 |
 |
| Plant Pathol J > Volume 36(5); 2020 > Article |
|
Abstract
Xanthomonas campestris pv. campestris (Xcc), the pathogen of black rot which is the most destructive disease of Brassica vegetables throughout the world. Here, we reported two novel sequence-characterized amplified region (SCAR) markers (i.e., XccR6-60 and XccR6-67) for the detection of Xcc race 6 via re-alignment of the complete genome sequences of Xcc races/strains/pathovars. The specificity of SCAR primer sets was verified by mean of PCR amplification using the genomic DNA template of Xcc races/strains/pathovars and two other plant infecting bacterial strains. The PCR result revealed that the XccR6-60 and XccR6-67 primer sets amplified 692-bp and 917-bp DNA fragments, respectively, specifically from race 6, while no visible amplification was detected in other samples. In addition, the SCAR primers were highly sensitive and can detect from a very low concentration of genomic DNA of Xcc race 6. However, the complete genome sequence of Xcc race 6 is not yet publicly available. Therefore, the cloning and sequencing of XccR6-60 and XccR6-67 fragments from race 6 provide more evidence of the specificity of these markers. These results indicated that the newly developed SCAR markers can successfully, effectively and rapidly detect Xcc race 6 from other Xcc races/strains/pathovars as well as other plant pathogenic bacteria. This is the first report for race-specific molecular markers for Xcc race 6.
Xanthomonas campestris pv. campestris (Xcc) is a gram negative rod-shaped proteobacterium which causes black rot disease, a global threat to Brassica crops. It can infect from the cultivated species (such as cabbage, cauliflower, broccoli, kohlrabi, Brussels sprouts, kale, mustard, radish, turnip and turnip greens) to the model plant Arabidopsis, ornamentals and weeds (An et al., 2011; Soengas et al., 2007; Vicente et al., 2001).
Xcc is a seed born, vascular pathogen which enters the plant through the hydathodes, wounds and insect transmission, and develops typical V-shaped lesions, as well as necrotic and darkened veins (Cook et al., 1952; Williams, 1980). This pathogen can spread through plant debris, rain drops, irrigation, agricultural implements, seed trade and weeds (Arias et al., 2000; Grimm and Vogelsanger, 1989; Lema et al., 2012; Schaad and Alvarez, 1993; Yuen et al., 1987). Black rot significantly reduces the yield (50-65%) and quality of cruciferous vegetables (Saha et al., 2016). Also, under a favorable condition, up to 100% yield loss has been reported in cabbage (Massomo et al., 2003; Williams, 1980). Traditionally, cabbage is one the most important (ranked third) Brassica vegetables in Republic of Korea and black rot is also identified as a major barrier for cabbage production (Kim, 1986; Park, 2006).
Xcc strains have been classified into different physiological races based on the interactions of pathogens with differential cultivars of Brassica species. To date, eleven physiological Xcc races have been noticed (Fargier and Manceau, 2007; Kamoun et al., 1992; Vicente et al., 2001). Vicente et al. (2001) isolated six Xcc races (race 1, 2, 3, 4, 5, and 6) in the United Kingdom. A total of seven races (race 1, 4, 5, 6, 7, 8, and 9) have been isolated from Brassica species in northwestern Spain (Lema et al. 2012). Five races (race 1, 4, 5, 6, and 7) were reported in Nepal while race 1, 4, and 6 were most common (Jensen et al., 2010). Four races (race 1, 3, 4, and 6) have been isolated in South Africa (Chidamba and Bezuidenhout, 2012). In 2017, two new Xcc races (race 10 and 11) have been reported in Portugal (Cruz et al., 2017). Among the Xcc races, race 1 and 4 are predominant in Brassica oleracea subspecies whereas race 6 is predominant in B. rapa (Lema et al., 2012; Vicente and Holub, 2013). Recently, race-specific molecular markers for Xcc race 1, 3, 4, and 5 have already been reported by our research group (Afrin et al., 2018, 2019; Rubel et al., 2017). In this study, we aimed to develop molecular markers specific to Xcc race 6.
Eight reference strains of Xcc races (races 1-8), eight race-unknown Xcc strains, six Xc pathovars, and two additional plant infecting-bacteria including Pseudomonas syringae pv. maculicola and Pectobacterium carotovora subsp. carotovora (described in our previous study by Afrin et al., 2019) were used in this study. The bacterial strains were cultured on KingŌĆÖs medium B at 30┬░C (48 h) (King et al., 1954).
The gDNA of the bacterial strains was isolated with a commercial kit (DNeasy Plant Mini Kit, Qiagen, Valencia, CA, USA) following the manufacturerŌĆÖs instructions. The intensity and purity of the gDNA were determined by using a ŌĆśNanoDrop spectrophotometerŌĆÖ (NanoDrop Technologies, Wilmington, DE, USA). Subsequently, the gDNA samples were stored at ŌłÆ20┬░C refrigerator until use.
The available complete genome sequences of Xcc races (races 1, 3, 4, and 9), race-unknown Xcc strains and Xc pathovars were retrieved from the NCBI (https://www.ncbi.nlm.nih.gov/genome/) (Table 1). To develop molecular markers, we aligned the complete genome of available Xcc races (race 1, 3, 4, and 9), Xcc race-unknown strains, and Xc pathovars (Table 1) with ŌĆśprogressiveMauveŌĆÖ tool (Darling et al., 2004). Further, this alignment was analyzed by the trial version of Geneious software program (http://www.geneious.com) and identified variable regions among the aligned genomes (Fig. 1).
