Carbamoyl Phosphate Synthase Subunit CgCPS1 Is Necessary for Virulence and to Regulate Stress Tolerance in Colletotrichum gloeosporioides
Article information
Abstract
Glomerella leaf spot (GLS) is a severe infectious disease of apple whose infective area is growing gradually and thus poses a huge economic threat to the world. Different species of Colletotrichum including Colletotrichum gloeosporioides are responsible for GLS. For efficient GLS control, it is important to understand the mechanism by which the cruciferous crops and C. gloeosporioides interact. Arginine is among one of the several types of amino acids, which plays crucial role in biochemical and physiological functions of fungi. The arginine biosynthesis pathway involved in virulence among plant pathogenic fungi is poorly understood. In this study, CgCPS1 gene encoding carbamoyl phosphate synthase involved in arginine biosynthesis has been identified and inactivated experimentally. To assess the effects of CgCPS1, we knocked out CgCPS1 in C. gloeosporioides and evaluated its effects on virulence and stress tolerance. The results showed that deletion of CgCPS1 resulted in loss of pathogenicity. The Δcgcps1 mutants showed slow growth rate, defects in appressorium formation and failed to develop lesions on apple leaves and fruits leading to loss of virulence while complementation strain (CgCPS1-C) fully restored its pathogenicity. Furthermore, mutant strains showed extreme sensitivity to high osmotic stress displaying that CgCPS1 plays a vital role in stress response. These findings suggest that CgCPS1 is major factor that mediates pathogenicity in C. gloeosporioides by encoding carbamoyl phosphate that is involved in arginine biosynthesis and conferring virulence in C. gloeosporioides.
Glomerella leaf spot (GLS) is a devastating contagious disease of apple which is caused by a ubiquitous fungal plant pathogen Colletotrichum gloeosporioides which is anamorph of Glomerella cingulate (Medeiros et al., 2010). Pathogenic fungi are involved in several crop diseases and cause enormous economic loss each year. The GLS pathogen mainly damages apple leaves causing dark spots followed by their dryness resulting in severe defoliation within 5-7 days of development (Hamada et al., 2019; Sutton and Sanhueza, 1998; Wang et al., 2012). In apple leaves and fruits, C. gloeosporioides causes slightly small lesions, which develop into bitter rot (Velho et al., 2015). Apple GLS caused by C. gloeosporioides may result in 75-90% defoliation under favourable conditions (Bogo et al., 2012).
The GLS is an emerging disease and only a few studies have reported the epidemiology and mechanism of this disease. In Colletotrichum spp., during conidia development the first step is landing and attachment on plant surface which is followed by conidial germination to form a germ tube with an appressorium at the terminal end. By applying mechanical force and cutinase like enzyme, appressorium penetrate plant cuticle and cell wall leading to development of primary and secondary hyphae (Prusky and Lichter, 2007). Molecular mechanistic studies to understand the basis of appressorial development and pathogenicity are required to formulate new approaches for effective control fungal diseases.
Previous studies showed that inhibition of arginine can affect fungal development and pathogenicity. Carbamoyl phosphate synthetase (CPS) and a polyprotein precursor of N-acetylglutamate kinase and N-acetylglutamyl-phosphate reductase are involved in arginine biosynthesis which is important in early stages of plant infection by C. higginsianum on the host plant Arabidopsis thaliana (Takahara et al., 2012). Disruption of the ARG 5, 6 gene containing sequences of acetyl glutamate kinase and acetyl glutamyl phosphate reductase results in arginine auxotrophy in Candida albicans (Negredo et al., 1997). In Magnaporthe oryzae, MoCpa2 plays a vital role in growth, conidia formation and virulence through arginine biosynthesis (Liu et al., 2016). L-arginine is involved in conidiation (asexual reproduction from spores) of the fungus Coniothyrium minitans (Gong et al., 2007). Zhang et al., in 2015, reported three genes involved in the different steps of biosynthesis of arginine i.e., MoARG5, 6 (2nd & 3rd step), MoARG7 (5th step), and MoARG1 (7th step) as shown in Supplementary Fig. 1. These genes are necessary for growth, sexual reproduction, and pathogenicity in M. oryzae (Zhang et al., 2015). Deletion of the gene in Fusarium oxysporum, encoding arginosuccinate lyase Arg4p results in reduced pathogenicity (Namiki et al., 2001).
