Molecular Characterization of Five Potyviruses Infecting Korean Sweet Potatoes Based on Analyses of Complete Genome Sequences
Article information
Abstract
Sweet potatoes (Ipomea batatas L.) are grown extensively, in tropical and temperate regions, and are important food crops worldwide. In Korea, potyviruses, including Sweet potato feathery mottle virus (SPFMV), Sweet potato virus C (SPVC), Sweet potato virus G (SPVG), Sweet potato virus 2 (SPV2), and Sweet potato latent virus (SPLV), have been detected in sweet potato fields at a high (~95%) incidence. In the present work, complete genome sequences of 18 isolates, representing the five potyviruses mentioned above, were compared with previously reported genome sequences. The complete genomes consisted of 10,081 to 10,830 nucleotides, excluding the poly-A tails. Their genomic organizations were typical of the Potyvirus genus, including one target open reading frame coding for a putative polyprotein. Based on phylogenetic analyses and sequence comparisons, the Korean SPFMV isolates belonged to the strains RC and O with >98% nucleotide sequence identity. Korean SPVC isolates had 99% identity to the Japanese isolate SPVC-Bungo and 70% identity to the SPFMV isolates. The Korean SPVG isolates showed 99% identity to the three previously reported SPVG isolates. Korean SPV2 isolates had 97% identity to the SPV2 GWB-2 isolate from the USA. Korean SPLV isolates had a relatively low (88%) nucleotide sequence identity with the Taiwanese SPLV-TW isolates, and they were phylogenetically distantly related to SPFMV isolates. Recombination analysis revealed that possible recombination events occurred in the P1, HC-Pro and NIa-NIb regions of SPFMV and SPLV isolates and these regions were identified as hotspots for recombination in the sweet potato potyviruses.
Sweet potato (Ipomoea batatas) is grown extensively throughout tropical and temperate regions worldwide and is the third most important root crop after potato and cassava. According to FAOSTAT data, it has been cultivated in 8.2 million ha worldwide with the production of 103 million metric tons in 2013. Almost 80% of all sweet potatoes are produced in Asia, particularly in China (~70%). Korea currently produces approximately 330,000 metric tons from 22,200 ha, and its production is increasing every year (FAOSTAT, 2013).
Viral diseases of sweet potato have become widespread, causing serious crop losses around the world. To date, more than 30 viruses have been identified to infect sweet potato (Brunt et al., 1996; Clark et al., 2012). The most severe disease in sweet potato is caused by coinfection with the whitefly-transmitted Sweet potato chlorotic stunt virus (SPCSV) and the aphid-transmitted Sweet potato feathery mottle virus (SPFMV), which results in the synergistic sweet potato virus disease (SPVD) causing yield losses of 70–100% (Gibson et al., 1998; Karyeija et al., 2000; Mukasa et al., 2006; Untiveros et al., 2008). Synergism has also been observed between SPCSV and the possibly whitefly-transmitted Sweet potato mild mottle virus (SPMMV) (Gutiérrez et al., 2003; Hahn, 1979). SPCSV caused synergistic diseases in sweet potato with many other sweet potato viruses (Untiveros et al., 2007).
In Korea, the following eight viruses has been reported to infect sweet potatoes; Sweet potato feathery mottle virus (SPFMV), Sweet potato virus C (SPVC), Sweet potato virus G (SPVG), Sweet potato virus 2 (SPV2), Sweet potato latent virus (SPLV), Sweet potato chlorotic fleck virus (SPCFV), Sweet potato leaf curl virus (SPLCV), and Sweet potato symptomless virus 1 (SPSMV-1) (Kwak et al., 2006; 2014). Among these eight viruses, five viruses (SPFMV, SPVC, SPVG, SPV2, and SPLV) are members of the genus Potyvirus, in the family Potyviridae (Adams et al., 2011; Clark et al., 2012). They had a high incidence of 96% on sweet potatoes collected nationwide in 2012, and SPFMV and SPVC were especially prevalent in Korea (Kwak et al., 2014).
SPFMV can be divided into three representative strains: Russet Crack (RC), Ordinary (O), and East Africa (EA; Abad et al., 1992; Kreuze et al., 2000; Kwak et al., 2007). Because SPFMV strain C has a relatively low homology with the other SPFMV strains, it was reclassified as a new species, SPVC, by the International Committee on Taxonomy of Viruses in 2010 (Untiveros et al., 2010). A previous study showed that Korean SPFMV isolates are similar to the RC and O strain isolates, but they are not similar to EA-like isolates (Kwak et al., 2007). The EA strain has been detected in East Africa as a distinct group (Kreuze et al., 2000; Mukasa et al., 2003). It has also been found in Peru (Untiveros et al., 2008), Vietnam (Ha et al., 2008), Easter Island (Rännäli et al., 2009) and, more recently, in China (Qin et al., 2013). The other potyviruses infecting sweet potato (e.g., SPVG, SPLV, and SPV2) have not been studied extensively, but their complete genome sequences were recently reported and compared to SPFMV (Ateka et al., 2007; Li et al., 2012; Rodriguez Pardina et al., 2012; Wang et al., 2013).