The variable regions among the genomes were considered for primer designing with the ŌĆśPCR Primer StatsŌĆÖ (http://www.bioinformatics.org/sms2/pcr_primer_stats.html) (Table 2). Further the designed primers were BLAST-searched (http://210.110.86.160/lab/home.html) to check the primer specificity. The PCR was performed with the bacterial gDNA. PCR reaction volume was total 20 ╬╝l consisting of 30 ng of gDNA as a template, 10 pmol of each primer, 0.25 mM dNTP mix, 5├Ś GoTaq buffer, 1 unit of GoTaq DNA Polymerase (Promega, Madison, WI, USA). The thermocycler conditions were as follows: 1├Ś (94┬░C, 5 min); 22├Ś (94┬░C, 30 s; 63┬░C, 40 s; 72┬░C, 45 s), 1├Ś (72┬░C, 5 min). Then the PCR products were loaded on 1.5% agarose gel followed by gel electrophoresis and visualized with a gel documentation system. Furthermore, the sensitivity of the sequence-characterized amplified region (SCAR) primer sets was determined by using a dilution series (50 ng/╬╝l, 5 ng/╬╝l, 0.5 ng/╬╝l, 0.05 ng/╬╝l, and 0.005 ng/╬╝l) of Xcc race 6 gDNA.
The genomic fragments (XccR6-60 and XccR6-67) specific to Xcc race 6 were amplified by PCR and purified with the Wizard SV gel and PCR cleanup system (Promega). Then, the purified PCR products were cloned with TOP cloner blunt kit (Enzynomics, Daejeon, Korea) following the manufacturerŌĆÖs instructions. The single colonies were checked by colony PCR and only positive clones were grown in a liquid LB media for overnight. Subsequently, the plasmid DNA was isolated and purified with a GeneAll kit (Seoul, Korea). The plasmid DNA samples were sequenced by ABI 3730XL DNA sequencer (Macrogen Inc., Seoul, Korea) using universal primers. The sequence alignment was carried out with the Clustal Omega online tool (https://www.ebi.ac.uk/Tools/msa/clustalo/).
Three-week-old cabbage plants were inoculated with eight reference races of Xcc (race 1-8) according to the method described by Afrin et al. (2018). Upon visible symptom of black rot, about 1 cm2 infected area of inoculated leaf was collected and sliced into small pieces. Thereafter, sliced pieces of leaf were placed in 250 ╬╝l sterile water (40 min) and 10 ╬╝l of the soaked water was used as a template for the bio-PCR.
The availability of the complete genome sequences of Xcc races, Xcc strains, Xc pathovars and their re-alignment enabled us to develop Xcc race specific molecular markers. In this study, we developed two SCAR markers XccR6-60 and XccR6-67, for rapid detection of race 6 from other Xcc races (Fig. 1). Since, the complete genome sequence of Xcc race 6 is not available yet, we analyzed the variable genomic regions among the aligned genomes. The result showed that the XccR6-60 and XccR6-67 fragments were specific to three race-unknown Xcc strains, including CN14, CN15, and CN16 (Fig. 1) and absent in the remaining genomes in the alignment. In the next step, we designed two sets of primers on XccR6-60 (693-bp) and XccR6-67 (923-bp) genomic fragments considering CN14, CN15, and CN16 as reference genome and amplified by the PCR.
The PCR analysis revealed that the XccR6-60 and XccR6-67 SCAR primer sets amplified about 692-bp and 917-bp fragments, respectively only from the gDNA of Xcc race 6 while no amplification was observed for the remaining tested samples (Fig. 2, Supplementary Fig. 1). Moreover, the sensitivity test of the SCAR primer sets revealed that they were sensitive enough to detect race 6 by PCR from total 0.05-0.5 ng template gDNA (Fig. 3).
Further, we cloned and sequenced the bands amplified by the SCAR primers. The sequence analysis showed that XccR6-60 primer set amplified fragment with 692-bp from Xcc race 6 (Supplementary Fig. 2A). Only 1-bp deletion at 515-bp position and two single nucleotide polymorphisms (SNPs) compared to the reference (CN14, CN15, and CN16). For XccR6-67, 6-bp deletions at 241-bp position 14 SNPs compared to the reference (CN14, CN15, and CN16) (Supplementary Fig. 2B).
The sequence obtained through the cloning of XccR6-60 and XccR6-67 fragments from Xcc race 6 was subjected to open reading frame (ORF) prediction with the ORF Finder tool of NCBI (https://www.ncbi.nlm.nih.gov/orffinder/). The results indicated that the XccR6-60 and XccR6-67 fragments contained 381-bp (129-aa) and 720-bp (239-aa) ORF, respectively (Fig. 4, Supplementary Figs. 3 and 4). Thereafter, BLASTp search was performed using these proteins in the NCBI. The result showed that 129-aa protein sequence of XccR6-60 had a significant homology (99% identity) with DUF4189 domain-containing protein of X. campestris (WP_076054737.1). In case of XccR6-67, 239-aa protein sequence had a significant homology with a hypothetical protein of X. campestris (WP_076054735.1) (Supplementary Figs. 3 and 4). In addition, the conserved domain analysis using these proteins revealed that the 129-aa protein of XccR6-60 contained a conserved domain called ŌĆśDomain of unknown function (DUF4189)ŌĆÖ while 239-aa protein of XccR6-67 contained a ŌĆśTrbL domain-containing proteinŌĆÖ (WP_076054735.1) (Supplementary Figs. 3 and 4).
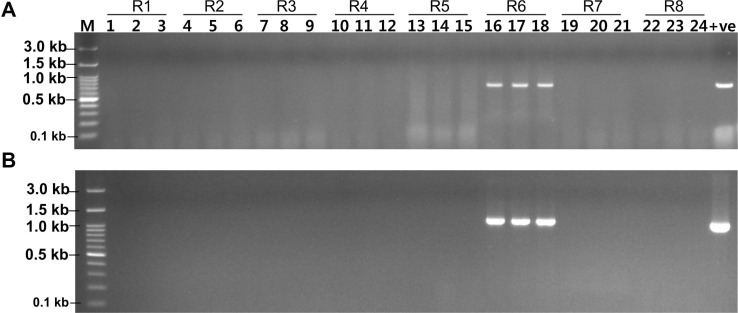
A bio-PCR was performed with artificially infected cabbage leaves with reference Xcc races (races 1-8) to detect the potentiality of race 6 specific primers (XccR6-60 and XccR6-67). The visible bands were only for those cabbage leaves inoculated with Xcc race 6 for both primer sets, while undetected for rest of the samples inoculated with other races (Fig. 5).