Arginine is one of the most important amino acids found in animals and affects their biochemical and physiological functions. Arginine performs key part in many biological processes including cellular growth, protein synthesis, hormonal metabolism, sexual reproduction, osmotic pressure, signal transduction, nitrogen metabolism, energy production, and urea synthesis (Bedford and Richard, 2005; Wu et al., 2009). The CPS enzyme is found in living organism consist of three isoforms CPS I, CPS II, and CPS III (Hong et al., 1994). Fungi contain CPS II with two different forms, one is pyrimidine specific and other is arginine specific. In prokaryotes and eukaryotes, CPS signifies the first step in pyrimidine and arginine biosynthesis (Holden et al., 1999; Raushel et al., 1999). CPS is used to synthesize carbamoyl phosphate using glutamine, bicarbonate, and two ATP molecules (Hilger et al., 1973; Piérard and Schröter, 1978). The de novo biosynthesis of arginine takes place through eight reactions which involves the synthesis of ornithine from glutamate followed by its reaction with carbamoyl phosphate to form citrulline. Finally, citrulline is converted into argininosuccinic acid which is then cleaved to form arginine using lyase.
Above reported studies could enable the understanding of molecular mechanism underlying appressorium formation. However, detailed, and advanced research on signaling cascades in C. gloeosporioides are not well studied at present. In a previous study, using Agrobacterium tumefaciens mediated transformation (ATMT), we have developed a library of C. gloeosporioides of wild type (WT) strain W16 containing 10,210 transformants (Wu et al., 2016). In the present study, we identified a T-DNA insertional mutant S58 which revealed non-pathogenicity to wounded apple leaves and fruits. The gene was found to play an important role during appressorial formation, pathogenicity as well as stress tolerance and conidial germination conidial germination. We designated it as CgCPS1 gene encoding CPS involved in arginine biosynthesis. The closest homolog of our query protein of 457 amino acids was explored by performing NCBI protein BLAST against non-redundant database using standard algorithm parameters. Apart from the CgCPS1 gene, other 13 homologous sequences of carbamoyl-phosphate synthase arginine-specific small chain (CPA1) were obtained by performing NCBI Nucleotide BLAST against the NR database of the GenBank. Then, the role of CgCPS1 in the appressorial formation and the infection progression of C. gloeosporioides was evaluated. The targeted gene deletion of CgCPS1 confirmed its involvement in cell wall integration, fungal growth, virulence, conidia germination and stress response to H2O2.
Materials and Methods
Fungal strains and culture conditions
C. gloeosporioides WT strain W16 (Wu et al., 2016; Zhou et al., 2017) which was used as model in this study was isolated as a plant pathogen from apple leaves and fruit (cv Gold delicious) in China. The fungal strain was maintained by frequent subculturing on potato dextrose agar (PDA) medium at 28°C using the previously described method (Xu et al., 2016). The process of ATMT was performed using Agrobacterium tumefaciens strain LBA4404. For gene manipulation in various plasmids Escherichia coli strain TG1 was used as a donor.
Screening of T-DNA integration site and cloning of CgCPS1
The mutant CgCPS1 sequence flanking T-DNA insertion was amplified by hiTAIL PCR as previously described (Liu and Chen, 2007). After purification and ligation into vector it was sequenced by Sangon Biotech company. The obtained sequences were analysed by BLAST using C. gloeosporioides genome database (http://genome.jgi.doe.gov/Gloci1/Gloci1.home.html) and the NCBI GenBank database.
Comparative Fungal Genomics Platform (CFGP) (http://cfgp.riceblast.snu.ac.kr/main.php) was used to analyze the sequences of CgCPS1 gene (Choi et al., 2013). The amplification of full-length CgCPS1 sequences was performed by using a pair of primers. (Table 1). InterProScan was used to analyze the domain of CgCPS1 (Hunter et al., 2012; Mitchell et al., 2015). Signal peptide in the protein sequences was identified by using Signal P 4.1 Server (Petersen et al., 2011).
Phylogenetic analysis of CgCPS1 gene from Colletotrichum gloeosporioides
Apart from the CgCPS1 gene, other 13 homologous sequences of CPA1 were obtained by performing NCBI Nucleotide BLAST against the NR database of the GenBank.