Members of the Potyviridae family are positive-sense single stranded RNA viruses, with a genome of approximately 10 kb. They are characterized by a 5′ untranslated region (UTR), a large open reading fame (ORF), and a 3′ UTR. The ORF consists of 10 functional proteins P1 (protein 1), HC-Pro (helper component proteinase), P3 (protein 3), 6K1 (6K protein 1), CI (cylindrical inclusion protein), 6K2 (6K protein 2), VPg (viral protein genome-linked), NIa-Pro (nuclear inclusion protein a-proteinase), NIb (nuclear inclusion protein b), and CP (coat protein) (Riechmann et al., 1992).
Studying genetic structure and diversity of viruses is important to understand the molecular evolutionary histories in relation to their virulence, dispersion, and emergence of epidemics. Sweet potato potyviruses are endemic in most sweet potato fields in Korea. Furthermore, co-infection of sweet potato potyviruses may result in the emergence of new viral strains as a result of genetic recombination, an important evolutionary force that has contributed to the divergence of several positive-sense RNA viruses, including potyviruses (Chare and Holmes, 2006; Lai, 1992). In the present study, we determined the complete genome sequences of eighteen isolates of five sweet potato potyviruses that were collected from sweet potato samples in Korea. We analyzed the genetic structure and variability of Korean isolates of sweet potato potyviruses to understand the evolutionary relationships among the isolates.
Materials and Methods
Isolate collection
To determine the full-length genome sequences, the following 18 potyvirus samples were obtained from 154 sweet potato samples collected in 10 regions of five Korean provinces in 2012 (Kwak et al., 2014): three samples (IS90, CW137 and GJ122) for SPFMV, three samples (HN52, UN202, CW135) for SPVC, three samples (SC11, IS103, HG167) for SPVG, five samples (SC6, SCN20, HN77, GJ118 and CW142) for SPV2, and four samples (SC5, HN76, HG181 and UN202) for SPLV (Fig. 1).
Reverse-transcription polymerase chain reaction (RT-PCR), cloning and sequencing
Total RNA was extracted from the infected sweet potato leaf, petiole and stem samples, using a Viral Gene-spin™ Viral DNA/RNA Extraction kit (Intron, Korea), according to the manufacturer’s instructions. RT-PCR was performed as a two-step procedure, which included RT, using AMV reverse transcriptase (Promega, USA), and PCR, using LA taq polymerase (TAKARA, Japan). Three or four large fragments covering the full-length genomes for sweet potato potyviruses were amplified using virus-specific primer pairs. Adjacent regions of these PCR fragments were overlapped by about 200 bp to ensure that they were from the same genome. End sequences of each RNA segment were obtained according to the 5′ and 3′ rapid amplification of cDNA ends (RACE) protocol (BM, Germany). Data on the composition of the primers set used in full-length genome sequencing are available on the supplementary file. RT-PCR amplified DNA fragments were purified using the MEGA quick-spin™ Kit (Intron, Korea) and cloned into the pGEM-T easy vector (Promega, USA), according to the manufacturer’s instructions, followed by transformation into the Escherichia coli strain DH5α. At least three clones of each fragment were completely sequenced. To verify sequence discords among the clones and to identify the dominant sequences in the viral population, we performed direct sequencing of RT-PCR products of entire genomes. Sequencing was performed by Genotech (Korea). The sequences were then assembled, using DNA Star v. 5.02, and they were then submitted to the GenBank database with the accession numbers listed in Table 1.
Sequence comparison and phylogenetic analyses
The complete nucleotide sequences and the deduced amino acid sequences were aligned using the Geneious and ClustalW methods in Geneious pro 8 and compared with those of the previously reported virus isolates SPFMV (RC, O, 10-O and Piu3), SPVC (Bungo, C1 and IL), SPVG (GWB-G, Jesus Maria and Z01001), SPV2 (GWB-2), and SPLV (TW) in Table 1. The ipomovirus SPMMV (Accession no. Z73124) was used as an outgroup. The SPMMV infecting sweet potato belongs to the genus Ipomovirus in the family Potyviridae and its complete genome sequence has the similar genetic structure with SPFMV-groups. Genius Pro 8 software was used to calculate the percentages of nucleotide and amino acid identities. Phylogenetic analyses were performed according to the maximum-likelihood method implemented in MEGA 6 (Tamura et al., 2013). Maximum likelihood phylogenetic trees were constructed using best fit nucleotide substitution models (GTR+G for full-length genome, GKY+G for 3′ UTR, and T92 for 5′ UTR) and best fit amino acid substitution models (JTT+G for P1 and VPg, JTT+G+I for CP, LG+G for HC-Pro, P3, 6K1, and 6K2, and LG+G+I for NIa-Pro and NIb). Bootstrap values were calculated using 1,000 random replication. All positions containing gaps and missing data were eliminated.
Recombination analysis
Recombinations between 30 sweet potato potyviruses and one ipomovirus SPMMV were analyzed using the RDP, GENECONV, BootScan, MaxChi, Chimaera, SiScan and 3Seq methods, implemented in the RDP4 software (Recombination Detection Program, ver. 4) with default settings and a Bonferroni corrected p-value cut-off of 0.01. To investigate the possibility of recombination, all recombination events identified by at least one of the methods were selected.