The present study was carried out to develop molecular marker for race-specific, rapid and precise detection of Xcc race 6 from other Xcc races/strains/pathovars. The new races of Xcc are emerging continuously in different brassica vegetable growing regions of the world. For example, five physiological races of Xcc (0-4) have been reported by Kamoun et al. (1992) based on differential interactions with different Brassica spp. Thereafter, Vicente et al. (2001) noticed six races of Xcc (race 1-6). A total of nine Xcc races have been reported by Fargier and Manceau (2007). More recently, Cruz et al. (2017) identified two new races of Xcc (race 10 and race 11). These races were determined by traditional method with the differential cultivars and the pathogens. Korean Xcc isolates need to be determined their race identity. However, the cultivar-based race determination method is more expensive and time consuming. Therefore, PCR-based simple, reliable and race-specific detect is crucial for the race determination of Korean isolates. So far, our research group have developed molecular markers for race-specific detection of Xcc race 1, 3, 4, and 5 (Afrin et al., 2018, 2019; Rubel et al., 2017).
In this study, we developed PCR-based SCAR markers (XccR6-60 and XccR6-67) specific to Xcc race 6 through re-alignment of available complete genome sequences of fifteen Xcc races/strains/pathovars (Fig. 1). As the complete genome sequence of Xcc race 6 is not available yet, the variable genomic regions of known Xcc genome sequences were considered as targets to develop the race 6-specific molecular markers. A similar approach was recently reported by Afrin et al. (2019) to develop Xcc race 5-specific molecular marker whose complete genome is not yet sequenced. The SCAR markers developed in this study were able to detect and identify Xcc race 6 from other Xcc races/strains/pathovars (Fig. 2, Supplementary Fig. 1). Further, these markers can detect Xcc race 6 from very low concentration of race 6 gDNA templates (total 0.05-0.5 ng) (Fig. 3). Moreover, the newly developed primer sets (XccR6-60 and XccR6-67) were highly sensitive as they were capable to detect race 6 directly from infected cabbage leaves within few hours by bio-PCR (Fig. 5). This suggests that newly developed SCAR markers provide a simple, specific, quick, low-cost and powerful method for identification of Xcc race 6. A similar bio-PCR technique was reported by Song et al. (2014) for X. oryzae pv. oryzae (Xoo) race K3a and by Afrin et al. (2018) for Xcc race 3. In the past, PCR-based molecular markers were successfully used for pathovar-specific detection of Xanthomonas. For example, hrp gene (hypersensitive reaction and pathogenicity) based PCR amplification technique has been used to distinguish X. campestris pathovars (Berg et al., 2005; Singh et al., 2014) and X. axonopodis pv. vesicatoria (Leite et al., 1994). Later, hrcC gene (encoding the outer membrane protein of the type III secretion system) based PCR assay for Xcc specific detection was also described (Zaccardelli et al., 2007). Moreover, Song et al. (2014) reported a SCAR marker for Xoo K3a race-specific detection. In addition, PCR-based molecular markers have also been reported for distinguishing fungal pathogens like Fusarium oxysporum f. sp. melonis race 2 and F. oxysporum f. sp. cubense race 4, which cause vascular wilt and panama disease in cucurbits and banana, respectively (Lin et al., 2009; Luongo et al., 2012). Wang et al. (2010) reported SCAR markers for race-specific detection of Puccinia striiformis f. sp. tritici Chinese races (CYR32 and CYR33), the causal agent of stripe rust of wheat.
The cloning and sequence analysis of XccR6-60 and XccR6-67 marker regions revealed that they have a considerably high homology with DUF4189 domain-containing protein (WP_076054737.1) and hypothetical protein (WP_076054735.1) of X. campestris, respectively. Further, domain analysis using protein sequence of XccR6-67 (239-aa) revealed that presence of TrbL domain (TrbL/VirB6 plasmid conjugal transfer protein, pfam04610) (Supplementary Fig. 4). VirB6 was reported as a core component of type IV secretion system (T4SS), which is involved in macromolecular translocation (Villamil Giraldo et al., 2012). Moreover, it has been reported that VirB6 domains direct the ordered export of a DNA substrate via the T4SS (Jakubowski et al., 2004).
The genomic fragments XccR6-60 and XccR6-67 were specific to race-unknown Xcc strain CN14, CN15, and CN16 but were not present in the other genomes analyzed (Fig. 1). Nevertheless, the primer sets designed to cover these regions were PCR amplified only from Xcc race 6 (Fig. 2). Besides, the NCBI BLAST search of the cloned sequence of XccR6-60 and XccR6-67 fragments from Xcc race 6 showed more than 99% and 97%, respectively identity with three race-unknown Xcc strains, CN14, CN15, and CN16 (Table 3). Therefore, it can be anticipated that these race-unknown Xcc strain (CN14, CN15, and CN16) might belong to race 6. However, more experimental evidences are required to support this hypothesis.
In conclusion, the newly developed SCAR markers (XccR6-60 and XccR6-67) could be useful for rapid, lowcost and reliable detection of race 6 from other Xcc races/strains/pathovars. Thus, these molecular markers would be valuable for early detection of this pathogen from infected Brassica crops worldwide.
Supplementary materials are available at Plant Pathology Journal website (http://www.ppjonline.org/).
Acknowledgments
This research work was supported by the Golden Seed Project (Center for Horticultural Seed Development) of the Ministry of Agriculture, Food and Rural Affairs in the Republic of Korea (MAFRA) under Grant no. 213007-05-4-CG100. We thank Dr. Joana G. Vicente, University of Warwick, UK for providing Xcc races (race 1-7). We also thank Dr. Pilar Soengas, Department of Plant Genetics, Misi├│n Biol├│gica de Galicia (MBG-CSIC), Pontevedra, Spain for providing Xcc race 8.
REFERENCES
Afrin, K. S., Rahim, M. A., Jung, H.-J., Park, J.-I., Kim, H.-T. and Nou, I.-S. 2019. Development of molecular marker through genome realignment for specific detection of Xanthomonas campestris pv. campestris race 5, the pathogen of black rot disease. J. Microbiol. Biotechnol. 29:785-793.