Sequences from the following organisms were used with accession numbers in parentheses; Colletotrichum aenigma (XM_037320351.1), Colletotrichum graminicola (XM_008093538.1), C. higginsianum (XM_018299653.1), Verticillium dahliae (XM_009651725.1), Scedosporium apiospermum (XM_016789182.1), Purpureocillium lilacinum (XM_018317856.1), Trichoderma citrinoviride (XM_024897429.1), Neurospora tetrasperma (XM_009850998.1), Sporothrix schenckii (XM_016728008.1), Fusarium graminearum (XM_011329858.1), Geosmithia morbida (XM_035466589.1), Pyricularia oryzae (XM_003714692.1), and Saccharomyces cerevisiae (NM_001183722.3). CPA1 from S. cerevisiae was added to compare the relative divergence from our subject organism.
Multiple sequence alignment of the 14 DNA sequences was performed in MEGA X (Kumar et al., 2018) by the ClustalW algorithm (Thompson et al., 2002). The evolutionary linkages were inferred using the Neighbor-Joining method (Saitou and Nei, 1987).
Targeted gene knockout and complementation
To assemble the CgCPS1 gene alternative vector pCPS1, 1.2-kb upstream and 1.3-kb downstream sequences of CgCPS1 were amplified with primer pairs S58F1/S58R1 and S58F2/S58R3, respectively. The PCR products were ligated into the pCambiaMX9 vector to make pCambia-F1 and pCambia-F2. The 2.0-kb gene hph cassette encoding hygromycin phosphotransferase was obtained from pTFCM and cloned in to pCambia-F2. The primers used are listed in Table 1. For the construction of complementation vector (N-terminal green fluorescent protein [GFP] tagging vector) pGFP-native promoter region of 3-kb full-length region of the Cgcps1 gene was amplified with primer pairs G08 F/G08 R. The PCR product was ligated into digested pGapneoR10. Then the plasmid was transformed into the Δcgcps (Δs58d10-2) through ATMT. The resulting mutants were selected using G-418 sulphate and analyzed through PCR amplification using the primers.
Fungal transformation
The targeted gene deletion was performed via homologous recombination using ATMT by transforming the pCambia-F2 into A. tumefaciens LBA4404 through electroporation as described previously (Lee and Bostock, 2006). The hygromycin resistant colonies were observed in 48-72 h. The colonies of transformants were shifted separately onto the PDA plates supplemented with hygromycin B (100 μg/ml) and kept at 28°C for 48 h. Then hyphal tips from each transformant were picked using sterilised needle and shifted to fresh PDA plates having hygromycin B and saved for future use.
DNA extraction and PCR amplification
The putative Δcgcps1 mutants were selected from PDA plates and stability of hygromycin resistance of transformants was tested by subculturing them five times on PDA media containing 100 μg/ml hygromycin B. Fungal genomic DNA was obtained using the cetyltrimethylammonium bromide procedure (He, 2000). The putative Δcgcps1 transformants were confirmed by PCR amplification with a pair of primer (Table 1).
Southern blotting
The genomic DNA of the C. gloeosporioides and selected mutants was prepared following standard method (Sambrook and Russell, 2001). Southern blot was carried out following Amersham ECL Direct Nucleic Acid Labelling and Detection system (GE Healthcare, Buckinghamshire, UK) according to manufacturer’s instructions (Zhang et al., 2014). For the mutant S58, 15-20 μg genomic DNA was digested with Hind III and the probe amplification was done with primers (Table 1), using pCamhybgfp1 as template to prepare a mutagenesis population of C. gloeosporioides.
Fungal growth and appressorium formation
The vegetative growth of fungal colonies was recorded by measuring colony diameter in plates of W16 cultured on complete medium (CM) medium at 25°C for 10 days. Colony colour and morphology was observed. Conidial growth was evaluated by collecting conidia from 10-day-old plate cultures. Conidial suspension concentration was quantified using a haemocytometer. The germination of conidia and formation of appressorium were quantified on hydrophobic coverslips at 25°C for 24 h and 48 h. The percentage of appressorium growth was evaluated through microscopic evaluation of at least 100 conidia or aspersoria. Onion epidermal cells kept in the centre of water agar (12%) were used for penetration assays. Conidial suspension adjusted to 1 × 105 spores/ml, was dropped on epidermal cells. Microscopic observation of onion epidermis was done at 16-hour culture (hc), 24 hc, and 32 hc. Each test was done at least three times.
