 |
 |
| Plant Pathol J > Volume 40(2); 2024 > Article |
|
Abstract
The microbiomes of two important rice cultivars in Vietnam which differ by their susceptibility to the bacterial leaf blight (BLB) disease were analyzed through 16S rRNA amplicon technology. A higher number of operational taxonomic units and alpha-diversity indices were shown in the BLB-resistant LA cultivar than in the BLB-susceptible TB cultivar. The BLB pathogen Xanthomonas was scantly found (0.003%) in the LA cultivar, whereas was in a significantly higher ratio in the TB cultivar (1.82%), reflecting the susceptibility to BLB of these cultivars. Of special interest was the genus Acholeplasma presented in the BLB-resistant LA cultivar at a high relative abundance (22.32%), however, was minor in the BLB-sensitive TB cultivar (0.09%), raising a question about its roles in controlling the Xanthomonas low in the LA cultivar. It is proposed that Acholeplasma once entered the host plant would hamper other phytopathogens, i.e. Xanthomonas, by yet unknown mechanisms, of which the triggering of the host plants to produce secondary metabolites against pathogens could be a testable hypothesis.
Rice is the main food for more than half of the global population and Asian countries account for about 90% of rice production and consumption (Chukwu et al., 2019). Today, rice cultivation is facing various infectious diseases, among which the bacterial leaf blight (BLB) caused by the pathogen Xanthomonas oryzae pv. oryzae (Xoo) has been considered one of the most severe (Mansfield et al., 2012). Being discovered first in Japan in 1908, the disease becomes more and more complicated, to different extents annually affecting various rice growing areas worldwide (Mew et al., 1993; Webb et al., 2010).
Approaching organic rice production by reducing the use of pesticides, special attention is given to the endophytes since they are supposed to control the host plant’s health via several direct and indirect beneficial effects (Raymaekers et al., 2020). The link between the endophytes and the susceptibility of the host plants against various phytopathogens is rather complex and so far, diverse mechanistic explanations have been reported. The most reasonable hypothesis is that the endophytes once successfully enter plant tissues, can directly compete with microbial pathogens for nutrients and space, excrete antagonistic compounds, or trigger the plant defense paths, by the way, establish a health-controlling system for the host plants (Dubey et al., 2020; Liu et al., 2017).
Rice bacterial endophytes showed great diversity (Moronta-Barrios et al., 2018; Walitang et al., 2019), and themost complex community information has been provided by metagenomic analyses (Kumar et al., 2021; Su et al., 2022). Pathogens can affect the microbiome differently, but generally reduce the overall diversity (Bulgari et al., 2014; Fadiji and Babalola, 2020). In specific cases, certain endophytic groups can have a strong relationship with the pathogens, either as an inhibitor or a mutualist (Brader et al., 2017).
This study aims to compare the microbiome of two important rice cultivars in Vietnam, the BLB-sensitive TB and the BLB-resistant LA to gain insight into the triangle relationship between endophytes-host plants-phytopathogens. The TB cultivar has been introduced to Vietnam since 1998, widely planted in the Red River Delta despite its high susceptibility to BLB disease, the annual yield loss accounts for 25-30% (occasionally 50%) (Vietnam Department of Crop Production). The LA cultivar is planted mainly in the Mekong Delta and has not been reported for pandemic BLB so far. It is therefore of special interest to have a closer look at the microbiomes of these two rice cultivars to understand the possible role of rice endophytes in supporting the host plants against the BLB disease.