Afrin, K. S., Rahim, M. A., Rubel, M. H., Natarajan, S., Song, J.-Y., Kim, H.-T., Park, J.-I. and Nou, I.-S. 2018. Development of race-specific molecular marker for Xanthomonas campestris pv. campestris race 3, the causal agent of black rot of crucifers. Can. J. Plant Sci. 98:1119-1125.

An, S.-Q., Lu, G.-T., Su, H.-Z., Li, R.-F., He, Y.-Q., Jiang, B.-L., Tang, D.-J. and Tang, J.-L. 2011. Systematic mutagenesis of all predicted gntR genes in Xanthomonas campestris pv. campestris reveals a GntR family transcriptional regulator controlling hypersensitive response and virulence. Mol. Plant.-Microbe. Interact. 24:1027-1039.


Arias, R. S., Nelson, S. C. and Alvarez, A. M. 2000. Effect of soil-matric potential and phylloplanes of rotation-crops on the survival of a bioluminescent Xanthomonas campestris pv. campestris. Eur. J. Plant Pathol. 106:109-116.
Berg, T., Tesoriero, L. and Hailstones, D. L. 2005. PCR-based detection of Xanthomonas campestris pathovars in Brassica seed. Plant Pathol. 54:416-427.

Bogdanove, A. J., Koebnik, R., Lu, H., Furutani, A., Angiuoli, S. V, Patil, P. B., Van Sluys, M.-A., Ryan, R. P., Meyer, D. F., Han, S.-W., Aparna, G., Rajaram, M., Delcher, A. L., Phillippy, A. M., Puiu, D., Schatz, M. C., Shumway, M., Sommer, D. D., Trapnell, C., Benahmed, F., Dimitrov, G., Madupu, R., Radune, D., Sullivan, S., Jha, G., Ishihara, H., Lee, S.-W., Pandey, A., Sharma, V., Sriariyanun, M., Szurek, B., VeraCruz, C. M., Dorman, K. S., Ronald, P. C., Verdier, V., Dow, J. M., Sonti, R. V., Tsuge, S., Brendel, V. P., Rabinowicz, P. D., Leach, J. E., White, F. F. and Salzberg, S. L. 2011. Two new complete genome sequences offer insight into host and tissue specificity of plant pathogenic Xanthomonas spp. J. Bacteriol. 193:5450-5464.



Bolot, S., Cerutti, A., Carr├©re, S., Arlat, M., Saux, M., Portier, P., Poussier, S., Jacques, M.-A. and No├½l, L. D. 2015. Genome sequences of the Race 1 and Race 4 Xanthomonas campestris pv. campestris strains CFBP 1869 and CFBP 5817. Genome Announc. 3:e01023-15.




Bolot, S., Guy, E., Carrere, S., Barbe, V., Arlat, M. and Noël, L. D. 2013a. Genome sequence of Xanthomonas campestris pv. campestris strain Xca5. Genome Announc. 1:e00032-12.