Infection assays
For pathogenicity test, mycelial plugs of the W16, Δcgcps1-1 and Δcgcps1-2 mutants, and the complemented transformant (CgCPS1-C) were inoculated on the intact apple leaves (cv Gold delicious), that were kept on plates having 12% water agar. Furthermore, a hyphal pellet of 1-mm diameter was injected into the injured fruits poked approximately 0.2-0.3 cm using sterilized toothpicks. Diseased lesions were observed at day 4, 6, and 10 post-inoculation (dpi). Each experiment was repeated thrice with three replicates each time.
Growth test/cell wall sensitivity assay
To assess the stress tolerance, the growth size and colony shapes were evaluated using fungal cultures grown on CM containing 200 μg/ml Congo Red (CR), 10 mM H2O2, 200 μg/ml Calcofluor White (CFW), 1 M NaCl, 1 M sorbitol, and 0.05% sodium dodecyl sulfate (SDS). All experiments were carried out in triplicates and repeated three times.
Subcellular localization
A fragment having the CgCPS1 and enhanced GFP was constructed to trace the subcellular localization of CgCPS1 in C. gloeosporioides in which CgCPS1-C::eGFP was expressed under the control of CgCPS1 native promoter. The fragment with native promoter was transferred into Δcgcps1 mutants through ATMT technology. The expression of GFP signals was spotted during different phases of conidium and appressorium development. Epifluorescence microscope (Leica DM5000B microscope, Jena, Germany) was used to record the GFP fluorescence.
Statistical analysis
Statistical assessment of data was carried out using GraphPad Prism version 7.0 (LaJolla, CA, USA). Results are presented as mean standard error of the mean. Two-way ANOVA and Tukey test was used to assess whether the results have significant difference and a P ≤ 0.05 was considered as significant.
Results
Identification of the T-DNA tagged gene CgCPS1 of C. gloeosporioides
To find the molecular basis of plant pathogenicity by C. gloeosporioides, T-DNA insertional mutagenesis library from our laboratory was used containing 10,210 transformants. To analyse the integration site of T-DNA in S58 mutant, genomic DNA was attained through hiTAIL PCR products and then sequencing was performed. The position of the mutant S58 was located at 399,910 bp which is a part of the first exon of a hypothetical-gene CPA1 (3,999,310-4,000,751 bp) located on contig 39 (Tan et al., 2021). It contained two exon and one intron. The targeted gene was named as CgCPS1 because it contained carbamoyl-phosphate small subunit protein. After sequencing, it was confirmed that the protein Cgcps1 contained one intron and 457 amino acid while the CgCPS1 gene from C. gloeosporioides is 1,374 bp.
Phylogenetic analysis of CgCPS1 gene from C. gloeosporioides
When compared with 13 homologous sequences of different fungal strains, they showed a very high percentage of identity among them. The phylogenetic tree shown in Fig. 1 arranges the most similar sequences in closer taxa. The CPA1 gene from C. gloeosporioides shares the highest identity with C. aenigma, followed by two other species, C. graminicola and C. higginsianum. The collective branch length of the Colletotrichum clade is 178.76 and is 1,363 units away from the S. cerevisiae CPA1 gene (Fig. 1).

Evolutionary relationships of carbamoyl-phosphate synthase arginine-specific small chain (CPA1) gene. The optimal tree with the sum of branch length 1,542 is shown. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree.
