 |
 |
| Plant Pathol J > Volume 34(1); 2018 > Article |
Abstract
Type VI secretion system (T6SS) has been discovered in a variety of gram-negative bacteria as a versatile weapon to stimulate the killing of eukaryotic cells or prokaryotic competitors. Type VI secretion effectors (T6SEs) are well known as key virulence factors for important pathogenic bacteria. In many Burkholderia species, T6SS has evolved as the most complicated secretion pathway with distinguished types to translocate diverse T6SEs, suggesting their essential roles in this genus. Here we attempted to detect and characterize T6SSs and potential T6SEs in target genomes of plant-associated and environmental Burkholderia species based on computational analyses. In total, 66 potential functional T6SS clusters were found in 30 target Burkholderia bacterial genomes, of which 33% possess three or four clusters. The core proteins in each cluster were specified and phylogenetic trees of three components (i.e., TssC, TssD, TssL) were constructed to elucidate the relationship among the identified T6SS clusters. Next, we identified 322 potential T6SEs in the target genomes based on homology searches and explored the important domains conserved in effector candidates. In addition, using the screening approach based on the profile hidden Markov model (pHMM) of T6SEs that possess markers for type VI effectors (MIX motif) (MIX T6SEs), 57 revealed proteins that were not included in training datasets were recognized as novel MIX T6SE candidates from the Burkholderia species. This approach could be useful to identify potential T6SEs from other bacterial genomes.
Secretion in bacterial species is a critical mechanism for survival and adaptation to their natural surrounding environmental stresses (Abby et al., 2016; Alfano and Collmer, 2004; Russell et al., 2014a). Many bacteria have evolved different secretion pathways termed from type I to type IX secretion systems to deliver effector molecules such as proteins, enzymes or toxins from a bacterial cell to its exterior (Abby et al., 2016; Tseng et al., 2009). Among them, the type VI secretion system (T6SS) is involved in both bacterial-host and inter-bacterial interactions, by delivering anti-eukaryotic or anti-bacterial effectors, respectively (Filloux et al., 2008; Russell et al., 2014a). The type VI secretion effectors (T6SEs) in plant-associated bacteria have been demonstrated to operate various functions such as inter-bacterial competition, quorum sensing, stress response, biofilm formation, and symbiosis (Bernal et al., 2017b; Cianfanelli et al., 2016; Hachani et al., 2016; Russell et al., 2014a; Ryu, 2015). Anti-bacterial effectors usually have corresponding immune proteins to help self-protect bacteria from the toxicity of their own secreted effectors. Notably, many components of T6SSs such as Hcp and VgrG proteins may serve as specialized effectors with toxic extension domains delivered into target cells along with cargo effectors (Cianfanelli et al., 2016; Hachani et al., 2016).
The T6SS were initially identified as imp genes in Rhizobium leguminosarum (Bladergroen et al., 2003), and then as evpA-H genes in Edwardsiella tarda (Rao et al., 2004). In 2006, structures of T6SS were elucidated in Vibrio cholera and Pseudomonas aeruginosa with 13 core proteins (Mougous et al., 2006; Pukatzki et al., 2006). So far, three main T6SS types including T6SS-i, T6SS-ii, and T6SS-iii have been discovered, in which the T6SS-i was classified into the distinct subtypes T6SS-i1, i2, i3, i4a, i4b, and i5, based on the organization and phylogenetic relationship of its core components (Barret et al., 2011; De Maayer et al., 2011; Li et al., 2015; Russell et al., 2014b). The T6SS-i type has been identified in a wide range of proteobacteria with 13 core components from TssA to TssM (Bingle et al., 2008; Russell et al., 2014a), whereas the T6SS-ii type has just been discovered in Francisella species with 17 core components (Broms et al., 2012; de Bruin et al., 2011). The T6SS-iii type was discovered later in Flavobacterium species with 12 core components (De Maayer et al., 2011; Russell et al., 2014b). Nowadays the mechanism and function of T6SS clusters among plant pathogenic and commensal bacteria have obtained much attention because of their important role to help bacteria getting advantages in adaptation and settlement through inter-bacterial and plant-bacterial interactions (Bernal et al., 2017b; Ryu, 2015).
