Rapid and Sensitive Detection of Lettuce Necrotic Yellows Virus and Cucumber Mosaic Virus Infecting Lettuce (Lactuca sativa L.) by Reverse Transcription Loop-Mediated Isothermal Amplification
Article information
Abstract
Cucumber mosaic virus (CMV) is damaging to the growth and quality of lettuce crops in Lanzhou, China. Recently, however, for the first time an isolate of lettuce necrotic yellows virus (LNYV) has been detected in lettuce crops in China, and there is concern that this virus may also pose a threat to lettuce production in China. Consequently, there is a need to develop a rapid and efficient detection method to accurately identify LNYV and CMV infections and help limit their spread. Reverse transcription loop-mediated isothermal amplification (RT-LAMP) assays were developed to detect the nucleoprotein (N) and coat protein (CP) genes of LNYV and CMV, respectively. RT-LAMP amplification products were visually assessed in reaction tubes separately using green fluorescence and gel electrophoresis. The assays successfully detected both viruses in infected plants without cross reactivity recorded from either CMV or LNYV or four other related plant viruses. Optimum LAMP reactions were conducted in betaine-free media with 6 mM Mg2+ at 65°C for LNYV and 60°C for 60 min for CMV, respectively. The detection limit was 3.5 pg/ml and 20 fg/ml using RT-LAMP for LNYV and CMV plasmids, respectively. Detection sensitivity for both RT-LAMP assays was greater by a factor of 100 compared to the conventional reverse transcription polymerase chain reaction assays. This rapid, specific, and sensitive technique should be more widely applied due to its low cost and minimal equipment requirements.
Lettuce (Lactuca sativa L.) is the most economically important vegetable crop belonging to the family Asteraceae (Krause-Sakate et al., 2001) and is typically eaten cold, raw, in salads, sandwiches, hamburgers, tacos as well as in many other dishes around the world (Fletcher et al., 2005; Wikipedia, 2019). In China, the economic importance of lettuce has also greatly increased during the past decade because of the rapid increase in demand for this crop, which is now exceeding half the world’s production. However, lettuce is susceptible to a number of viral pathogens, including Cucumber mosaic virus (genus: Cucumovirus; family: Bromoviridae). Cucumber mosaic virus (CMV) is one of the most widespread plant viruses with an extensive host range, infecting approximately 1,000 species in over 100 plant families that include cereals, fruits, vegetables and ornamentals, and can cause significant economic losses for numerous vegetable and horticultural crops (Roossinck, 1999). In lettuce plants symptoms caused by CMV include plant stunting, leaf chlorosis, and mosaic as well as poor head formation (Bruckart and Lorbeer, 1975; El-Borollosy and Waziri, 2013). The virus is readily transmitted in a non-persistent manner by more than 75 species of aphids (Palukaitis et al., 1992). It may also be transmitted mechanically and can be seed-borne in a number of hosts.
In 2018, a preliminary field survey was conducted in a major lettuce cultivation area in Lanzhou City, Gansu Province, China. This survey detected lettuce necrotic yellows virus (LNYV) for the first time in fields around Shichuan Town (Gaolan County, Lanzhou City, Gansu Province, China; 36°10″N, 103°59″E). The main symptoms observed were plant stunting, leaf chlorosis, leaf roughness and occasional necrosis within leaf tissue. Mosaic symptoms were also present and attributed to a mixed LNYV infection of both CMV and lettuce mosaic virus (LMV) (Fig. 1). LNYV is a member of the genus Cytorhabdovirus, family Rhabdoviridae (Tordo et al., 2005). Infection by members of the genus Cytorhabdoviridae is characterized by the accumulation of enveloped virions in the cytoplasm of host cells (Jackson et al., 2005). LNYV causes a serious disease of lettuce crops grown in both Australia and New Zealand, with the first reports published in 1963 and 1973, respectively (Fry et al., 1973; Stubbs and Grogan, 1963), and it continues to cause crop losses of up to 70% in some cultivated areas (Fletcher et al., 2017; unpublished report). There also have been isolated reports of similar lettuce virus infections in Spain (Rubio-Huertos and Garcia-Hidalgo, 1982), Italy (Ragozzino et al., 1989), and Great Britain (Blancard et al., 2006). LNYV is transmitted in a persistent, propagative manner by the blackcurrant-sowthistle aphid (Hyperomyzus lactucae L.) (Higgins et al., 2016). Ragozzino et al. (1989) also determined that the currant-lettuce aphid (Nasonovia ribisnigri) was the primary vector of LNYV disease outbreaks in Italy.