Rice plants of the two domestic cultivars Bacthom (TB) and IR4625 (LA) at their tillering stage were collected in the Summer-Autumn season of 2018-2019 from the Red River Delta (Vu Thu district, Thai Binh province, 20°21′28.8″N, 106°17′15.8″E) and the Mekong Delta (Thu Thua district, Long An province, 10°34′27.6″N, 106°21′57.2″E), respectively (Supplementary Table 1). The plant samples were carefully collected from the fields to prevent any damage to the roots, kept in clean plastic bags, and brought in ice-cool boxes back to the laboratory for further processing.
The plant samples were washed carefully with tap water, then rinsed in distilled sterile water and let to air dry. Plant parts roots and stems were cut into 4-5 cm fragments, whereas the leaves were cut at the green parts at a 1-2 cm distance from the BLB-symptomatic areas of the diseased plants, and at the same parts of the healthy plants. The plant materials of 1 g each were macerated in liquid nitrogen using sterile mortars and pestles.
The total DNA was then extracted using DNeasy PowerSoil DNA Pro Kit (Qiagen, Germantown, MD, USA) following the manufacturer’s instructions. The V3-V4 hypervariable region of the 16S rRNA gene was amplified using barcoded primers and PCR conditions following Illumina Inc.’s protocol (Illumina, San Diego, CA, USA). Briefly, individual barcoded libraries were directly prepared by PCR using long primers (Klindworth et al., 2013) incorporating the Illumina adapter sequences (16S_Amplicon_PCR_Fw: TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG;16S_Amplicon_PCR_Rv: GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAATCC). Afterward, the amplicons were purified with the AMPure XP bead clean-up (A63880l, Beckman Coulter, Brea, CA, USA) and checked for size, integrity, and purity by using the Bioanalyzer equipment (Agilent, Santa Clara, CA, USA). Before sequencing, the library concentration was quantified fluorimetrically by using Qubit 2 (Invitrogen, Carlsbad, CA, USA). Library sequencing was carried out with the Illumina Miseq system using a 250bp paired-end strategy, meanwhile, the parameters of the system were set to provide output as a separate FastQ file containing index reads. The raw FastQ dataset was demultiplexed using QIIME 1.9.1. and then processed with DADA2 v1.4.0 for adapter removal and quality filtering. Sequence trimming and chimer removing were performed by using the FilterAndTrim and the removeBimeraDenovo functions, respectively. The effective sequences were then clustered into operational taxonomic units (OTUs) at 97% similarity using the assigned Taxonomy function. Finally, the Ribosomal Database Project classifier was used to assign representative sequences to the microbial taxa. All statistical analyses were performed in R using the vegan package (v2.6.4) (Oksanen et al., 2016) and the Phyloseq package (v1.42) (McMurdie and Holmes, 2013). Relative abundances of OTUs between samples and α-diversity indices (Observed, Chao1, Shannon, and Simpson) were measured for estimation of the species richness and community composition in the samples. Microbial abundance differences between cultivars were visualized using non-metric multidimensional scaling (NMDS) based on Bray dissimilarity on normalized data and tested for significance using the ANOSIM algorithm (999 permutations). The package DESeq2 (v1.38.3) was used to determine statistically significant differences in OTU bacterial relative abundance levels (Love et al., 2014). Plots were generated using the ggplot2 R package (v3.4.2).