Results
Genome characterization of Korean sweet potato potyviruses
The full-length genome sequences of 18 isolates of five sweet potato-infecting potyviruses collected in 2012 were obtained (Table 1). The disease symptoms included chlorosis, purpling, chlorotic ring spot, vein banding, mottle, puckering patterns on the infected leaves (Fig. 2). The complete genomes ranged from 10810 to 10819 nt for SPFMV, 10829 to 10830 nt for SPVC, 10800 nt for SPVG, 10730 to 10732 nt for SPV2, and 10081 nt for SPLV, excluding the poly-A tails. Their genomic organizations were typical of members of the Potyvirus genus. They putatively encoded a single large polyprotein of 3,490 or 3,493 aa for SPFMV, 3,481 aa for SPVC, 3,488 aa for SPVG, 3,466 aa for SPV2, and 3,247 aa for SPLV. This polyprotein potentially composed of 10 proteins (P1, HC-Pro, P3, 6K1, CI, 6K2, VPg, NIa-Pro, NIb and CP; Fig. 3).

Representative virus symptoms in sweet potato. (A) mottle and puckering; (B) Chlorosis and vein banding; (C) chlorotic ring spot; (D) chlorotic local lesion and purpling in a cultivated field.

Genome organization of the SPFMV group of potyviruses. P1; protein 1, HC-Pro; helper component proteinase, P3; protein 3, 6K1 (6K protein 1), CI;cylindrical inclusion protein, 6K2 (6K protein 2), VPg; viral protein genome-linked, NIa-Pro; nuclear inclusion protein a-proteinase, NIb; nuclear inclusion protein b, CP; coat protein, PIPO; Pretty Interesting Potyviridae ORF, and PISPO; Pretty Interesting Sweetpotato Potyvirus.
The P1 protein of SPFMV Korean isolates consisted of 664 aa, which was similar to the SPFMV-S and SPFMV-10-O isolates and smaller than SPFMV-O (689 aa) and SPFMV-Piu3 (724 aa). P1 proteins of the isolates of SPVC, SPVG and SPV2 consisted of 654, 618 and 618 aa, respectively. For SPLV, all the isolates had a P1 protein of 442 aa, lacking the P1-N terminal part of the sweet potato potyviruses. The sizes of HC-Pro, P3, CI, 6K2 and NIa-Pro were 458, 352, 643, 53, and 243 aa, respectively, which were identical in all isolates. The size of the 6K1 protein was 52 aa, with the exception of the SPVG isolates (53 aa). The sizes of the VPg proteins were variable; 191 aa for SPFMV-IS90, 192 aa for the remaining SPFMV isolates, SPVC, and SPVG, 193 aa for SPLV and 194 aa for SPV2 isolates. The size of NIb was 521 aa, with the exception of SPLV (518 aa). The size of the CP protein was highly variable, from 293 to 355 aa. SPVG isolates had the longest CP protein (355 aa), followed by SPV2 (332 aa), SPFMV (315 aa), SPVC (313 aa), and SPLV (293 aa.) The 5′ UTR was 113–162 nt long, and the 3′ UTR was 194–222 nt long.
In addition to large polyprotein-encoding proteins, two small ORFs (+2 frame relative to polyprotein) were identified. A small Pretty Interesting Potyviridae ORF (PIPO), which is conserved throughout the Potyviridae family (Chung et al., 2008), was identified in the P3 protein of all the Korean sweet potato potyvirus isolates. The typical size of PIPO for SPFMV, SPVC, SPVG, SPV2, and SPLV was confirmed as 70, 70 (except 76 aa for SPVC-Bungo), 76, 80, and 83 (except 84 aa for SPLV-UN202, 79 aa for SPLV-TW) aa, respectively. Another ORF, Pretty Interesting Sweetpotato Potyvirus ORF (PISPO), has been detected in P1 proteins of potyviruses infecting sweet potato (Clark et al., 2012). PISPO was identified in the isolates of four viruses, the exception being SPLV. The typical sizes of PISPO for SPFMV, SPVC, SPVG, and SPV2 were confirmed as 249, 240, 230, and 231 aa, respectively. In the 5′ terminal region of PISPO and PIPO, a highly conserved motif, G1-2A6, which is known to be involved in ribosomal frameshift (Chung et al., 2008), was found in all isolates. The full-length genome sequences are available in the GenBank database, with the accession numbers listed in Table 1.
Pairwise sequence comparison and phylogenetic analysis
The complete nucleotide and deduced amino acid sequences of 18 sweet potato potyvirus isolates were compared to those of previously reported virus isolates. Phylogenetic analysis and pairwise sequence comparisons, based on the complete nucleotide sequences (Figs. 4, 5), indicated that Korean SPFMV isolates belonged to the RC and O strains. The Korean SPFMV isolates CW137 and IS90 had 98–99% nucleotide (nt) sequence identity with the RC strain, and the SPFMV-GJ122 isolate had 96–98% nt sequence identity with the O strain. Korean SPVC isolates had higher (99%) nt sequence identity with the Japanese isolate Bungo than with the Peruvian isolate C1. Korean SPVC isolates had 70–71% nt sequence identity with SPFMV. Korean SPVG isolates had 99% nt sequence identity with the three previously reported SPVG isolates. Korean SPV2 isolates had 97–98% nt sequence identity with the SPV2 GWB-2 isolate from the USA, and were more similar to SPVG than any other sweet potato virus. For SPLV, Korean isolates showed 88–89% nt sequence identity with the Taiwanese isolate TW. The nt sequence identities between the Korean SPLV isolates were 92–98%.