Deletion and complementation of CgCPS1
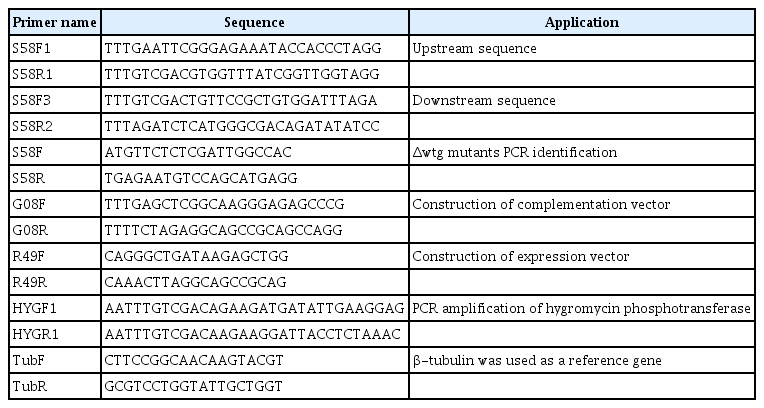
The deletion of CgCPS1 was executed to detect the pathogenicity of this marker as displayed in Fig. 2A. The selection of putative Δcgcps1 mutants was performed by growing on PDA media supplemented with hygromycin. The further confirmation was done by Southern blot analysis and reverse transcription polymerase chain reaction (RT-PCR). The Δcgcps1 mutants (Δcgcps1-1 and Δcgcps1-2) were first confirmed by qualitative PCR. No band was identified in the Δcgcps1 mutants, while a band of 1.3 kb was detected in WT (Fig. 2B). Southern blot was performed for confirmation of two selected deletion mutants (Δcgcps1-1 and Δcgcps1-2) (Fig. 2C). A complementation strain containing the native promoter CgCPS1-C was generated by reintroducing the gene CgCPS1 to Δcgcps1-2 mutants followed by validation with RT-PCR (Fig. 2). The schematic deletion of target gene was shown in Supplementary Fig. 1 which is involved in arginine biosynthesis. Disruption of target gene inhibited the biosynthesis of arginine.
Construction of replacement vector and verification of CgCPS1 deletion (A) The scheme for CgCPS1 deletion. hph, hygromycine phosphotransferase gene. The enzyme used are E (EcoRI), H (HindIII), and S (SalI). (B) The ∆cgcps1 mutants were confirmed through PCR using marker (2,000, 1,500, 1,000, and 500 bp). (C) Southern blot for confirmation of wild-type (WT) and ∆cgcps1 mutants. Genomic DNA of WT and ∆cgcps1 mutants were digested using HindIII. (D) Detection of CgCPS1 in WT, the ∆cgcps1 mutants, and the CgCPS1-C through reverse transcription polymerase chain reaction. Also used β-tubulin gene as a reference gene.
CgCPS1 involved in arginine biosynthesis
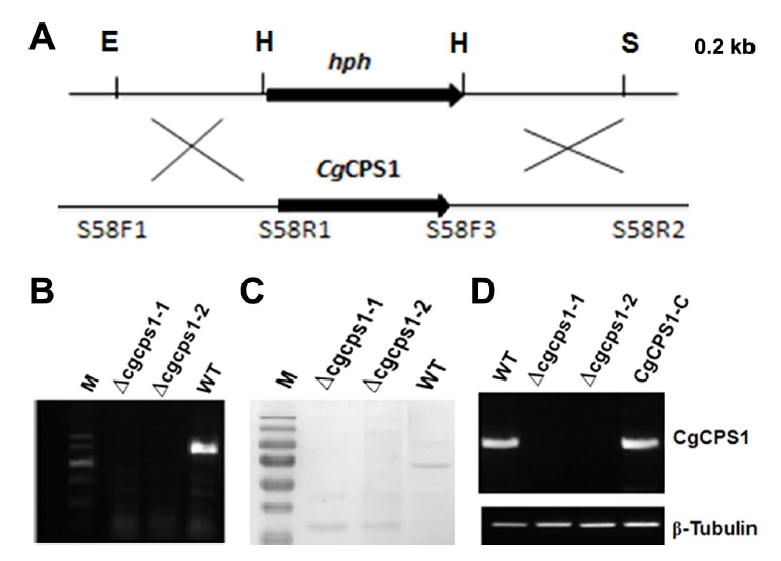
At first, we recorded the vegetative growth of W16, Δcgcps1-1 mutant, and CgCPS1-C on CM and minimal media (MM) media to identify the function of CgCPS1 in C. gloeosprioides. No significant change in colony diameter was observed on CM media after 7 days of incubation. There was reduced growth of W16 and CgCPS1-C on MM media but Δcgcps1-1 mutant could not grow (Fig. 3A). In C. orbiculare and yeast CAP1 is involved in arginine biosynthesis, so MM plates with different concentration of arginine were used to observe the growth of Δcgcps1 mutants. Exogenous supply of arginine restored the slow growth, but no aerial hyphal growth. As ornithine is precursor of arginine, so to observe the growth of mutant MM was used containing different concentrations of ornithine. The findings indicated that exogenous ornithine was not able to reinstate the growth defects of Δcgcps1 mutants (Fig. 3B). These results showed that CgCPS1 was involved in arginine utilization.