Bolot, S., Roux, B., Carrere, S., Jiang, B.-L., Tang, J.-L., Arlat, M. and Noël, L. D. 2013b. Genome sequences of three atypical Xanthomonas campestris pv. campestris strains, CN14, CN15, and CN16. Genome Announc. 1:e00465-13.



Chidamba, L. and Bezuidenhout, C. C. 2012. Characterisation of Xanthomonas campestris pv. campestris isolates from South Africa using genomic DNA fingerprinting and pathogenicity tests. Eur. J. Plant Pathol. 133:811-818.

Cook, A. A., Walker, J. C. and Larson, R. H. 1952. Studies on the disease cycle of black rot of crucifers. Phytopathology. 42:162-167.
Cruz, J., Tenreiro, R. and Cruz, L. 2017. Assessment of diversity of Xanthomonas campestris pathovars affecting cruciferous plants in Portugal and disclosure of two novel X. campestris pv. campestris races. J. Plant Pathol. 99:403-414.
da Silva, A. C. R., Ferro, J. A., Reinach, F. C., Farah, C. S., Furlan, L. R., Quaggio, R. B., Monteiro-Vitorello, C. B., Van Sluys, M. A., Almeida, N. F., Alves, L. M. C., Amaral, A. M., Bertolini, M. C., Camargo, L. E. A., Camarotte, G., Cannavan, F., Cardozo, J., Chambergo, F., Ciapina, L. P., Cicarelli, R. M. B., Coutinho, L. L., Cursino-Santos, J. R., El-Dorry, H., Faria, J. B., Ferreira, A. J. S., Ferreira, R. C. C., Ferro, M. I. T., Formighieri, E. F., Franco, M. C., Greggio, C. C., Gruber, A., Katsuyama, A. M., Kishi, L. T., Leite, R. P., Lemos, E. G. M., Lemos, M. V. F., Locali, E. C., Machado, M. A., Madeira, A. M. B. N., Martinez-Rossi, N. M., Martins, E. C., Meidanis, J., Menck, C. F. M., Miyaki, C. Y., Moon, D. H., Moreira, L. M., Novo, M. T. M., Okura, V. K., Oliveira, M. C., Oliveira, V. R., Pereira, H. A., Rossi, A., Sena, J. A. D., Silva, C., de Souza, R. F., Spinola, L. A. F., Takita, M. A., Tamura, R. E., Teixeira, E. C., Tezza, R. I. D., de dos Santos, M., Truffi, D., Tsai, S. M., White, F. F., Setubal, J. C. and Kitajima, J. P. 2002. Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature. 417:459-463.


Darling, A. C. E., Mau, B., Blattner, F. R. and Perna, N. T. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. G. enome Res. 14:1394-1403.

Desai, D., Li, J.-H., de Jong, E., Braun, R., Pitman, A., Visnovsky, S., Hampton, J. and Christey, M. 2015. Draft genome sequences of two New Zealand Xanthomonas campestris pv. campestris isolates, ICMP 4013 and ICMP 21080. Genome Announc. 3:e01247-15.




Fargier, E. and Manceau, C. 2007. Pathogenicity assays restrict the species Xanthomonas campestris into three pathovars and reveal nine races within X. campestris pv. campestris. Plant Pathol. 56:805-818.

Grimm, R. and Vogelsanger, J. eds. by Z. Klement, 1989. Black rot disease on cabbage, irrigation and spreading. In: Proceeding of the 7th International Conference on Plant Pathogenic Bacteria, Akademiai Kiado, Budapest, Hungary. 225-229.
Jakubowski, S. J., Krishnamoorthy, V., Cascales, E. and Christie, P. J. 2004. Agrobacterium tumefaciens VirB6 domains direct the ordered export of a DNA substrate through a type IV secretion system. J. Mol. Biol. 341:961-977.



Jensen, B. D., Vicente, J. G., Manandhar, H. K. and Roberts, S. J. 2010. Occurrence and diversity of Xanthomonas campestris pv. campestris in vegetable Brassica fields in Nepal. Plant Dis. 94:298-305.


Kamoun, S., Kamdar, H. V., Tola, E. and Kado, C. I. 1992. Incompatible interactions between crucifers and Xanthomonas campestris involve a vascular hypersensitive response: role of the hrpK locus. Mol. Plant-Microbe Interact. 5:22-33.

Kim, B. S. 1986. Testing for detection of Xanthomonas campestris pv. campestris in crucifer seeds and seed disinfection. Korean J. Plant Pathol. 2:96-101.
King, E. O., Ward, M. K. and Raney, D. E. 1954. Two simple media for the demonstration of pyocyanin and fluorescin. J. Lab. Clin. Med. 44:301-307.

Leite, R. P. J, Minsavage, G. V., Bonas, U. and Stall, R. E. 1994. Detection and identification of phytopathogenic Xanthomonas strains by amplification of DNA sequences related to the hrp genes of Xanthomonas campestris pv. vesicatoria. Appl. Environ. Microbiol. 60:1068-1077.



Lema, M., Cartea, M. E., Sotelo, T., Velasco, P. and Soengas, P. 2012. Discrimination of Xanthomonas campestris pv. campestris races among strains from northwestern Spain by Brassica spp. genotypes and rep-PCR. Eur. J. Plant Pathol. 133:159-169.