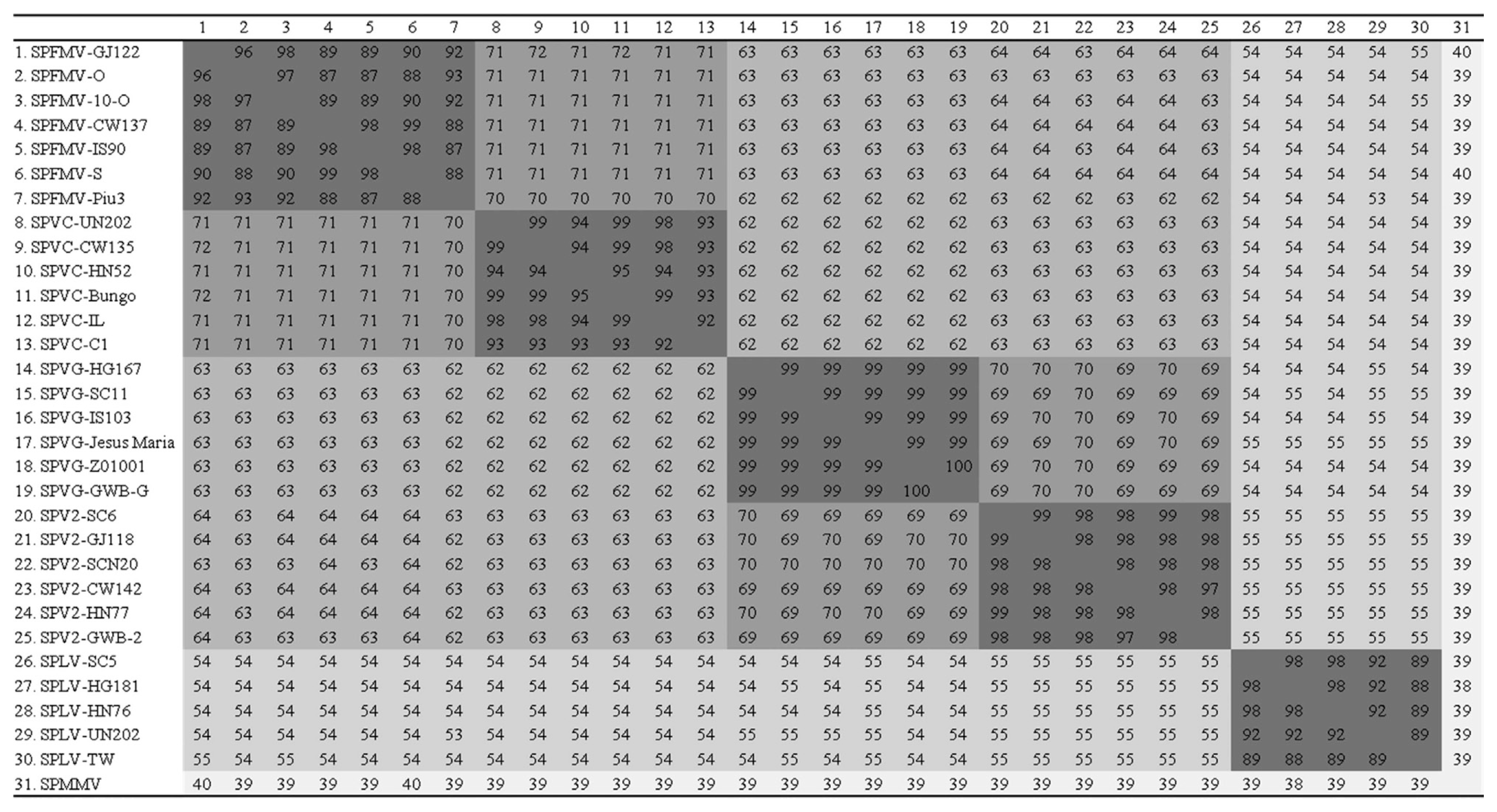
Pairwise sequence comparison of complete genome nucleotide sequences (% identity). Sweet potato feathery mottle virus (SPFMV), Sweet potato virus C (SPVC), Sweet potato virus G (SPVG), Sweet potato virus 2 (SPV2), Sweet potato latent virus (SPLV).

Phylogenetic trees reconstructed using the complete nucleotide sequences of the sweet potato potyvirus isolates. Phylogenetic trees were reconstructed using the maximum-likelihood method in MEGA 6. Numbers at each node indicate bootstrap percentages based on 1,000 replicates in maximum-likelihood tree. Horizontal branch length is drawn to scale with the bar indicating 2 nt replacements per site. SPMMV(Z73124) was used as the outgroup. Sweet potato feathery mottle virus (SPFMV), Sweet potato virus C (SPVC), Sweet potato virus G (SPVG), Sweet potato virus 2 (SPV2), Sweet potato latent virus (SPLV).