In total 30 rice plants of the LA and TB cultivars (15 samples each) were collected (Supplementary Table 1). The BLB-diseased TB plants (8 samples) were selected according to the appearance of tannish gray to white lesions along the veins, while the healthy plants should have no visible infection despite growing in the same fields as the symptomatic counterparts. For the LA cultivar, no BLB-diseased sample was detected at the sampling trips. From each plant, different compartments of roots, stems, and leaves were processed separately, thus yielding a total of 43 samples that qualified for the microbiome analyses (Supplementary Table 1). After quality check and removal of reads classified as chloroplasts and mitochondria, the number of reads was 234,803 and the resulting sequences were arranged in 232 OTUs, comprising 172 OTUs from the TB cultivar and 216 OTUs from the LA cultivar (Supplementary Table 1).
Comparing the alpha diversity of the two rice cultivars, all three indices Chao1, Shannon, and Simpson in the LA cultivar were higher than those of the TB cultivar (Fig. 1A). Within the TB cultivar, the two indices Shannon and Simpson, but not the Chao1 index, were decreased significantly in the diseases samples as compared to the healthy samples, showing the effect of the pathogen Xanthomonas to the microbiome diversity (Fig. 1B). Regarding plant compartments, the Shannon index (showing the overall diversity) in leaves and roots of the LA cultivar was higher than those of the TB cultivar, but in stems this index in the TB cultivar was higher (Fig. 1C). The other indices Chao1 and Simpson confirmed this data (Supplementary Table 2). NMDS showed a partial overlapping between microbiomes of the TB and LA cultivars (P = 0.023) (Supplementary Fig. 1). Considering plant compartments, the TB cultivar showed higher variability (P = 0.001) than the LA cultivar (P = 0.02). In both cultivars, the microbiome of leaves and stems showed intimacy, whereas the microbiome of roots clustered separately. Microbiome of the diseased TB samples did not change remarkably (P = 0.062) but made a subcluster within the microbiome of healthy samples (Supplementary Fig. 1B).
At the phylum level (Fig. 2A), Proteobacteria was found most abundant, making 82.80% and 60.90% for the TB and LA cultivars, respectively. The phylum Actinobacteria accounted for 15.22% in the TB, and 13.59% in the LA, standing for the second and third most abundant phyla in these cultivars, respectively. As a notable distinctive feature, the phylum Tenericutes was detected at a high relative abundance, most in stems and leaves (Supplementary Fig. 2A), representing the second most abundant group in the LA cultivar (22.32%), however, was scant in the TB cultivar (0.09%). At far lower abundance levels (1% and below) several phyla like Firmicutes, Deinococcus-Thermus, Bacteroidetes, and Fibrobacteres were detected in both cultivars.
At the class level (Fig. 2B), Alpha-proteobacteria was the most abundant in both TB and LA cultivars (79.90% and 60.10%, respectively), followed by Actinobacteria (15.22% and 13.59%, respectively). Corresponding to the distinguished phylum Tenericutes found in the LA cultivar, the class Mollicutes was found solely in this cultivar at a high abundance of 22.32%, most associated with leaves and stems (Supplementary Fig. 2B). The third most abundant class in the TB cultivar was Gamma-proteobacteria (2.57%), whereas in the LA cultivar was Clostridia (1.40%). Regarding the minor groups in the microbiome (1% and below), there exist certain differentiation between the two cultivars, i.e Plantomycetia presented only in the TB cultivar, whereas several classes such as Armatimonadia, Chloroflexia, Epsilon-proteobacteria, Ktedonobacteria, and Nitrospirae presented only in the LA cultivar.