Lin, Y.-H., Chang, J.-Y., Liu, E.-T., Chao, C.-P., Huang, J.-W. and Chang, P.-F. L. 2009. Development of a molecular marker for specific detection of Fusarium oxysporum f. sp. cubense race 4. Eur. J. Plant Pathol. 123:353-365.


Liu, Y.-C., Wang, S.-C., Yu, Y.-J., Fung, K.-M., Yang, M.-T., Tseng, Y.-H., Tsai, S.-F., Sun, H. S., Lyu, P.-C. and Chou, S.-H. 2015. Complete genome sequence of Xanthomonas campestris pv. campestris strain 17 from Taiwan. Genome Announc. 3:e01466-15.


Luongo, L., Vitale, S., Haegi, A. and Belisario, A. 2012. Development of SCAR markers and PCR assays for Fusarium oxysporum f. sp. melonis race 2-specific detection. J. Plant Pathol. 94:193-199.
Massomo, S. M. S., Nielsen, H., Mabagala, R. B., Mansfeld-Giese, K., Hockenhull, J. and Mortensen, C. N. 2003. Identification and characterisation of Xanthomonas campestris pv. campestris strains from Tanzania by pathogenicity tests, Biolog, rep-PCR and fatty acid methyl ester analysis. Eur. J. Plant Pathol. 109:775-789.
Qian, W., Jia, Y., Ren, S.-X., He, Y.-Q., Feng, J.-X., Lu, L.-F., Sun, Q., Ying, G., Tang, D.-J., Tang, H., Wu, W., Hao, P., Wang, L., Jiang, B.-L., Zeng, S., Gu, W.-Y., Lu, G., Rong, L., Tian, Y., Yao, Z., Fu, G., Chen, B., Fang, R., Qiang, B., Chen, Z., Zhao, G.-P., Tang, J.-L. and He, C. 2005. Comparative and functional genomic analyses of the pathogenicity of phytopathogen Xanthomonas campestris pv. campestris. Genome Res. 15:757-767.



Roux, B., Bolot, S., Guy, E., Denanc├®, N., Lautier, M., Jardinaud, M.-F., Saux, M., Portier, P., Jacques, M.-A., Gagnevin, L., Pruvost, O., Lauber, E., Arlat, M., Carr├©re, S., Koebnik, R. and No├½l, L. D. 2015. Genomics and transcriptomics of Xanthomonas campestris species challenge the concept of core type III effectome. BMC Genomics. 16:975



Rubel, M. H., Robin, A. H. K., Natarajan, S., Vicente, J. G., Kim, H.-T., Park, J.-I. and Nou, I.-S. 2017. Whole-genome realignment facilitates development of specific molecular markers for races 1 and 4 of Xanthomonas campestris pv. campestris, the cause of black rot disease in Brassica oleracea. Int. J. Mol. Sci. 18:2523



Saha, P., Kalia, P., Sharma, M. and Singh, D. 2016. New source of black rot disease resistance in Brassica oleracea and genetic analysis of resistance. Euphytica. 207:35-48.

Schaad, N. W. and Alvarez, A. eds. by J. G. Swings and E. L. Civerolo, 1993. Xanthomonas campestris pv. campestris: cause of black rot of crucifers. In: Xanthomonas, Chapman and Hall, London, UK. 51-55.
Singh, D., Raghavendra, B. T., Singh, P., Singh, H., Raghuwanshi, R. and Singh, R. P. 2014. Detection of black rot disease causing pathogen Xanthomonas campestris pv. campestris by bio-PCR from seeds and plant parts of cole crops. Seed Sci. Technol. 42:36-46.

Soengas, P., Hand, P., Vicente, J. G., Pole, J. M. and Pink, D. A. C. 2007. Identification of quantitative trait loci for resistance to Xanthomonas campestris pv. campestris in Brassica rapa. Theor. Appl. Genet. 114:637-645.


Song, E.-S., Kim, S.-Y., Noh, T.-H., Cho, H., Chae, S.-C. and Lee, B.-M. 2014. PCR-based assay for rapid and specific detection of the new Xanthomonas oryzae pv. oryzae K3a race using an AFLP-derived marker. J. Microbiol. Biotechnol. 24:732-739.


Thieme, F., Koebnik, R., Bekel, T., Berger, C., Boch, J., B├╝ttner, D., Caldana, C., Gaigalat, L., Goesmann, A., Kay, S., Kirchner, O., Lanz, C., Linke, B., McHardy, A. C., Meyer, F., Mittenhuber, G., Nies, D. H., Niesbach-Kl├Čsgen, U., Patschkowski, T., R├╝ckert, C., Rupp, O., Schneiker, S., Schuster, S. C., Vorh├Člter, F.-J., Weber, E., P├╝hler, A., Bonas, U., Bartels, D. and Kaiser, O. 2005. Insights into genome plasticity and pathogenicity of the plant pathogenic bacterium Xanthomonas campestris pv. vesicatoria revealed by the complete genome sequence. J. Bacteriol. 187:7254-7266.



Vicente, J. G., Conway, J., Roberts, S. J. and Taylor, J. D. 2001. Identification and origin of Xanthomonas campestris pv. campestris races and related pathovars. Phytopathology. 91:492-499.


Vicente, J. G. and Holub, E. B. 2013. Xanthomonas campestris pv. campestris (cause of black rot of crucifers) in the genomic era is still a worldwide threat to brassica crops. Mol. Plant Pathol. 14:2-18.