The reconstructed phylogenetic trees, based on amino acid sequences of five mature proteins (HC-Pro, P3, CI, NIa-Pro, and CP), were consistent with the nucleotide sequence-based trees, but the trees of P1, 6K1, 6K2, VPg, and NIb amino acids showed different groupings compared to the nucleotide sequence-based trees (Figs. 6, 7, 8, and 9). Regarding the P1 protein, SPLV was highly divergent from viruses of the SPFMV lineage (SPFMV, SPVC, SPVG and SPV2). The P1 protein of Korean SPLV isolates had about 70% homology with the Taiwanese SPLV isolate.

Phylogenetic trees reconstructed using the amino acid sequences of proteins P1, HC-Pro, and P3. Phylogenetic trees were reconstructed using the maximum-likelihood method in MEGA 6. Numbers at each node indicate bootstrap percentages based on 1,000 replicates in maximum-likelihood tree. Horizontal branch length is drawn to scale with the bar indicating 0.2 nt or 0.5 nt replacements per site. SPMMV(Z73124) was used as the outgroup. Sweet potato feathery mottle virus (SPFMV), Sweet potato virus C (SPVC), Sweet potato virus G (SPVG), Sweet potato virus 2 (SPV2), Sweet potato latent virus (SPLV). Other abbreviations: P1; protein 1, HC-Pro; helper component proteinase, P3; protein 3.

Phylogenetic trees reconstructed using the amino acid sequences of proteins 6K1, CI, and 6K2. Phylogenetic trees were reconstructed using the maximum-likelihood method in MEGA 6. Numbers at each node indicate bootstrap percentages based on 1,000 replicates in maximum-likelihood tree. Horizontal branch length is drawn to scale with the bar indicating 0.2 nt or o.5 nt replacements per site. SPMMV(Z73124) was used as the outgroup. Sweet potato feathery mottle virus (SPFMV), Sweet potato virus C (SPVC), Sweet potato virus G (SPVG), Sweet potato virus 2 (SPV2), Sweet potato latent virus (SPLV). Other abbreviations: 6K1 (6K protein 1), CI;cylindrical inclusion protein, 6K2 (6K protein 2).

Phylogenetic trees reconstructed using the amino acid sequences of proteins VPg, NIa-Pro, and NIb. Phylogenetic trees were reconstructed using the maximum-likelihood method in MEGA 6. Numbers at each node indicate bootstrap percentages based on 1,000 replicates in maximum-likelihood tree. Horizontal branch length is drawn to scale with the bar indicating 0.2 nt or 0.5 nt replacements per site. SPMMV(Z73124) was used as the outgroup. Sweet potato feathery mottle virus (SPFMV), Sweet potato virus C (SPVC), Sweet potato virus G (SPVG), Sweet potato virus 2 (SPV2), Sweet potato latent virus (SPLV). Other abbreviations: VPg; viral protein genome-linked, NIa-Pro; nuclear inclusion protein a-proteinase, NIb; nuclear inclusion protein b.
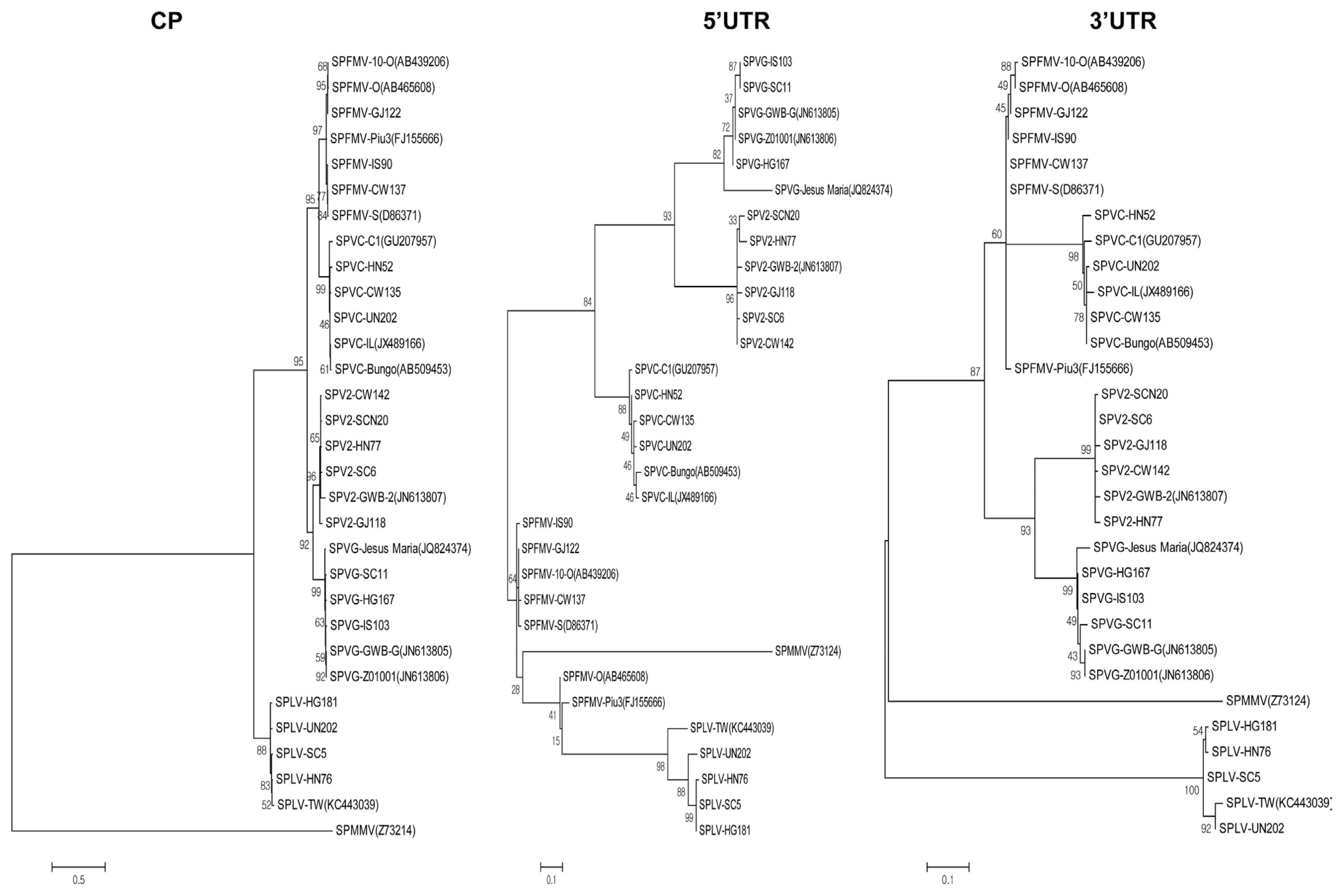