Healthy lettuce plant (A) and lettuce plant (B) infected with lettuce necrotic yellows virus (LNYV) exhibiting symptoms of plant stunting, leaf chlorosis, leaf roughness and intermittent necrosis within the leaf tissue. (C) Mosaic symptoms also present were attributed to the mixed infection of LNYV with both cucumber mosaic virus (CMV) and lettuce mosaic virus (LMV).
Viral disease management is dependent upon an accurate detection procedure which is convenient, reproducible and scalable for a wide range of samples (Viswanathan et al., 2013). Currently available methods commonly used for detecting viruses that affect lettuce include enzyme-linked immunosorbent assay (ELISA), reverse transcription (RT)–polymerase chain reaction (PCR) for LNYV and CMV (Callaghan and Dietzgen, 2005; Deyong et al., 2005; Dietzgen and Francki, 1988; Koenig, 1981) and immunocapture (IC)-RT-PCR for CMV (El-Borollosy and Waziri, 2013). These methods are valuable for identifying latent infections at most stages of infection. However, false negative results and low sensitivity are common problems for the ELISA method. Moreover, PCR-based methods are expensive in terms of time and equipment and hence impractical for large-scale use. The IC-RT-PCR technique omits the need to isolate RNA while providing a more rapid and less costly approach to prepare templates for amplification (Gambley et al., 2009); however, we have found IC-RT-PCR sensitivity insufficient in detecting low CMV concentrations (Zhang et al., 2017).
On the other hand, the loop-mediated isothermal amplification of nucleic acids (LAMP) assay, is not only sensitive and relatively inexpensive to use but can also be routinely conducted at a constant temperature between 58–70°C for approximately 1 h in a water bath or on a heating block. The technique can also be used for amplifying RNA by adding reverse transcriptase to the LAMP reaction mixture (often referred to as RT-LAMP) (Bhat et al., 2013). Both LAMP and RT-LAMP use four to six primers along with DNA polymerization with strand-displacing activity to generate amplification products. These products can be visually detected using agarose gel electrophoresis or through color changes visualized by adding SYBR Green I, ethidium bromide or calcein to the reaction mixture (Notomi et al., 2000; Parida et al., 2008). Both methods have been successfully used in the detection of many DNA- (Bhat et al., 2013; Ravindran et al., 2012) and RNA-based (He et al., 2016; Liu et al., 2010; Zhao et al., 2010) plant pathogens. In this study, we developed and optimized a simple, rapid, reliable and visual RT-LAMP method for the accurate detection of LNYV and CMV infections in lettuce plants.
Materials and Methods
Plant sample collection and reference virus isolates
Within the cultivation areas of Shichuan and Donggang towns, Lanzhou City, China, we collected naturally infected lettuce leaf samples that displayed typical viral symptoms (i.e., plant stunting, leaf chlorosis, leaf roughness and intermittent necrosis in leaf tissues). From field experience gained in New Zealand and from employing known (tested) positives of viruses from New Zealand used here as references (Fletcher et al., 2017; unpublished), lettuce leaves were tested using RT-PCR to confirm their respective virus infection status. Lettuce leaf samples from individual plants that tested positive for LNYV or CMV served as a reference source of each virus and were stored at −70°C prior to analysis. Other virus reference sources for our experiments, each isolated from lily (Lilium oriental cv. Sorbonne), included lily symptomless virus (LSV), a CMV lily isolate, lily mottle virus (LMoV), and arabis mosaic virus (ArMV) (Zhang et al., 2017; Zhang, unpublished data).
RT-LAMP primer design
The conserved regions of nucleoprotein (N) and coat protein (CP) genes were used as the basis for the design of the RT-LAMP primers for LNYV and CMV detection. This was done by establishing sequence alignments (multiple sequence alignment) of N and CP gene sequences of LNYV and CMV isolated from lettuce available in GenBank (accession Nos. AJ746190.1 and AJ810253), respectively (Callaghan and Dietzgen, 2005; Deyong et al., 2005). Primer design was conducted using PrimerExplorer version 5 software (Eiken Chemical Co., Ltd., Japan) with the default settings. Two LAMP external primers (F3 and B3) and two internal primers (FIP [F1c + F2] and BIP [B1c + F2]) as well as a pair of loop primers (F-loop and B-loop) and a single loop primer (B-loop) for LNYV and CMV detection were respectively designed and were capable of detecting a total of eight sequences of the N gene of LNYV and seven sequences of the CP gene of CMV (sequences shown in Table 1).