At the genus level (Fig. 3C), lower diversity was recorded in the TB cultivar (172 genera) than in the LA cultivar (216 genera). Common genera with significant relative abundance in both cultivars included Sphingomonas, Rhizobium, Methylobacterium, and Curtobacterium, among which the genus Sphingomonas was the most abundant, represented 40.24% in the TB and 21.99% in the LA cultivar. The other genera Rhizobium, Methylobacterium, and Curtobacterium were found similarly in both cultivars at lower ratios of 8.56%, 13.40%, and 4.29% for TB, and 10.20%, 13.20%, and 3.02% for LA, respectively. These features of rice microbiomes were consistent with and documented in previous studies (Krishnamoorthy et al., 2021; Kunda et al., 2018). Higher variability was observed among the taxa presented in the microbiomes at far lower ratios (1% and below), depending on the plant compartments (Supplementary Fig. 2C).
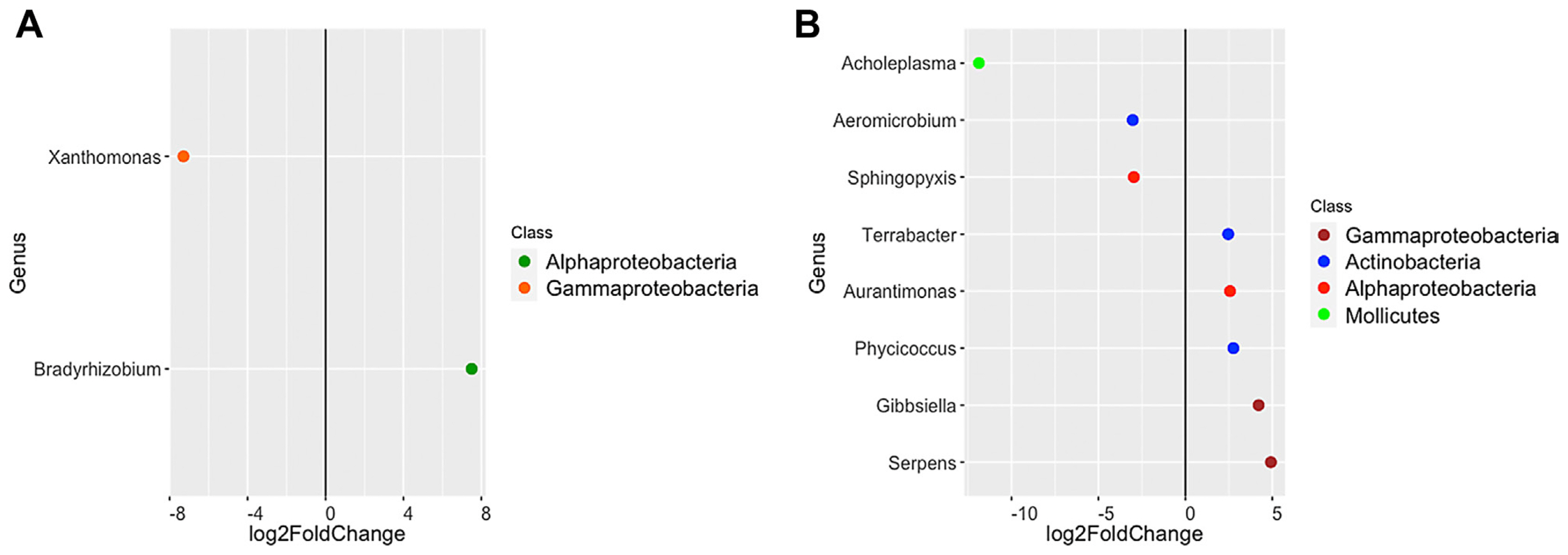
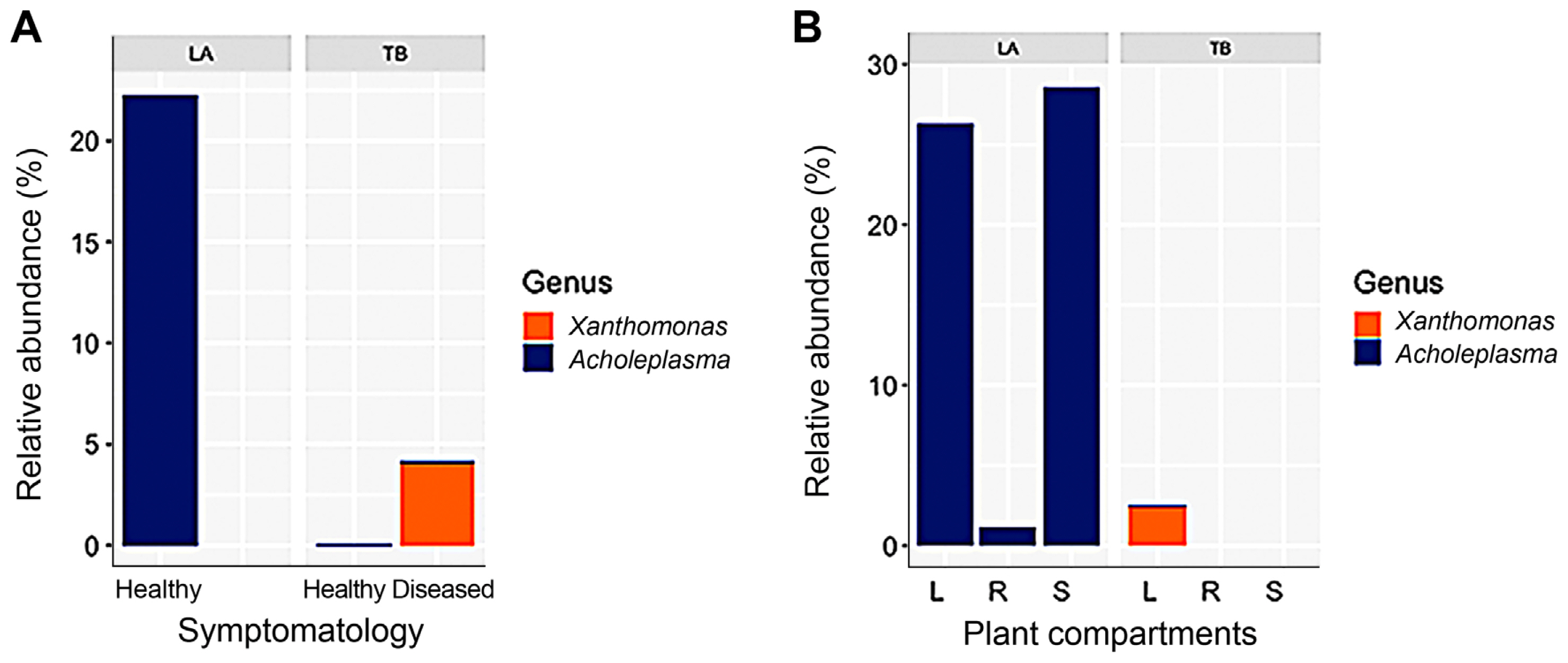
Further, linear discriminant analyses between the microbiome of diseased and healthy samples within the TB cultivar and between healthy samples of the two cultivars were performed, showing that the diseased TB samples differed by the abundance of Xanthomonas (Fig. 3A), whereas the healthy samples of the two cultivars differed most notably by the Acholeplasma abundance (Fig. 3B). Xanthomonas was detected exclusively in the diseased TB samples (at a relative abundance of 4.56%), accumulating in the leaves, whereas was almost negligible in the LA cultivar (0.003%) (Fig. 4A). On the other hand, Acholeplasma was exclusively detected in the LA cultivar at a high relative abundance (22.32%), accumulating inleaves and stems (Fig. 4B), however presented at a very low relative abundance (0.09%) in the TB cultivar (Fig. 4A). The high abundance of Acholeplasma in the LA cultivar versus the high abundance of Xanthomonas in the TB cultivar (the diseased samples) was supposed to respectively correlate with the resistance and sensitivity of these cultivars to the BLB disease.
The plant microbiome variability is affected by different factors, including the plant growth stages, soil characteristics, and several environmental conditions (Kunda et al., 2018). Regarding plant compartments, the root microbiome depends largely on soil characteristics, whereas the phyllosphere microbiome is more influenced by environmental conditions such as climate, pathogens, and cultivation practices (Bez et al., 2021; Compant et al., 2019). In this study, the microbiome dissimilarity of the two rice cultivars was more significant in the phylosphere than in the roots, implying that the leaf-associated pathogen such as Xoo would have stronger influences.
The presence of phytopathogens in the host plants generally leads to a decrease in microbiome diversity (Bulgari et al., 2014; Fadiji and Babalola, 2020). Vice versa, a higher diversified microbiome would better support the host plants fighting against pathogens (Podolich et al., 2015). In this study, the high ratio of Xanthomonas in the BLB-diseased TB samples (Fig. 4) is proposed to be a factor leading to less diversified microbiomes compared to that of the healthy TB and LA samples (Fig. 1A and B). It is supposed that both factors the low Xanthomonas content and the high diversity of microbiome have correlated to the BLB resistance of the LA cultivar.
Acholeplasma to our knowledge has not been reported in other studies on rice microbiomes, i.e. neither in wild rice O. longistaminata (Peng et al., 2021), nor in other rice cultivars of O. sativa planted in different climate conditions (Bez et al., 2021; Wang et al., 2021), indicating that their presence in LA cultivar is a unique characteristic. Further, as they were abundantly present in the BLB-resistant LA cultivar but absent in the BLB-sensitive TB cultivar, raising a question about their roles in controlling the Xanthomonas ratio, and correspondingly, the resistance to BLB disease of the LA cultivar. Acholeplasama is found ubiquitously as saprophytes and commensals in vertebrates, insects, and plants (Siewert et al., 2014). They have limited biosynthetic capabilities, however owing to a wide variety of transporters, they thrive well, freely or in association with hosts, by utilizing external carbon and energy sources (Lazarev et al., 2011).