Phylogenetic trees reconstructed using the amino acid sequence of protein CP and nucleotide sequences of the 5′ UTR and 3′ UTR. Phylogenetic trees were reconstructed using the maximum-likelihood method in MEGA 6. Numbers at each node indicate bootstrap percentages based on 1,000 replicates in maximum-likelihood tree. Horizontal branch length is drawn to scale with the bar indicating 0.1 nt or 0.5 nt replacements per site. SPMMV(Z73124) was used as the outgroup. Sweet potato feathery mottle virus (SPFMV), Sweet potato virus C (SPVC), Sweet potato virus G (SPVG), Sweet potato virus 2 (SPV2), Sweet potato latent virus (SPLV). Other abbreviations: CP; coat protein, UTR; untranslated region.
Sequence comparison
The nucleotide and amino acid sequence identities between sweet potato-infecting potyvirus isolates are summarized in Table 2. SPFMV-GJ122 had 98% and 96% nucleotide sequence identities with SPFMV-10-O and SPFMV-O, respectively, which belong to the O strain of SPFMV. SPFMV-GJ122 exhibited 89–90% and 92% nucleotide sequence identities with the RC and EA strains of SPFMV, respectively. SPFMV-GJ122 had 71–72% nucleotide sequence identity with SPVC isolates, 63% with SPVG isolates, 63–64% with SPV2, and 54–55% with SPLV isolates. The comparison of the polyproteins for SPFMV-GJ122 with all other potyvirus isolates showed higher amino acid identity than nucleotide identity, with the exception of SPLV isolates (48%).
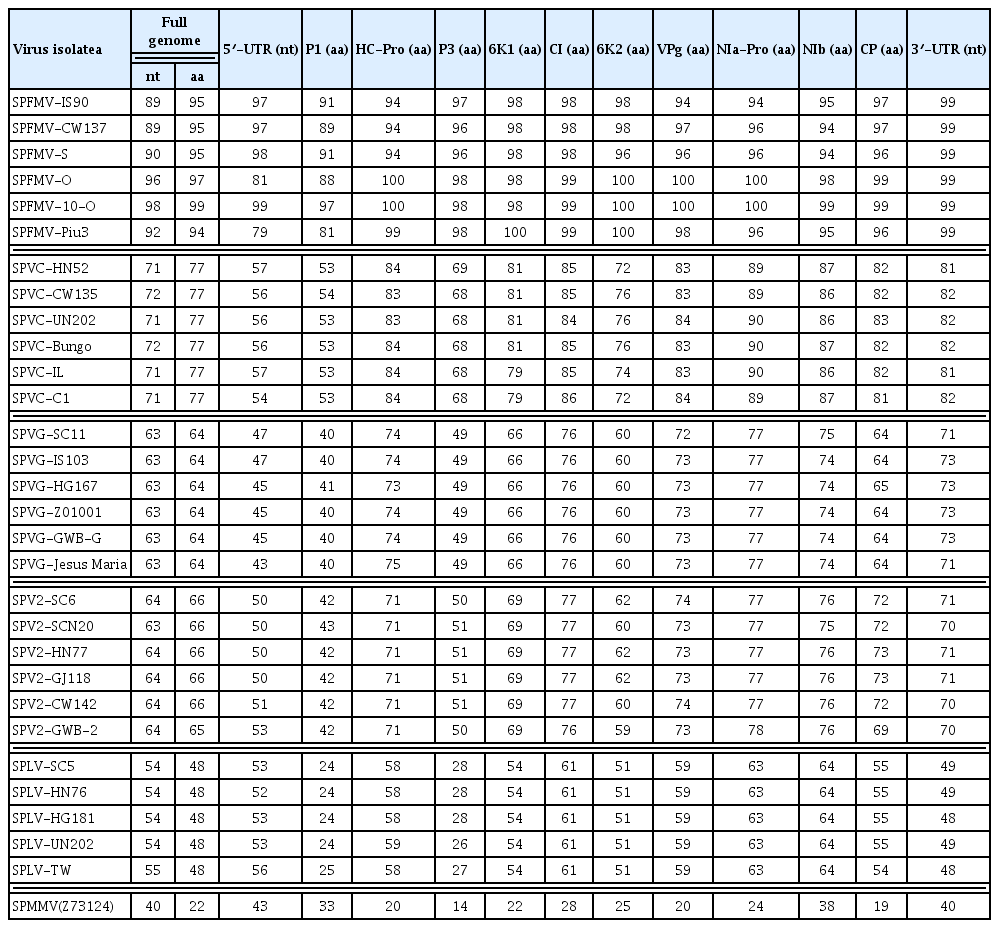
Nucleotide and amino acid sequence identities (%) between the Korean isolates SPFMV-GJ122 and other potyvirus isolates infecting sweet potato
For 10 mature proteins cleaved from a large polyprotein, the amino acid sequence identities between SPFMV-GJ122 and all other sweet potato-infecting potyvirus isolates were 24–97% for P1, 58–100% for HC-Pro, 27–98% for P3, 54–100% for 6K1, 61–99% for CI, 51–100% for 6K2, 59–100% for VPg, 63–100% for NIa-Pro, 64–99% for Nib, and 54–99% for CP. The amino acid identities of the P1 and P3 proteins were lower than those found for the other proteins. P1 proteins of SPLV isolates had the lowest amino acid identities (18–25%) with viruses of the SPFMV lineage. SPFMV-GJ122 and the SPFMV-10-O isolate, which belonged to the O strain of SPFMV, showed higher identity with the RC strain of SPFMV in the P1 protein and 5′ UTR. In addition, the nucleotide sequence identities between SPFMV-GJ122 and the other isolates for the 5′ and 3′ UTRs were 43–98% and 48–99%, respectively.
Recombination analysis