RNA extraction
Total RNA was extracted from each lettuce leaf sample of about 100 mg (fresh weight) using the RNAprep Pure Kit (For Plant) (Tiangen Biotech Co. Ltd., Beijing, China) according to the manufacturer’s instructions. The final elution step was conducted using 50 μl of RNAse-free H2O. The concentration of the RNA sample was measured with a NanoDrop ND-1000 Spectrophotometer (NanoDrop Technologies LLC, Wilmington, DE, USA).
Cloning, sequencing, and sequence analysis
The first-strand viral cDNA was synthesized with the PrimeScript RT Reagent Kit with gDNA Eraser (Takara Biotechnology Co., Ltd., Dalian, China) using random hexamers. PCR was carried out in 12.5 μl reaction volumes containing 1.0 μl of the cDNA products, 1.25 μl of a 10× PCR buffer (100 mmol/l Tris-HCl [pH 8.3], 500 mmol/l KCl, 15 mmol/l MgCl2), 0.4 mmol of a dNTP mixture, 0.3 U of Taq polymerase (TaKaRa Ex Taq, Takara) and 0.1 μM of each LNYV-F and LNYV-R (or CMV-F and CMV-R) primers (Table 1). Both PCR amplification consisted of 35 cycles at 94°C for 30 s, 52°C for 45 s and 72°C for 1 min.
Amplified PCR products were purified using the Agarose Gel DNA Fragment Recovery Kit (Takara) and purified fragments were cloned into the pMD18-T vector (Takara). The ligates were transformed into competent cells of the Escherichia coli strain DH5α. Plasmid DNA preparations were obtained using the TaKaRa MiniBEST Agarose Gel DNA Extraction Kit (Takara). The cloned plasmid was denoted either pMD18-LNYV or pMD18-CMV, and the presence of the inserted PCR products was confirmed by means of agarose gel electrophoresis and a sequence assay. Sequencing was performed at the Sangon Biotech (Shanghai) Co., Ltd. (Shanghai, China). Sequences were analyzed using the Basic Local Alignment Search Tool (BLAST) for virus identification within species.
LAMP and its optimization
The LAMP reaction was conducted in 12.5 μl reaction volumes that contained 0.5 μl of the cDNA products (50 ng), 1.25 μl of a 10×ThermpoPol buffer (New England BioLabs, Ipswich, MA, USA), a final concentration of 6 mM MgSO4, 0.8 M betaine (Sigma-Aldrich, St. Louis, MO, USA), 1.4 mM dNTPs, 2 μM each of the external primers (F3 and B3), 1.6 μM each of the internal primers (FIP and BIP), 0.4 μM each of the loop primers and 4 U of Bst DNA polymerase (New England BioLabs). Betaine has typically been used as an additive for isothermal nucleic acid amplification reactions because it lowers the melting temperature (Tm) of DNA; however, there is some doubt as to its efficacy for all LAMP reaction systems (Ma et al., 2017). Following these steps, we determined the optimum incubation temperature, duration and concentration of MgSO4 and betaine in each primer set of LNYV and CMV used for the RT-LAMP assays.
To determine the optimum temperature, the reaction was conducted for 100 min at seven different temperatures: 56, 58, 60, 62, 65, 68, and 70°C followed by 80°C for 10 min in a thermal cycler (Bio-Rad, Hercules, CA, USA). To determine the optimum duration (time), the reaction was conducted at 58°C and 60°C under five different time periods (20, 40, 60, 80, and 100 min) followed by 80°C for 10 min for LNYV and CMV detection, respectively.
To determine the optimum MgSO4 concentration, the reaction was conducted at 58°C and 60°C for 60 min at eight different MgSO4 concentrations: 0, 2, 4, 6, 8, 10, 12, and 14 mM for LNYV and CMV detection, respectively. To determine the optimum betaine concentration, the reaction was conducted at 58°C and 60°C for 60 min at seven different betaine concentrations: 0, 0.4, 0.8, 1.2, 1.6, 2.0, and 2.4 M for LNYV and CMV detection, respectively.
LAMP amplification products (5 μl) were analyzed and evaluated by direct visual inspection of the reaction tube after the addition of SYBR Green I (Invitrogen, Carlsbad, CA, USA) (1:1,000 TE; v/v). In addition, LAMP amplification products (5 μl) were routinely detected using agarose gel electrophoresis (2.0% agarose; TAE).