Acholeplasma species have been reported associating with plants, e.g., A. florum from lemon and grapefruit flowers (McCoy et al., 1984), A. brassicae from the surface of broccoli, and A. palmae from crown tissues of the coconut palm (Tully et al., 1994b), however, they have not been described as primary phytopathogens (Kube et al., 2014). Under unfavorable circumstances, Acholeplasma may transform into nano-cells that have a higher pathogenicity for plants (Chernov et al., 2006). In particular, some Acholeplasma can infect plants like its close relative Phytoplasma (Strauss, 2009). Extracellular enzymes degrading plant cell walls like cellulase and amylase were found in Acholeplasma, which might support them to enter the host plant tissues (Kube et al., 2014). Further, Acholeplasma has been detected in association with diverse insect species that feed on plant tissues (Tully et al., 1994a), and by the way can be transmitted into plants (Bonnet et al., 1991). Apparently, in the LA cultivar, Acholeplasma lives symbiotically inside host plants’ tissues without harming the hosts. However, the lipopolysaccharides produced by Acholeplasma might act as an elicitor triggering the host plants to produce secondary metabolites against further invasion of other pathogens (Fadiji and Babalola, 2020). Specifically, it is proposed that the notably high relative abundance of Acholeplasma in stems and leaves of the LA cultivar could hamper the invasion of Xanthomonas, making this cultivar resistant to the BLB disease. These results strongly encourage future studies on the role of Acholeplasma in BLB disease resistance.
Acknowledgments
The study was supported by Vingroup Innovation Foundation (Grant VINIF.2021.TS.111).
Electronic Supplementary Material
Supplementary materials are available at The Plant Pathology Journal website (http://www.ppjonline.org/).
Fig. 1
Alpha diversity estimation of the microbiome associated with BLB-resistant LA and BLB-sensitive TB rice cultivar. (A) The three indices Chao1, Shannon, and Simpson showing the microbiome diversity of the LA and TB cultivars (all healthy samples). (B) The three indices Chao1, Shannon, and Simpson showing microbiome diversity of healthy and diseased TB samples. (C) The Shannon index of the microbiome in plant compartments (L, leaf; R, root; S, stem) of the LA and TB cultivars.
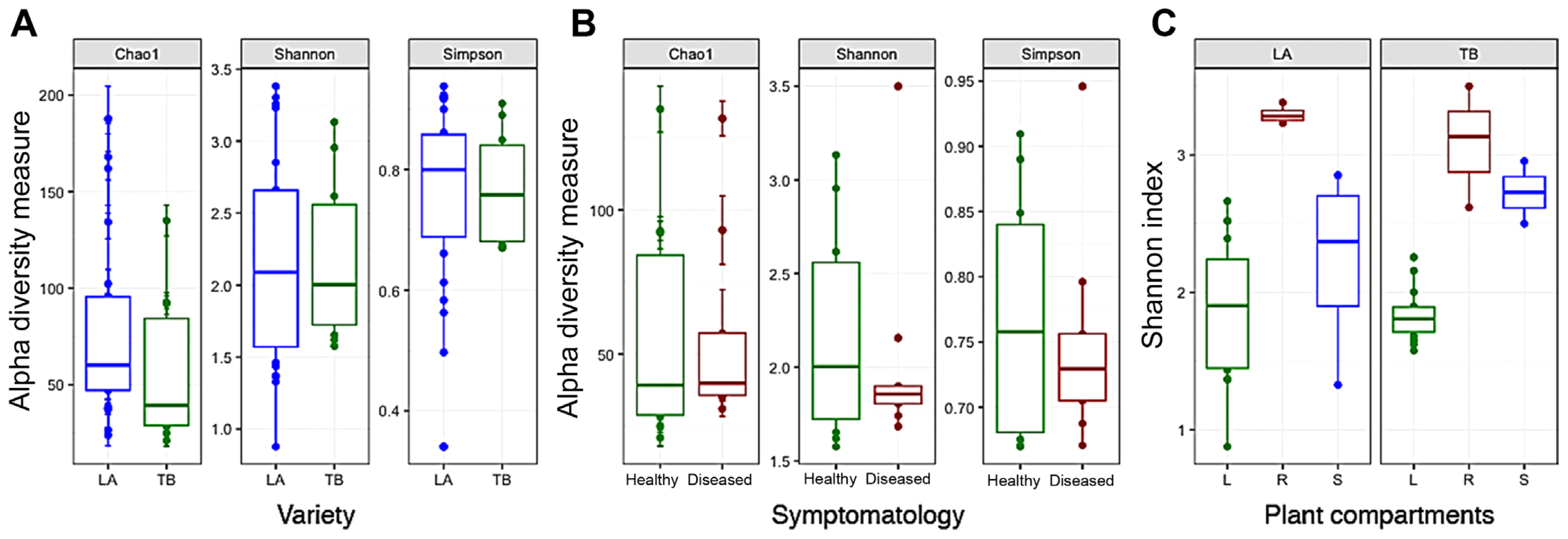
Fig. 2
Relative abundance of the predominant taxa in the two rice cultivars microbiomes at the phylum (A), class (relative abundance > 0.01) (B), and genus (relative abundance > 0.05) (C) levels.
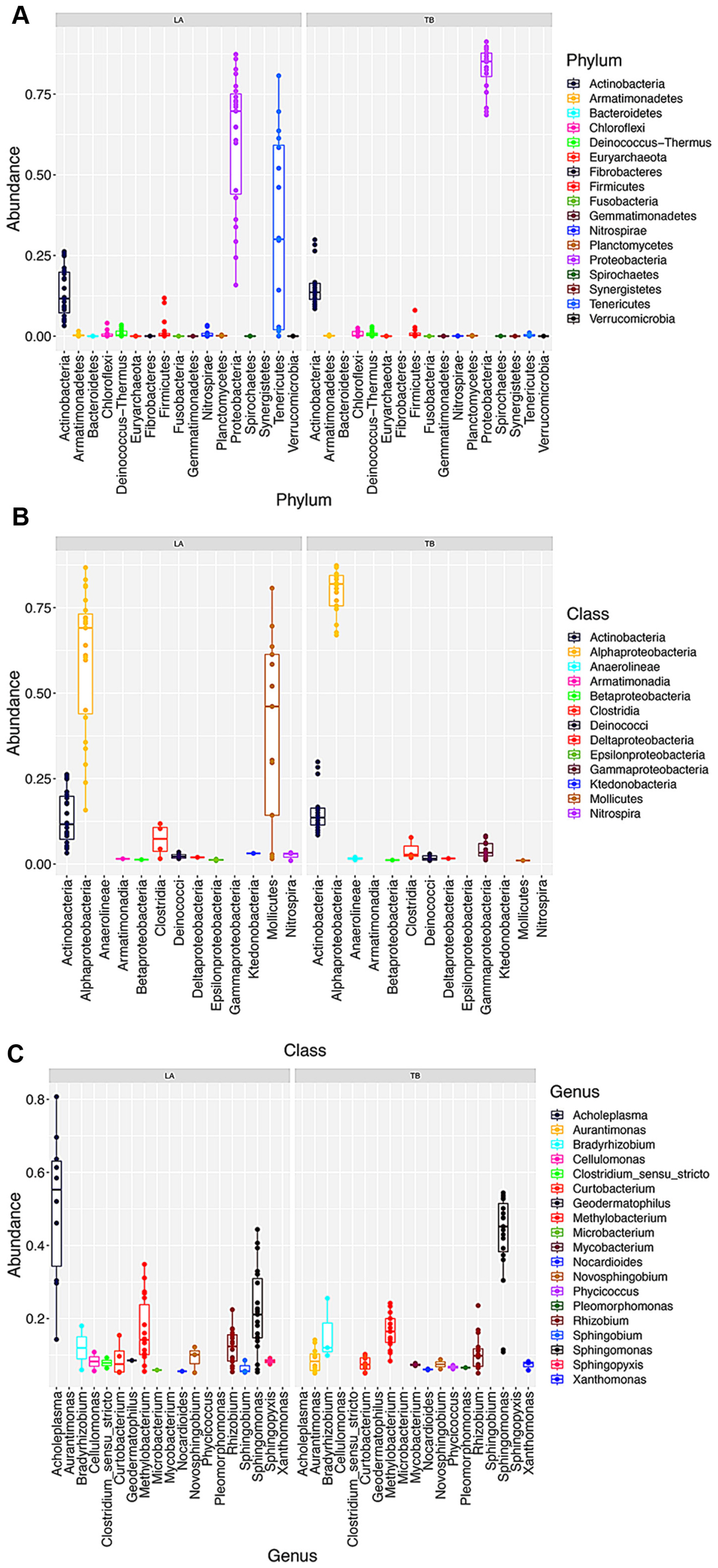
Fig. 3
Differential abundance analysis (P < 0.05) based on DESeq2. Each data point represents a genus-level operational taxonomic units (y-axis) identified as significantly different between healthy and sick samples of the TB cultivar (A) and healthy samples of the LA and TB cultivars (B). (A) The negative values of log2 fold change (x-axis) indicate higher relative abundance in the sick samples of TB cultivar and positive values indicate higher relative abundance in the healthy samples of TB cultivar. (B) The negative values of log2 fold change (x-axis) exhibit higher relative abundance in the LA cultivar and positive values exhibit higher relative abundance in the TB cultivar.
References
Bez, C., Esposito, A., Thuy, H. D., Hong, M. N., Val, G., Licastro, D., Bertani, I., Piazza, S. and Venturi, V. 2021. The rice foot rot pathogen Dickeya zeae alters the in-field plant microbiome. Environ. Microbiol. 23:7671-7687.