To examine whether recombination occurred in the sweet potato potyviruses, the full-length nucleotide sequences of 30 sweet potato potyviruses were examined using different recombination detecting programs in RDP4. In total, 16 potential recombinant types were detected by at least one of the methods (Table 3). However, most isolates seemed to be ‘tentative’ recombinants, as they were supported by less than four methods or one of the parental isolates was labeled as ‘unknown’. Other than this ‘tentative’ recombination, only four possible recombination types (type 3, 4, 12 and 15) were detected in 5 sweet potato potyvirus isolates (Table 3). In particular, recombination type 4, supported by all seven methods with the highest p-value of 2.232×10−101, was found to have occurred in the isolates SPFMV-GJ122 and SPFMV-10-O. These were detected as recombinants between SPFMV-O as a major parent and SPFMV-S as a minor parent. In this recombination event, a genomic region (10,764–1,130 nt, 5′ UTR and P1-N terminal regions) was replaced with the homologous region of SPFMV-S. Recombination type 3, which covers NIa-Pro, NIb, CP, and 3′ UTR, was unique to the Peruvian isolate, SPFMV-Piu3, which belonged to the EA strain and was reported previously (Untiveros et al., 2010; Yamasaki et al., 2010). In addition, two possible recombination type 12 and 15 were detected in the Korean SPLV isolates SPLV-UN202 and -SC5, respectively. These recombinants had the same parental isolates, SPLV-TW and SPLV-HN76. Most recombination types (12 of 16 types) were labeled as ‘tentative’ recombination, and involved mainly interspecies recombination. The P1, HC-Pro and NIa-NIb regions were identified as hotspots for recombination in the sweet potato potyviruses.
Discussion
In this study, to characterize Korean sweet potato potyviruses at molecular level, we determined the complete genome sequences of 18 isolates of five sweet potato potyviruses collected from sweet potato in 2012. We analyzed their genetic diversity by comparing their sequences with the sequences of 12 previously reported sweet potato potyvirus isolates (Table 1).
The complete genomes of 18 isolates consisted of from 10,081 to 10,830 nt, excluding the poly-A tail. They encoded a single large polyprotein of 3,247–3,493 aa. Their genomic organizations, encoding 10 mature proteins, were typical of members of the Potyvirus genus. The recently reported PIPO, which is conserved throughout the Potyviridae family (Chung et al., 2008), was identified in the P3 region of all of the Korean isolates. PIPO is reported as an essential gene encoding the P3-PIPO fusion product via ribosomal frameshifting or transcriptional slippage at a highly conserved G1-2A6 motif (Chung et al., 2008). Another novel ORF, PISPO, which has been detected in the P1 region of potyviruses infecting sweet potato (Clark et al., 2012), was identified in the isolates of four viruses, the exception being SPLV. The expression and function of PISPO are understood only incompletely.