Giraldo, A. M., Sivanesan, D., Carle, A., Paschos, A., Smith, M. A., Plesa, M., Coulton, J. and Baron, C. 2012. Type IV secretion system core component VirB8 from Brucella binds to the globular domain of VirB5 and to a periplasmic domain of VirB6. Biochemistry. 51:3881-3890.


Vorh├Člter, F.-J., Schneiker, S., Goesmann, A., Krause, L., Bekel, T., Kaiser, O., Linke, B., Patschkowski, T., R├╝ckert, C., Schmid, J., Sidhu, V. K., Sieber, V., Tauch, A., Watt, S. A., Weisshaar, B., Becker, A., Niehaus, K. and P├╝hler, A. 2008. The genome of Xanthomonas campestris pv. campestris B100 and its use for the reconstruction of metabolic pathways involved in xanthan biosynthsis. J. Biotechnol. 134:33-45.

Wang, B., Hu, X., Li, Q., Hao, B., Zhang, B., Li, G. and Kang, Z. 2010. Development of race-specific SCAR markers for detection of Chinese races CYR32 and CYR33 of Puccinia striiformis f . sp. tritici. Plant Dis. 94:221-228.

Fig.┬Ā1
Whole genome alignment of the publicly available genome sequences of Xanthomonas campestris pv. campestris (Xcc) races, strains and pathovars. The syntenic blocks are connected among the Xcc races/strains/Xc pathovars. The variable region among the Xcc races/strains/Xc subspecies are shown with the line diagram in the bottom of the alignment. Genomic blocks are shown as colored rectangles.
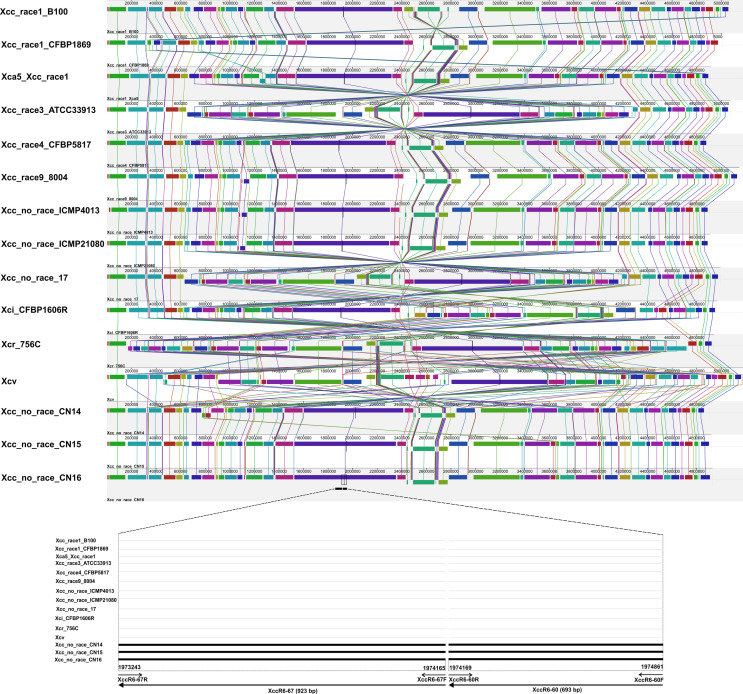
Fig.┬Ā2
Gel electrophoresis (1.5% agarose) of the PCR products of Xanthomonas campestris pv. campestris (Xcc) race 6 specific sequence-characterized amplified region primer sets (A, XccR6-60, 692-bp; B, XccR6-67, 917-bp) amplified from genomic DNA of Xcc races and pathovars. Lane M, 100 bp plus DNA ladder; lanes 1-8, Xcc race 1-8; lane Xci, X. campestris pv. incane (Xci); lane Xcr, X. campestris pv. raphanin (Xcr); lane Xcv, X. campestris pv. vesicatoria (Xcv); lane Xag, X. axonopodis pv. glycines (Xag); lane Xad, X. axonopodis pv. dieffenbachiae (Xad); lane Xcz, X. campestris pv. zinniae (Xcz); lane Psm, Pseudomonas syringae pv. maculicola (Psm); lane Pcc, Pectobacterium carotovora subsp. carotovora (Pcc).
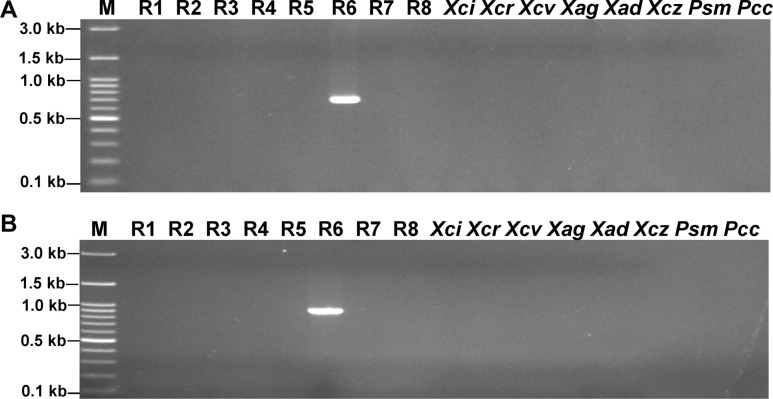
Fig.┬Ā3
Sensitivity of the Xanthomonas campestris pv. campestris (Xcc) race 6 specific SCAR primers (XccR6-60 and XccR6-67). (A) XccR6-60. (B) XccR6-67. Lane M, 100 bp plus DNA ladder; lane 1, 50 ng/╬╝l gDNA; lane 2, 5 ng/╬╝l; lane 3, 0.5 ng/╬╝l; lane 4, 0.05 ng/╬╝l; lane 5, 0.005 ng/╬╝l; lane 6, negative control. For the PCR amplification, 1 ╬╝l DNA of Xcc race 5 from each dilutions was used as a template.