RT-PCR
The primer pair designated LNYV-F and LNYV-R (or CMV-F and CMV-R) was based on the conserved region of the LNYV N gene (or the CMV CP gene) and used for RT-PCR analysis. Primer pair sequences are provided in Table 1. As described above, total RNA was extracted from lettuce plants that tested positive for LNYV or CMV using the RNAprep Pure Kit (For Plant). Subsequently, RT was performed to synthesize the first strand of cDNA according to the procedures described above, while PCR was conducted in 12.5 μl reaction volumes according to the procedures described above. A 5.0 μl volume of the PCR product was verified by electrophoresis on a 2.0% agarose gel and were of the expected sizes (i.e., 614 and 587 bp for LNYV and CMV, respectively).
Analysis of RT-LAMP specificity
Total RNA extracts from lily and lettuce leaves infected with LSV, the CMV lily isolate, LMoV, ArMV, LNYV, and the CMV lettuce isolate were used to evaluate the cross reactivity of the RT-LAMP assay. A 0.5 μl volume of total RNA (100 ng/μl) was added to the reaction mixture. Water and healthy lettuce leaves were used as the blank and negative controls, respectively.
Comparison between RT-LAMP and RT-PCR sensitivities
To compare the sensitivity of RT-LAMP to RT-PCR, the pMD18-LNYV or pMD18-CMV plasmids were serially diluted 10-fold from 3.5 × 105 to 3.5 × 10−4 ng/ml or from 2.0 × 105 to 2.0 × 10−6 ng/ml, respectively. A 0.5 μl volume of each dilution was added to the LAMP or PCR mixture.
Evaluation of RT-LAMP using field-collected samples
To validate the RT-LAMP assays, 16 leaf samples were collected from lettuces at different growth stages including seedling, rosette or lotus, and heading. Specimens exhibited virus-like symptoms and were collected from seven fields located in two towns, Shichuan and Donggang around Lanzhou City, Gansu Province, China, and tested for LNYV and CMV using RT-LAMP assays. RT-LAMP amplification products (5 μl) were detected using agarose gel electrophoresis (2.0% agarose; TAE) and by direct visual examination with the addition SYBR Green I. Also, for comparison, each leaf sample was tested using RT-PCR according to the above method.
Results
Confirmation of virus identities
Sequences from samples of each virus analyzed using the nucleotide-nucleotide BLAST (Blastn) tool showed that cloned LNYV and CMV fragments shared 98.7% and 98% homology, respectively, to a known LNYV lettuce isolate (GenBank No. KP109949.1) and to a known CMV lettuce isolate (GenBank No. AJ810253). The sequence of cloned LNYV fragment was submitted to GenBank at the National Center for Biotechnology Information (NCBI) under accession number (MK850384).
Optimization of LAMP reactions
The optimum temperatures for detecting LNYV and CMV using RT-LAMP were confirmed through 100 min RT-LAMP reactions at seven different temperatures, between 56°C and 70°C. After a 100 min incubation period, reaction products were analyzed by electrophoresis. No amplification was seen at 56°C; whereas amplification was evident at 58, 60, 62, 65, 68, and 70°C however the strongest reaction was observed at 65°C for LNYV detection (Fig. 2A, upper). Similarly for CMV, no amplification was seen at 56 °C and 58°C but was evident at 60, 62, 65, 68, and 70°C but with no obvious difference (Fig. 2A, lower). Therefore, the optimum temperature was selected as 65°C for LNYV and 60°C for CMV. To determine the minimum reaction time of LNYV and CMV detection, LAMP was conducted at 65°C and 60°C for 20, 40, 60, 80, and 100 min. Typical ladder-like patterns were clearly observed at 60, 80, and 100 min; however, no amplification was observed at 20 and 40 min for either virus (Fig. 2B). Consequently, the minimum reaction time was determined as 60 min for both viruses.