Defect of the Δcgcps1 mutant in fungal growth and restoration by exogenous arginine but not ornithine. (A) Morphology of fungal colony of the wild type (WT) W16, Δcgcps1 mutant and complemented transformant on complete medium and minimal media (MM) media. (B) The Δcgcps1 mutant was inoculated on MM with and without varied concentrations of arginine or ornithine and culturing at 28°C for 7 days.
Mutation in CgCPS1
The gene was knocked out to detect the potential role and contribution of CgCPS1 in pathogenicity. The Δcgcps1 mutants showed that these mutants were unable to form appressoria on artificial conditions of hydrophobic surfaces. To confirm the loss of pathogenicity by disruption of CgCPS1, the WT, Δcgcps1, and CgCPS1-C strains were inoculated on separated apple leaves and fruits. At 7 dpi, no symptom was observed on leaves and fruits. The Δcgcps1 mutants failed to infect the wounded fruits (Fig. 4A), but the WT and complemented strains produced a large number of invasive hyphae at 3 dpi. The growth of Δcgcps1 mutants was found slower as compared to the WT. The Δcgcps1 mutants lost virulence completely as they were unable to develop mature appressoria on detached leaves (Fig. 4B). The Δcgcps1 mutants were grown on PDA medium and it was observed that colony diameter was also reduced than that of the WT (Fig. 4C). Microscopic studies also revealed that the germ tubes of Δcgcps1 mutants were increased in size but could not enter the apple plant cells. On the other hand, in the WT, many conidia established appressorium which entered epidermal cells and finally developed into swollen invading hyphae developed into swollen invading-hyphae finally (Fig. 4D). We also incubated conidial suspension on a glass slide and on onion epidermal cells. The Δcgcps1 mutants failed to form appressorium on epidermal cells (Fig. 4E). Our results have revealed that the WT strain and complementation transformant produced typical GLS lesions on both apple leaves and fruits. However, analysis of Δcgcps1 were nonpathogenic for susceptible the apple leaves and fruits.

The ∆cgcps1 mutants are unable to cause disease in both apple fruits and leaves. (A) Leaf inoculation assay. (B) Fruit inoculation assay. (C) Growth of wild type, ∆cgcps1 mutants, and complementation transformant CgCPS1-C. (D) Examined appressoria and secondary structure under light microscopy. Scale bars = 10 µm. (E) Using light microscopy investigated development of secondary conidia and invasion of hyphae on onion epidermis. Scale bars = 10 µm. Red arrow, conidium; Blue arrow, appressorium; Green arrow, invasive hyphae; white arrow, secondary conidium.
Subcellular localization of CgCPS1
Fluorescence by GFP was recorded at different stages of fungal life cycle. The GFP signals were identified in the active expression of the native promoter and whole CgCPS1 sequence which were recorded during conidia, germ tube, and appressorium development (Fig. 5). In this study, it was observed that fluorescence signal was distributed in the cytoplasm, especially in the mitochondria. Furthermore, it was observed that expression of GFP signals was much higher during nascent conidia and germ tube development than infection hyphae. These findings that CgCPS1 is induced during conidia development than appressorium formation.
Effect of stress assay on cell wall
In this study, to investigate the stress responses of WT strain, CgCPS1 complementary strain and Δcgcps1 mutants to cell wall inhibitors were observed. All these strains were grown on CM containing 200 μg/ml CR, 10 mM H2O2, 200 μg/ml CFW, 1 M NaCl, 1 M sorbitol, and 0.05% SDS. The effects showed that Δcgcps1 mutants were more sensitive to CR, 10 mM H2O2, 200 μg/ml CFW, 1 M NaCl, 1 M sorbitol, and 0.05% SDS and inhibition growth rate was significantly higher as compared to WT and complementary strain (Fig. 6A). These findings suggest that CgCPS1 is not only intricate in response to high osmotic stress but also significant for maintenance of cell wall integrity. Data analysis showed the action of inhibiting growth rate of Δcgcps1 mutants was higher than W16 and CgCPS1-C (Fig. 6B).

(A) Stress response assays of wild type (WT) strain W16, ∆cgcps1 mutants (∆cgcps1-1 and ∆cgcps1-2), and the CgCPS1-C. W16, the ∆cgcps1 mutants (∆cgcps1-1 and ∆cgcps1-2) and the CgCPS1-C were allowed to grow on potato dextrose agar (PDA) medium containing 200 µg/ml Calcofluor White, and 200 μg/ml Congo Red, 1 M sorbitol, 1 M NaCl, and 0.05% sodium dodecyl sulfate (SDS). (B) The growth inhibition of W16, the ∆cgcps1 mutants (∆cgcps1-1 and ∆cgcps1-2), and the CgCPS1-C strains under different chemical components represented by the colony diameter. Means and standard errors were calculated from three replicates.