Fig.┬Ā4
Nucleotide and amino acid sequences of the XccR6-60 (A) and XccR6-67 (B) genomic fragments cloned and sequenced from Xanthomonas campestris pv. campestris race 6. The open reading frames are presented in grey highlighted area: XccR6-60, 381-bp and XccR6-67, 720-bp. The deduced amino acid sequences are shown in the above of the nucleotide sequences: XccR6-60, 126-aa and XccR6-67, 239-aa. The nucleotide sequence is shown as reverse complement.

Fig.┬Ā5
Detection of Xanthomonas campestris pv. campestris (Xcc) race 6 by bio-PCR with artificially Xcc (races 1-8, R1-R8) inoculated cabbage leaves with XccR6-60 (A) and XccR6-67 (B) primer sets. Lanes 1-24, artificially inoculated leaf samples with Xcc races (lanes: 1-3, race 1; 4-6, race 2; 7-9, race 3; 10-12, race 4; 13-15, race 5; 16-18, race 6; 19-21, race 7; 22-24, race 8; +ve, gDNA of Xcc race 5 as a positive control; -ve, negative control; M, 100 bp plus DNA ladder).
Table┬Ā1
Publicly available complete genome sequences of Xcc races, Xcc strains, and Xc subspecies, and their accession numbers
| Sl. No. | Bacterial strains | GenBank accession | Races | Origin | Host | Disease | Genome size (bp) | Gene | Protein | References |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | B100 (Xcc) | AM920689 | 1 | Italy | Brassica oleracea var. botrytis | Black rot | 5,079,002 | 4,542 | 4,236 | Vorh├Člter et al. (2008) |
| 2 | CFBP1869 (Xcc) | NZ_CM002545 | 1 | C├┤te dŌĆÖIvoire | Brassica oleracea | Black rot | 5,011,687 | 4,489 | 4,166 | Bolot et al. (2015) |
| 3 | Xca5 (Xcc) | NZ_HF570389 | 1 | America | Brassicaceae | Black rot | 4,916,437 | 4,486 | 4,169 | Bolot et al. (2013a) |
| 4 | ATCC 33913 (Xcc) | AE008922 | 3 | - | Cabbage | Black rot | 5,076,187 | 4,240 | 4,179 | da Silva et al. (2002) |
| 5 | CFBP 5817(Xcc) | NZ_CM002673 | 4 | Chile | Brassica oleracea var. botrytis | Black rot | 4,918,955 | 4,447 | 4,122 | Bolot et al. (2015) |
| 6 | Strain 8004 (Xcc) | NC_007086 | 9 | Sussex, UK | Cauliflower | Black rot | 5,148,708 | 4,584 | 4,297 | Qian et al.(2005) |
| 7 | ICMP 4013 (Xcc) | CP012146 | - | New Zealand | Crucifers | Black rot | 4,908,383 | 4,389 | 4,040 | Desai et al. (2015) |
| 8 | ICMP 21080 (Xcc) | CP012145 | - | New Zealand | Crucifers | Black rot | 4,911,121 | 4,389 | 4,039 | Desai et al. (2015) |
| 9 | CN14 (Xcc) | CM001871 | - | Guilin, China | B. juncea, B. rapa | Black rot | 4,908,255 | 4,445 | 4,127 | Bolot et al. (2013b) |
| 10 | CN15 (Xcc) | NZ_CM001890 | - | Guilin, China | B. juncea, B. rapa | Black rot | 4,920,230 | 4,496 | 4,162 | Bolot et al. (2013b) |
| 11 | CN16 (Xcc) | NZ_CM001897 | - | Guilin, China | B. juncea, B. rapa | Black rot | 4,928,563 | 4,496 | 4,162 | Bolot et al. (2013b) |
| 12 | Strain 17 (Xcc) | CP011946 | - | Taiwan | Crucifers | Black rot | 4,994,426 | 4,398 | 4,086 | Liu et al. (2015) |
| 13 | CFBP 1606R (Xci) | NZ_CM002635 | - | France | Matthiola incana | Leaf spot | 4,967,288 | 4,423 | 4,071 | Roux et al. (2015) |
| 14 | Strain 756C (Xcr) | NC_017271 | - | - | Raphanus sativus | Bacterial spot | 4,941,214 | 4,372 | 4,055 | Bogdanove et al. (2011) |
| 15 | Strain 85-10 (Xcv) | NC_007508 | - | Florida, USA | Pepper | Bacterial spot | 5,178,466 | 4,900 | 4,799 | Thieme et al. (2005) |
Table┬Ā2
List of Xcc race 6 specific primers used for in this study




 PDF Links
PDF Links PubReader
PubReader Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Supplement
Supplement Print
Print