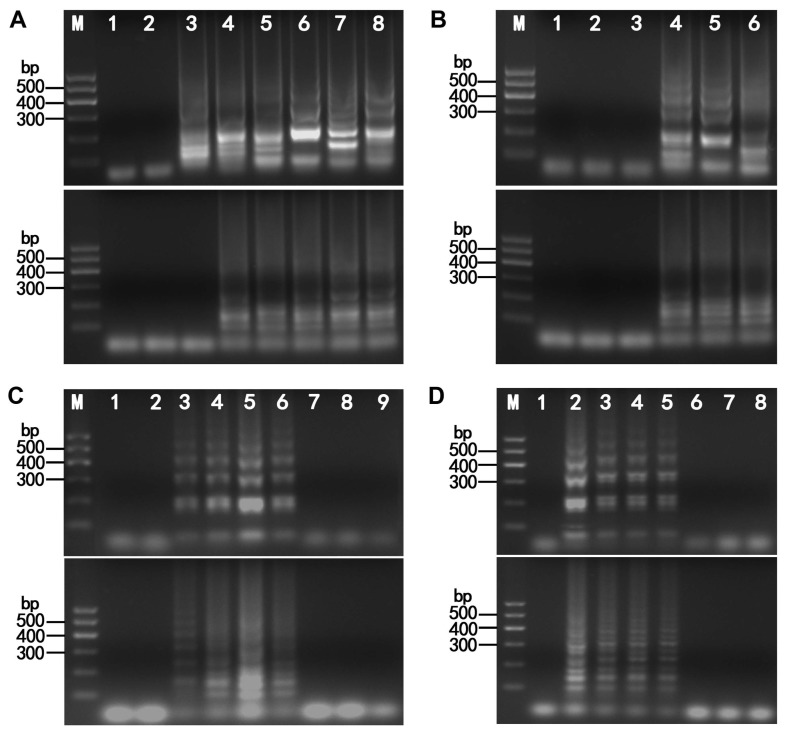
Agarose gel electrophoresis showing the effect of amplification temperatures (A), reaction durations (B), different concentrations of MgSO4 (C) and of betaine (D) on the detection of lettuce necrotic yellows virus (LNYV) and cucumber mosaic virus (CMV) using reverse transcription loop-mediated isothermal amplification (RT-LAMP). (A) Temperature. Lanes loaded with LNYV (upper) and CMV (lower) products of RT-LAMP assays conducted at different temperatures. Lane M: DL600 marker; lane 1: negative control; lanes 2–8: 56, 58, 60, 62, 65, 68, and 70°C. (B) Time duration. Lanes loaded with LNYV (upper) and CMV (lower) products of RT-LAMP assays conducted over different durations. Lane M: DL600 marker; lane 1: negative control; lanes 2–6: 20, 40, 60, 80, and 100 min. (C) MgSO4. Lanes loaded with LNYV (upper) and CMV (lower) products of RT-LAMP assays conducted using different concentrations of MgSO4. Lane M: DL600 marker; lane 1: water control; lanes 2–9: 0, 2, 4, 6, 8, 10, 12, and 14 mM. (D) Betaine. Lanes loaded with LNYV (upper) and CMV (lower) products of RT-LAMP assays conducted using different concentrations of betaine. Lane M: DL600 marker; lane 1: water control; lanes 2–8: 0, 0.4, 0.8, 1.2, 1.6, 2.0, and 2.4 M.
The optimum MgSO4 concentration in our experiment was 6 mM for both viruses (Fig. 2C). The optimum betaine concentration for LAMP detection of LNYV and CMV was determined by testing a range of betaine concentrations (i.e., from 0 to 2.4 M). Typical ladder-like patterns were clearly observed by agarose gel electrophoresis without betaine. Ladder-like patterns were observed at 0.4, 0.8, and 1.2 M betaine; however, no amplification was observed at 1.6, 2.0, and 2.4 M betaine for either virus (Fig. 2D). Consequently, we determined that the optimal conditions for our subsequent RT-LAMP analyses were 65°C and 60°C for 60 min using 6 mM MgSO4 and no betaine for LNYV and CMV detection, respectively.
Specificity of RT-LAMP assays for LNYV and CMV detection
To determine the specificity of the RT-LAMP assays, we tested them using all RNA extracts from lily leaves infected with LSV, CMV from lily, LMoV, and ArMV as references, as well as lettuce leaves infected with LNYV and CMV. Following the application of RT-LAMP, only the product of the LNYV-infected lettuce sample displayed a typical ladder-like pattern using agarose gel electrophoresis. No bands were observed in healthy lettuce sample, or in the samples infected with LSV, the CMV lily isolate, LMoV, ArMV or the CMV lettuce isolate (Fig. 3A, left). Similarly, only the product of the CMV-infected lettuce sample displayed a typical ladder-like pattern, with no bands observed in healthy lettuce sample, or in the samples infected with LSV, the CMV lily isolate, LMoV, ArMV or LNYV (Fig. 3B, left).