Effect on growth and appressorium formation
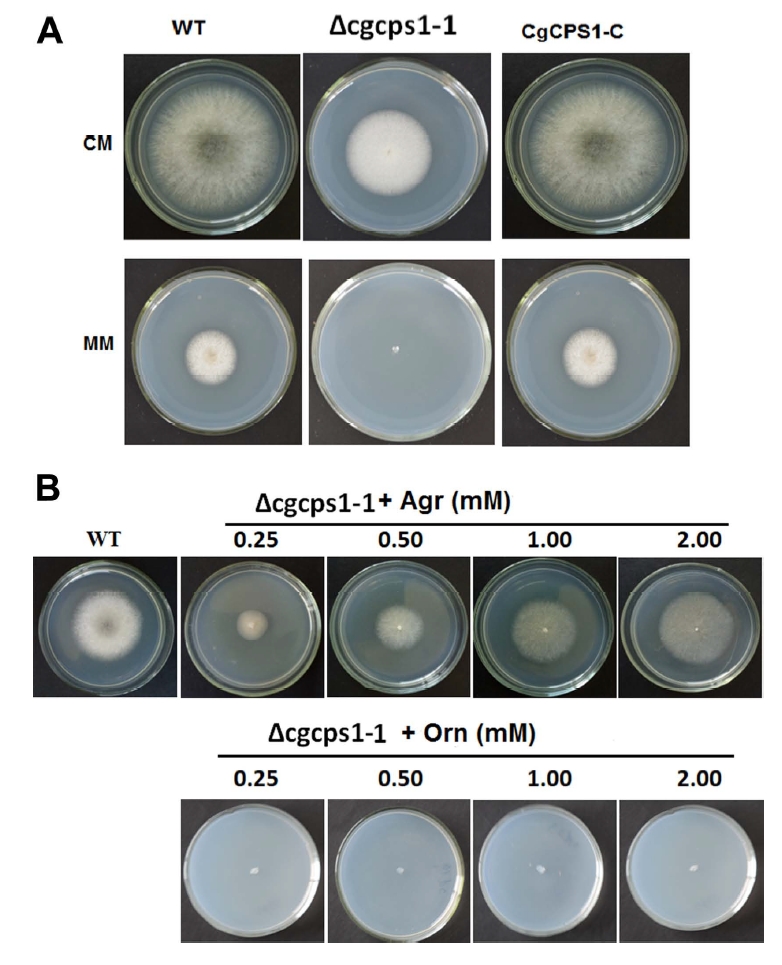
The null mutant grew slower than the WT and complementary strains on the PDA plates. The colony diameter was also reduced as compared to that of WT. We have investigated the role of CgCPS1 in conidia germination and appressorium formation incubating on the cellophane membrane. The WT, Δcgcps1, and complementation strains after germination continued to lengthen the germ tube, but Δcgcps1 could not form any appressorium at 12 h (Fig. 7).
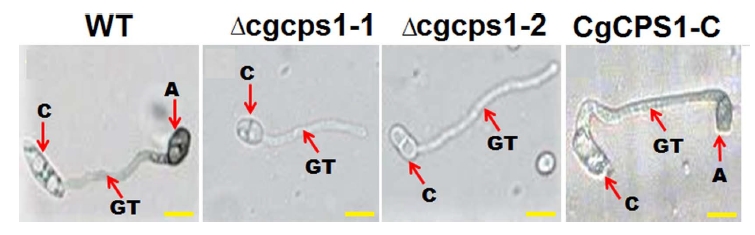
CgCPS1 is essential for appressorium formation. A droplet of freshly harvested conidial suspension. W16, the ∆cgcps1 mutants (∆cgcps1-1 and ∆cgcps1-2), and the CgCPS1-C were grown on hydrophobic surface of the Gel Bond membrane at 25°C at 12 h. Scale bars = 10 µm. A, appressorium; GT, germ tube; C, conidium.