Bonnet, F., Saillard, C., Vignault, J. C., Garnier, M., Carle, P., Bove, J. M., Rose, D. L., Tully, J. G. and Whitcomb, B. F. 1991.
Acholeplasma seiffertii sp. nov., a mollicute from plant surfaces. Int. J. Syst. Bacteriol. 41:45-49.

Brader, G., Compant, S., Vescio, K., Mitter, B., Trognitz, F., Ma, L.-J. and Sessitsch, A. 2017. Ecology and genomic insights into plant-pathogenic and plant-nonpathogenic endophytes. Annu. Rev. Phytopathol. 55:61-83.


Bulgari, D., Casati, P., Quaglino, F. and Bianco, P. A. 2014. Endophytic bacterial community of grapevine leaves influenced by sampling date and phytoplasma infection process. BMC Microbiol. 14:198.




Chernov, V. M., Moukhametshina, N. E., Gogolev, Y. V., Nesterova, T. N., Trushin, M. V. and Chernova, O. A. 2006. Adaptation to unfavorable conditions of growth: pathogenicity of Acholeplasma laidlawii PG8. Electron. J. Biomed. 3:11-15.
Chukwu, S. C., Rafii, M. Y., Ramlee, S. I., Ismail, S. I., Hasan, M. M., Oladosu, Y. A., Magaji, U. G., Akos, I. and Olalekan, K. K. 2019. Bacterial leaf blight resistance in rice: a review of conventional breeding to molecular approach. Mol. Biol. Rep. 46:1519-1532.