Specificity of reverse transcription loop-mediated isothermal amplification (RT-LAMP) for lettuce necrotic yellows virus (LNYV) (A) and cucumber mosaic virus (CMV) (B). Amplified products from RT-LAMP were visualized by agarose gel electrophoresis (left) and by the naked eye with the addition of SYBR Green I (right). (A) Lane M: DL600 marker; lane 1: negative control; lane 2: healthy lettuce; lane 3: lily symptomless virus (LSV)-infected lily; lane 4: CMV-infected lily; lane 5: lily mottle virus (LMoV)-infected lily; lane 6: arabis mosaic virus (ArMV)-infected lily; lane 7: CMV-infected lettuce; lane 8: LNYV-infected lettuce. (B) Lane M: DL600 marker; lane 1: negative control; lane 2: healthy lettuce; lane 3: LSV-infected lily; lane 4: CMV-infected lily; lane 5: LMoV-infected lily; lane 6: ArMV-infected lily; lane 7: LNYV-infected lettuce; lane 8: CMV-infected lettuce.
The reaction products of RT-LAMP were further confirmed using direct observation under green fluorescence by adding SYBR Green I. Only the products of LNYV- and CMV-infected lettuce samples showed green fluorescence (Fig. 3A, right and 3B, right).
Sensitivity of RT-LAMP versus RT-PCR
To compare the sensitivity between RT-LAMP and RT-PCR, 10-fold serial dilutions of pMD18-LNYV or pMD18-CMV plasmids were used as templates. The detection limits for LNYV were determined to be 3.5 × 10−3 ng/ml, namely, 3.5 pg/ml using RT-LAMP (Fig. 4A, upper) and 3.5 × 10−1 ng/mL using RT-PCR (Fig. 4A, lower). The detection limits for CMV were determined to be 2.0 × 10−5 ng/ml, namely, 20 fg/ml using RT-LAMP (Fig. 4B, upper) and 2.0 × 10−3 ng/ml using RT-PCR (Fig. 4B, lower). Thus, the RT-LAMP assays were more sensitive than the RT-PCR assays by a factor of 100 for both viruses. In addition, the reaction products of RT-LAMP were also directly observed using SYBR Green I fluorescence. Both methods yielded similar detection limit results for both viruses (Fig. 4A, middle and 4B, middle).
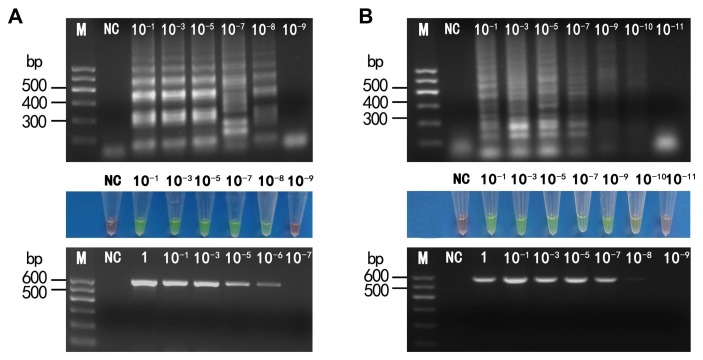
Comparison of the sensitivity of detection of lettuce necrotic yellows virus (LNYV) (A) and cucumber mosaic virus (CMV) (B) by reverse transcription loop-mediated isothermal amplification (RT-LAMP) and reverse transcription polymerase chain reaction (RT-PCR). The LNYV or CMV plasmid was diluted 10-fold from the initial concentration (3.5 × 105 ng/ml or 2.0 × 105 ng/ml) respectively and used as a template to assess RT-LAMP and RT-PCR sensitivity for each virus. (A) LNYV. Amplified products from RT-LAMP were visualized by agarose gel electrophoresis (upper) and by the naked eye with the addition of SYBR Green I (middle). Lane M: DL600 marker; NC: negative control; lanes 3–8: LNYV plasmid dilutions; amplified products from RT-PCR were visualized by agarose gel electrophoresis (lower). (B) CMV. Amplified products from RT-LAMP were visualized by agarose gel electrophoresis (upper) and by the naked eye with the addition of SYBR Green I (middle). Lane M: DL600 marker; NC: negative control; lanes 3–9: CMV plasmid dilutions; amplified products from RT-PCR were visualized by agarose gel electrophoresis (lower).