Discussion
Plant diseases caused by pathogenic fungi result in significant expenditures to combat these diseases to ensure food security. According to an estimate, about 30% of the harvest of the world is wasted each year due to plant diseases (Fisher et al., 2012). Therefore, seeking a permanent solution for these plant diseases is necessary for sustainable plant production. Glomerella leaf spot is a widespread disease in apple plant which is tremendously involved in decreasing the yield of this crop. In present study, an attempt was made to understand and explore genetic mechanism involved in the development of virulence in the host plant by a pathogenic organism C. gloeosporioides. The CgCPS1 gene was identified and disrupted experimentally to assess its significance in arginine biosynthesis and virulence. It was found that this gene exhibits a high homology to small subunit arginine-specific CPS). It was considered that the arginine-specific CgCPS1 is involved in conidiation, appressorial development and fungal pathogenesis in C. gloeosporioides. For the functional analysis of CgCPS1, the comparison of pathogenicity of mutants and complementary transformants was carried out. Complementary transformant, when tested on apple fruits and leaves showed that its pathogenicity is restored while Δcgcps1 mutants had lost their pathogenicity at all (Fig. 4). We identified that CPS small subunit CgCPS1 in C. gloeosporioides and observed that deletion mutants are unable to grow on MM plates (Fig. 3). Moreover, this defect was inhibited by supplying exogenous arginine. When Δcgcps1 mutants were grown on media containing different concentration of arginine, they had grown gradually, but were unable to grow on CM plates containing ornithine. On the basis above findings, we suggest that the presence of CgCPS1 gene is exclusively needed for maintaining the pathogenic character in C. gloeosporioides. On the basis above findings, we suggest that the presence of CgCPS1 gene is exclusively needed for maintaining the pathogenic character in C. gloeosporioides.
A few similar studies carried out for other genes had provided evidences that arginine-specific subunit is necessary for fungal disease cycle and virulence (Liu et al., 2016; Namiki et al., 2001; Zhang et al., 2015). Since CgCPS1 contains the sequences like arginine-specific subunit which is already proved to be involved in conidia formation and disease progression, we performed further assays to prove its role in GLS. The CgCPS1 mutant was unable to develop necrotic lesions when wounded site of apples was exposed to the conidia or hyphal pellets (Fig. 4A). This demonstrated that CgCPS1 is crucial for growth of pathogen in the host tissues.
The deletion of CgCPS1 resulted in significant imperfection in appressorial formation. The phenotypic study revealed that ∆cgcps1 did not develop appressorium (Fig. 4D). C. gloeosporioides strains were inoculated on apple leaves and fruits to validate the role of CgCPS1 in penetration and infection process (Fig. 4). The results have shown that both WT and complementation strains caused lesions but Δcgcps1 mutants were completely unable to cause damage on both apple leaves and poked fruits. The results support the hypothesis that CgCPS1 is prerequisite for penetration and pathogenicity. Subcellular localization showed that CgCPS1 induced during conidia formation (Fig. 5).
Rho GTPases are important regulators that participate not only in diverse function but also critical for processes such as morphogenesis, development, and host infection (Nesher et al., 2011; Zhang et al., 2013). Moreover, it was also found that an intracellular level of cAMP in the C. gloeosporioides vegetative hyphae was controlled by Rho GTPases (CgRhoB) (Auyong et al., 2015). CgRhoB has role in cell wall integrity, conidial germination, and pathogenicity (Xu et al., 2016). To determine whether CgCPS1 gene is important to maintain cell wall integrity, the Δcgcps1 mutants, WT and complementary strains were grown on CM media complemented with different cell wall inhibitors. The Δcgcps1 mutants resulted in significant osmotic stress to 200 μg/ml CR, 10 mM H2O2, 200 μg/ml CFW, 1 M NaCl, 1 M sorbitol, and 0.05% SDS (Fig. 6A).
Collectively results of this study support an important role of CgCPS1 in arginine biosynthesis, which is crucial for fungal development, virulence and mediates stress tolerance in C. gloeosporioides. The study showed new insight into primary mechanism of arginine biosynthesis that may help in disease controling.
Notes
Conflicts of Interest
No potential conflict of interest relevant to this article was reported.
Electronic Supplementary Material
Supplementary materials are available at Plant Pathology Journal website (http://www.ppjonline.org/).
Strains used in current study
The presumed pathway of arginine biosynthesis and targeted gene deletion in Colletotrichum gloeosporioides.