Compant, S., Samad, A., Faist, H. and Sessitsch, A. 2019. A review on the plant microbiome: ecology, functions, and emerging trends in microbial application. J. Adv. Res. 19:29-37.



Dubey, A., Malla, M. A., Kumar, A., Dayanandan, S. and Khan, M. L. 2020. Plants endophytes: unveiling hidden agenda for bioprospecting toward sustainable agriculture. Crit. Rev. Biotechnol. 40:1210-1231.


Fadiji, A. E. and Babalola, O. O. 2020. Elucidating mechanisms of endophytes used in plant protection and other bioactivities with multifunctional prospects. Front. Bioeng. Biotechnol. 8:467.



Klindworth, A., Pruesse, E., Schweer, T., Peplies, J., Quast, C., Horn, M. and Glöckner, F. O. 2013. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 41:e1.



Krishnamoorthy, A., Gupta, A., Sar, P. and Maiti, M. K. 2021. Metagenomics of two gnotobiotically grown aromatic rice cultivars reveals genotype-dependent and tissue-specific colonization of endophytic bacterial communities attributing multiple plant growth promoting traits. World J. Microbiol. Biotechnol. 37:59.



Kube, M., Siewert, C., Migdoll, A. M., Duduk, B., Holz, S., Rabus, R., Seemüller, E., Mitrovic, J., Müller, I., Büttner, C. and Reinhardt, R. 2014. Analysis of the complete genomes of Acholeplasma brassicae, A. palmae and A. laidlawii and their comparison to the obligate parasites from ‘Candidatus Phytoplasma’. J. Mol. Microbiol. Biotechnol. 24:19-36.

Kumar, M., Kumar, A., Sahu, K. P., Patel, A., Reddy, B., Sheoran, N., Charishma, K., Rajashekara, H., Bhagat, S. and Rathour, R. 2021. Deciphering core-microbiome of rice leaf endosphere: revelation by metagenomic and microbiological analysis of aromatic and non-aromatic genotypes grown in three geographical zones. Microbiol. Res. 246:126704.


Kunda, P., Dhal, P. K. and Mukherjee, A. 2018. Endophytic bacterial community of rice (Oryza sativa L.) from coastal saline zone of West Bengal: 16S rRNA gene based metagenomics approach. Meta Gene 18:79-86.

Lazarev, V. N., Levitskii, S. A., Basovskii, Y. I., Chukin, M. M., Akopian, T. A., Vereshchagin, V. V., Kostrjukova, E. S., Kovaleva, G. Y., Kazanov, M. D., Malko, D. B., Vitreschak, A. G., Sernova, N. V., Gelfand, M. S., Demina, I. A., Serebryakova, M. V., Galyamina, M. A., Vtyurin, N. N., Rogov, S. I., Alexeev, D. G., Ladygina, V. G. and Govorun, V. M. 2011. Complete genome and proteome of Acholeplasma laidlawii. J. Bacteriol. 193:4943-4953.




