng, M. J. 2016. Development of reverse transcription thermostable helicase-dependent DNA amplification for the detection of Tomato spotted wilt virus. J. AOAC Int 99:1596-1599.



- TOOLS
-
METRICS

-
- 1 Crossref
- 0 Scopus
- 1,230 View
- 89 Download
- ORCID iDs
- Related articles




 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Supplement
Supplement Print
Print