In the previous reports, SPFMV was classified into four distinct strains, russet crack (RC), ordinary (O), common(C), and East Africa (EA) based on their pathogenecities, occurred area, and 3′-terminal nucleotide sequences including the CP genes (Kreuze et al., 2000; Untiveros et al., 2008). However, classifying isolates into strains based on 3′-terminal nucleotide sequence identities didn’t result in groupings related to symptom severity (Gutierre et al., 2003). Further analysis using complete nucleotide sequences have provided a better understanding of the taxonomy and biological properties of SPFMV (Yamasaki et al., 2010). Based on sequence comparison and phylogenetic analysis of the complete nucleotide sequences, Korean SPFMV isolates were assigned to the RC and O strains. The Korean SPFMV isolate, SPFMV-GJ122, had over 97% nucleotide sequence identity with SPFMV-10-O. It was grouped into the O strain of SPFMV, but had high identity with the RC strain of SPFMV in the 5′ UTR and P1 regions due to recombination. This result could explain why SPFMV-GJ122 and SPFMV-10-O were classified into the same group as the SPFMV-RC strain in the 5′UTR nucleotide-based phylogenetic tree (Fig. 9) and had higher nucleotide and amino acid sequence identities with SPFMV-S than with SPFMV-O in the 5′ UTR and P1 protein regions (Table 2). The isolate SPFMV-10-O was reported as a triple recombinant of strains O, EA, and RC of SPFMV (Untiveros et al., 2010). In comparison with the biological properties of SPFMV strains, SPFMV-RC causes russet cracks and severe skin discoloration of roots, and SPFMV-O induces mild skin discoloration. On the other hand, SPFMV-10-O, a recombinant of SPFMV-O and –RC strains, induces no russet crack or obvious root skin discoloration, and protects sweet potato plants from SPFMV-S infection by cross-protection (Yamasaki et al., 2009).
Korean SPVC isolates had 99% nucleotide sequence identity with the Japanese isolate Bungo, while, 70–71% sequence identity with SPFMV. These results support that SPVC is a separate species within the Potyvirus genus (Tairo et al., 2005; Untiveros et al., 2010). Korean SPVG isolates had 99% nucleotide sequence identity with the three previously reported SPVG isolates. SPVG has been divided into two groups, with <90% identity based on nucleotide sequences of the coat protein (Rännäli et al., 2009). Korean SPVG isolates presented in this study were clustered with the Argentinean and USA isolates. Korean SPV2 isolates had 97–98% nucleotide sequence identities with the SPV2 GWB-2 isolate from the USA, and were more closely related to SPVG than to any other sweet potato virus. In the case of SPLV, Korean isolates had 88–89% sequence identity with the Taiwanese isolate TW. The sequence identities between Koran SPLV isolates were 92–98%. On the other hand, Korean SPLV isolates not belonging to the SPFMV lineage were determined to be phylogenetically distantly related.
In order to understand the relationship of sweet potato potyviruses, we further analyzed the amino acid sequences for each mature protein. The constructed phylogenetic trees based on amino acid sequences of mature proteins had groupings similar to those of the nucleotide sequence-based trees. In the case of the P1 protein, SPLV was highly divergent from viruses of the SPFMV lineage. The P1 protein was the most variable region within and between species of potyviruses infecting sweet potato. The amino acid sequence identities of the P1 protein between SPFMV and other viruses were 24–54%, and the sequence identities of the P1 proteins of SPFMV strains were 81–91% (Table 2). This region might be important for recombination in sweet potato potyviruses (Untiveros et al., 2010; Yamasaki et al., 2010).
When plants are co-infected with multiple viral strains or species, the possibility of emergence of new viral strains due to genetic recombination cannot be ruled out (Chare and Holmes, 2006). Recombination has been shown to be an important force in the evolution of plant viruses, including several members of Potyviridae (Jonson et al., 2009; Seo et al., 2009). Identifying genomic regions that are hotspots for recombination is, therefore, crucial in understanding the putative role of recombination in the evolution and emergence of new viral strains. In this study, we analyzed recombination using the full-length nucleotide sequences of 30 sweet potato potyviruses and one ipomovirus. A total of 16 potential recombinant events were detected by at least one of the methods (Table 3). However, the potential recombination events, with the exception of tentative recombination, occurred in the P1, HC-Pro and NIa-NIb regions within the SPFMV and SPLV species. These regions were identified as hotspots for recombination in the sweet potato potyviruses. Especially, P1 protein, as the most divergent region with regard to both length and amino acid sequences region (Adams et al., 2005), is under looser evolutionary constraint and vulnerable to the recombination in the family Potyviridae (Nguyen et al., 2013; Seo et al., 2009; Valli et al., 2007).
As previously reported in the literature (Li et al., 2012), we also did not detect plausible recombination in SPVC, SPVG, or SPV2. Most recombination events were described as ‘tentative’, and were mainly interspecies recombination. Further analysis of the complete sequences of different strains of SPVG, SPV2, and SPVC is required to better understand the evolution among the isolates of each virus and other viruses in the SPFMV lineage. Our findings have also revealed that recombination is the major driving force in the emergence of new variants of sweet potato potyviruses.
Understanding the molecular variation of viruses is essential to design knowledge-based control strategies. This study provides the first report of the complete genome sequences of Korean sweet potato potyviruses (SPFMV, SPVC, SPVG, SPV2, and SPLV) and our full-genome sequence-based molecular analyses will facilitate the development of strategies for the control of sweet potato potyviruses.
Acknowledgments
This research was supported by a grant (iPET: No. 110034-05-5-HD110) from Agricultural Biotechnology Development Program, Ministry of Agriculture, Food and Rural Affairs of Republic of Korea.