Evaluation of RT-LAMP using field-collected samples
We analyzed 16 field samples using the RT-LAMP and RT-PCR assays. Of the 16 samples, six were positive for LNYV, which were able to be detected using both RT-LAMP and RT-PCR (Fig. 5A, upper and middle). For CMV, 10 out of the 16 tested positive using RT-LAMP (Fig. 5B, upper) whereas only eight were positive using RT-PCR (Fig. 5B, middle). We observed clearer ladder-like patterns in lanes 4, 5, 7, 8, and 9 using the RT-LAMP (Fig. 5B, upper), whereas the products of RT-PCR in lanes 4, 5, 7, 8, and 9 were weak (Fig. 5B, middle). Five samples were negative for both LNYV and CMV.

By using reverse transcription loop-mediated isothermal amplification (RT-LAMP) and reverse transcription polymerase chain reaction (RT-PCR), lettuce necrotic yellows virus (LNYV) (A) and cucumber mosaic virus (CMV) (B) were detected in field-collected samples from the Lanzhou City district. Amplification products using RT-LAMP (upper), RT-PCR (middle) were visualized by agarose gel electrophoresis and by the naked eye with the addition of SYBR Green I (lower). Lane M: DL600 marker; lane 1: negative control; lanes 2–17: lettuce leaf samples.
Products from both methods analyzed using SYBR Green I fluorescence and agarose gel electrophoresis yielded similar results for LNYV and CMV detection to those above (Fig. 5A, lower and 5B, lower).
Discussion
Although different molecular methods have been developed for LNYV and CMV detection, many require technical expertise and specialized equipment (Callaghan and Dietzgen, 2005; Deyong et al., 2005). In this study, we successfully developed a simple, sensitive and reliable method to detect both LNYV and CMV using RT-LAMP assays that can be performed at temperatures between 58–70°C and 60–70°C at approximately 60 min for LNYV and CMV respectively. We found that the RT-LAMP and RT-PCR methods yielded similar results using field-collected samples for LNYV detection (Fig. 5A). When RT-LAMP and RT-PCR analyses were used to detect CMV in field-collected samples, the reaction products were invisible in lanes 2 and 10 using RT-PCR (Fig. 5B, middle) and were therefore considered CMV-negative. However, their amplicons did show clear ladder-like patterns using the RT-LAMP method (Fig. 5B, upper). In addition, the RT-PCR products of five samples in lanes 4, 5, 7, 8, and 9 also showed weak bands (Fig. 5B, middle), but again their amplicons showed clear ladder-like patterns using RT-LAMP (Fig. 5B, upper). Because the field lettuce samples were collected at different growth stages including early seedling, rosette or lotus and heading there may well have been differences in virus concentration. We therefore speculate that the virus titer of CMV in samples 2, 4, 5, 7–10 might be relatively low and not so easily detected using RT-PCR. Further survey work needs to be undertaken to verify this conclusion. We believe these preliminary results demonstrate that our RT-LAMP assay is a reliable method with greater sensitivity than RT-PCR. The negative result in our assays of specimens 3, 6, 14, 16, 17 might be explained either by the presence of other viruses not tested in these experiments (e.g., luteovirus) or a nutrient imbalance caused by low nitrogen or iron concentrations.
We believe the key step for the success of the LAMP method is the design of the primers (Hardinge et al., 2018; Zhao et al., 2018). In this study, we designed two external and two internal primer pairs for LNYV and CMV detection. In addition, a pair of loop primers for LNYV and a single loop primer for CMV detection, were designed, which are capable of recognizing eight sequences of the N gene of LNYV and seven sequences of the CP gene of CMV. Our results indicated that the combination of these primers contributed to the high detection sensitivity. We found the sensitivities of both RT-LAMP assays were greater by a factor of 100 compared to the conventional RT-PCR assays (Fig. 4). With respect to the sensitivity of LAMP-based assays, our results for LNYV and CMV were consistent with those obtained by other researchers (Bhat et al., 2013; Liu et al., 2010; Zhao et al., 2018).
Standardization of the LAMP reaction mixture (especially the concentration of MgSO4 and betaine) as well as the optimum temperature and duration are also important for successful pathogen detection (Bhat et al., 2013). Our experiments served to standardize all parameters used for LNYV and CMV detection. Of particular interest was finding that the betaine-free treatment was far superior to that of the widely used addition of 0.8 M betaine (Fig. 2D). This is consistent with results from Ma et al. (2017). An explanation for this might be that betaine inhibited the reaction efficiency of isothermal amplification reactions as found by Ma et al. (2017).