Liu, H., Carvalhais, L. C., Crawford, M., Singh, E., Dennis, P. G., Pieterse, C. M. J. and Schenk, P. M. 2017. Inner plant values: diversity, colonization and benefits from endophytic bacteria. Front. Microbiol. 8:2552.



Love, M. I., Huber, W. and Anders, S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550.




Mansfield, J., Genin, S., Magori, S., Citovsky, V., Sriariyanum, M., Ronald, P., Dow, M., Verdier, V., Beer, S. V., Machado, M. A., Toth, I., Salmond, G. and Foster, G. D. 2012. Top 10 plant pathogenic bacteria in molecular plant pathology. Mol. Plant Pathol. 13:614-629.



McCoy, R. E., Basham, H. G., Tully, J. G., Rose, D. L., Carle, P. and Bové, J. M. 1984.
Acholoplasma florum, a new species isolated from plants. Int. J. Syst. Bacteriol. 34:11-15.
McMurdie, P. J. and Holmes, S. 2013. Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8:e61217.



Mew, T. W., Alvarez, A. M., Leach, J. E. and Swings, J. 1993. Focus on bacterial blight of rice. Plant Dis. 77:5-12.

Moronta-Barrios, F., Gionechetti, F., Pallavicini, A., Marys, E. and Venturi, V. 2018. Bacterial microbiota of rice roots: 16S-based taxonomic profiling of endophytic and rhizospheric diversity, endophytes isolation and simplified endophytic community. Microorganisms 6:14.



Oksanen, J., Blanchet, F. G., Kindt, R., Legendre, P., Minchin, P. R., O’Hara, R. B., Simpson, G. L., Solymos, P., Stevens, M. H. H. and Wagner, H. 2016. Vegan: Community ecology package. R package version 2.3-5. R Foundation, Vienna, Austria.
Peng, X., Xie, J., Li, W., Xie, H., Cai, Y. and Ding, X. 2021. Comparison of wild rice (Oryza longistaminata) tissues identifies rhizome-specific bacterial and archaeal endophytic microbiomes communities and network structures. PLoS ONE 16:e0246687.



Podolich, O., Ardanov, P., Zaets, I., Pirttilä, A. M. and Kozyrovska, N. 2015. Reviving of the endophytic bacterial community as a putative mechanism of plant resistance. Plant Soil 388:367-377.


Raymaekers, K., Ponet, L., Holtappels, D., Berckmans, B. and Cammue, B. P. A. 2020. Screening for novel biocontrol agents applicable in plant disease management: a review. Biol. Control 144:104240.

Siewert, C., Hess, W. R., Duduk, B., Huettel, B., Reinhardt, R., Büttner, C. and Kube, M. 2014. Complete genome determination and analysis of Acholeplasma oculi strain 19L, highlighting the loss of basic genetic features in the Acholeplasmataceae. BMC Genomics 15:931.




Su, P., Wicaksono, W. A., Li, C., Michl, K., Berg, G., Wang, D., Xiao, Y., Huang, R., Kang, H., Zhang, D., Cernava, T. and Liu, Y. 2022. Recovery of metagenome-assembled genomes from the phyllosphere of 110 rice genotypes. Sci. Data 9:254.




Tully, J. G., Rose, D. L., Carle, P., Bové, J. M., Hackett, K. J. and Whitcomb, R. F. 1994a.
Acholeplasma entomophilum sp. nov. from gut contents of a wide range of host insects. Int. J. Syst. Bacteriol. 38:164-167.

Tully, J. G., Whitcomb, R. F., Rose, D. L., Bové, J. M., Carle, P., Somerson, N. L., Williamson, D. L. and Eden-Green, S. 1994b.
Acholeplasma brassicae sp. nov. and Acholeplasma palmae sp. nov., two non-sterol-requiring mollicutes from plant surfaces. Int. J. Syst. Bacteriol. 44:680-684.


Walitang, D., Samaddar, S., Choudhury, A. R., Chatterjee, P., Ahmed, S. and Sa, T. 2019. Diversity and plant growth-promoting potential of bacterial endophytes in rice. In: Plant growth promoting rhizobacteria (PGPR): prospects for sustainable agriculture, eds. by R. Z. Sayyed, M. S. Reddy and S. Antonius, pp. 3-17. Springer, Singapore.


Wang, P., Kong, X., Chen, H., Xiao, Y., Liu, H., Li, X., Zhang, Z., Tan, X., Wang, D., Jin, D., Deng, Y. and Cernava, T. 2021. Exploration of intrinsic microbial community modulators in the rice endosphere indicates a key role of distinct bacterial taxa across different cultivars. Front. Microbiol. 12:629852.



- TOOLS
-
METRICS

-
- 0 Crossref
- 0 Scopus
- 697 View
- 44 Download
- ORCID iDs
-
Hang Thuy Dinh

https://orcid.org/0000-0002-9399-3720 - Related articles



 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Supplement1
Supplement1 Print
Print