The optimum temperature for the LAMP reaction was initially achieved using a thermal cycler to maintain a constant reaction temperature. Subsequently, two further options were evaluated to ascertain their suitability for this purpose: a water bath and a heat block. We found that all three methods used to incubate RT-LAMP reactions yielded similar amplification results (data not shown), which confirmed that any device that can accurately maintain a set temperature is adequate for RT-LAMP amplification.
The products of RT-LAMP were routinely confirmed using agarose gel electrophoresis. Similarly, to other studies (Bhat et al., 2013; He et al., 2016), our experiments were also able to confirm that RT-LAMP products could be detected through a simple visual examination under green fluorescence using SYBR Green I. Both visual methods (gel electrophoresis and green fluorescence) yielded similar results in specificity and detection limits as well as with field-collected samples (Figs. 3–5). From our results, we believe green fluorescence does appear to be a suitable examination method for conducting RT-LAMP experiments in laboratories with limited facilities. However, further studies are required to develop a more practical and rapid field-based LAMP procedure, such as IC-RT-LAMP for lettuce crops.
The ability to detect a broad range of isolates is usually desirable for a general diagnostic assay; however, our RT-LAMP assay for CMV did not react with the CMV lily isolate (Fig. 3). In a reciprocal assay conducted using a LAMP assay developed specifically for the CMV lily isolate (Zhang, unpublished data) we found no reactivity with the lettuce samples infected with CMV (data not shown). This difference may be related to viral evolution; namely, the CMV lily isolate may have evolved into a new host subgroup within the CMV subgroup I, becoming more closely adapted to lily plants while losing their capacity to infect other hosts (Liu et al., 2004; Masuta et al., 2002). Subsequent research confirmed that the host range of the CMV lily isolate was indeed narrow. While the CMV lily isolate could infect cucumbers, Datura stramonium and Phaseolus vulgaris, it could not infect normal indicator hosts of CMV include Nicotiana tabacum, Nicotiana glutinosa, and Chenopodium amaranticolor (Liang et al., 2008). A comparison between sequence data further confirmed that the sequence identity of CP sequences at a nucleotide level were only 77.7% between the CMV lettuce isolate (Gen-Bank No. AJ810253) and the CMV lily isolate (GenBank No. DQ 767971). Further investigation is needed to explore how limited the host range is for the CMV lily isolate, particularly its capacity to infect lettuce and other plant species.
This study further confirmed CMV infection in lettuce crops in Lanzhou City and is the first to confirm the presence of LNYV in lettuce in China. Even with our limited collection of field samples, we were able to detect a proportion of viral infections among the 16 samples, with 6 (37.5%) and 10 (62.5%) testing positive for LNYV and CMV, respectively, using our RT-LAMP assays. In addition, we found LNYV + CMV mixed infections in 3 (18.8%) of plants tested (Fig. 5). Similarly we also found LNYV + CMV + LMV mixed infections in 2 (12.5%) of plants tested (Zhang, unpublished data). From our primary field survey of lettuce conducted in 2018, and from discussions with growers, we determined that CMV and LMV infections appear to have contributed to a reduction in yield in Lanzhou City lettuce crops. LNYV also appeared to contribute to yield losses in these crops. There is concern that once established within the main areas of production LNYV, alone or in combination CMV and LMV may cause serious economic losses to the lettuce industry in China. Consequently, it is important for lettuce growers in Gansu Province and other lettuce growing regions to consider adopting improved practical measures for virus control. The low cost, convenient and simple RT-LAMP method, being high in sensitivity and specificity, which we successfully developed for the detection of LNYV and CMV and reported here may offer growers an effective tool to maintain the production of healthy seedlings, identify sources of virus infection in alternative hosts and facilitate the timely application of insecticides for vector control.
Acknowledgments
This study was supported by the Key Technology Talent Program of the Chinese Academy of Sciences (CAS) (Grant No. 2016-65), the Lanzhou Talent Innovation and Entrepreneurship Project (Grant No. 2019-HLJC-9), the Lanzhou Chengguan District Science and Technology Project (Grant No. 2019-6-2), the Science and Technology Service Network Initiative of CAS (KFJ-STS-QYZD-120), and the Northwest Institute of Eco-Environment and Resources, CAS (Y855Z11001). We would like to thank The New Zealand Institute for Plant and Food Research, Lincoln, New Zealand, for hosting Dr Yubao Zhang as a research scholar working with John D Fletcher during 2017-2018. The authors have no conflict of interest to declare.