The Burkholderia genus belongs to the β-subclass of proteobacteria with approximately over 60 species dwelling in diverse niches (Coenye and Vandamme, 2003; Compant et al., 2008). Of the plant pathogenic Burkholderia species, B. glumae, B. gladioli, and B. plantarii are emergent representatives that cause essential rice diseases such as panicle blight, sheath rot, and seedling blight (Estrada-de los Santos et al., 2013; Ham et al., 2011; Jeong et al., 2003; Solis et al., 2006; Ura et al., 2006). Besides, some Burkholderia species have been recently discovered as plant-associated beneficial bacteria (e.g., B. phytofirmans PsJN, Burkholderia sp. KJ006, B. phenoliruptrix BR3459a) that may promote plant-growth, enhance disease resistance, or foster nitrogen fixation in plants (Estrada-de los Santos et al., 2013; Mitter et al., 2013). In many Burkholderia species, the T6SS machinery has evolved as the most complicated system with distinct types; remarkably, it has specialized functions and can transport exclusively specific effectors (Burtnick et al., 2011; Schell et al., 2007; Schwarz et al., 2010), suggesting its essential role in this genus. However, the lack of apparent information regarding determinants of passage substrates through the T6SS apparatus and the fact that this secretion pathway may be suppressed under standard laboratory conditions (Bernard et al., 2010; Silverman et al., 2011), have caused difficulties in both the experimental and computational identification of novel T6SEs. So far, several bioinformatics approaches have been proposed to recognize T6SEs based on heretical methods and sequence homology searching (Li et al., 2015; Russell et al., 2012; Salomon et al., 2014).
In this report, we attempted to detect and characterize T6SS clusters and T6SE candidates in target genomes of plant-associated or enviromental Burkholderia species based on computational analyses. The component proteins of each cluster were manually checked in detail and phylogenetic trees of three components (TssC, TssD, and TssL) were established to elucidate the relationship among annotated T6SS clusters. The potential T6SEs in the bacterial genomes of Burkholderia species were inferred based on homology searching. Furthermore, we attempted to reveal a T6SEs containing marker for type six (MIX T6SEs) using a screening approach based on the profile hidden Markov model (pHMM) method.
Annotations of available T6SS clusters and their components in some bacterial genomes were obtained from the SecreT6 database (http://db-mml.sjtu.edu.cn/SecReT6/) (Li et al., 2015). To annotate T6SS clusters in the remaining uncharacterized genomes, we extracted nucleotide sequences from the public sequence database at National Center for Biotechnology Information NCBI (https://www.ncbi.nlm.nih.gov/genome/) and utilized the CD-easy tool in SecReT6 to annotate the open reading frames (ORFs) along with their corresponding amino acid sequences. Next, the T6SS-BLASTP program with default values in SecreT6 was used to detect the T6SS component proteins in target genomes and loci with at least five components were considered as potential T6SS clusters. The component genes of all clusters were then manually checked in detail. Furthermore, we compared the T6SS clusters obtained here with those detected by using TXSScan MacSyFinder program (Abby et al., 2016; Abby and Rocha, 2017), to carry out the optimized validation for potential functional systems. In final, the clusters that either were detected by both programs or contained at least 10 core components explored by SecReT6 were further used in distribution and phylogenetic analyses because of their highly confident presence and functional ability in genomes. Three important component proteins (e.g., TssB, TssC, TssL) were utilized for constructing phylogenetic trees to elucidate the phylogenetic relationship among the detected T6SS clusters. Protein sequences were aligned using the ClustalW algorithm (Thompson et al., 1994) in MEGA6 (http://www.megasoftware.net/) (Tamura et al., 2013), and phylogenetic trees were constructed based on the Neighbor-Joining (NJ) method (Saitou and Nei, 1987) with 1000 bootstrap replicates and other default parameters (phylogenetic reconstruction, substitution type: amino acids, model/methods: Jones-Taylor-Thornton (JTT) model (Jones et al., 1992), rates among sites: uniform rates and gap missing data treatment: partial deletion).
The complete sequenced genomes and protein sequences of Burkholderia and other reference species were downloaded from the NCBI genome database. In total, 105 experimentally verified T6SEs in different bacterial species were extracted from the integrated database SecReT6 and existing literature (Bernal et al., 2017a; Li et al., 2015). The evidential effectors were then used as query sequences to recover homologous proteins from the complete genomes of Burkholderia bacterial strains using BLASTP search (https://blast.ncbi.nlm.nih.gov/Blast.cgi) (Altschul et al., 1990) with identity ≥ 30% and E-value < 10−6. Next, the amino acid sequences of the collected proteins were checked manually. Proteins with length less than 100 residues or containing a signal peptide checked by the SignalP program (http://www.cbs.dtu.dk/services/SignalP/) (Petersen et al., 2011) were removed. To avoid overlapping results obtained from the similarity of short domains, we also checked the length skew between query sequences and the identified protein sequences in all hits; hits with a length skew greater than 200 amino acids were then eliminated. For hits with identity ranging from 30% to lower than 50%, only proteins that were detected in at least three hits by three different query effectors or having an annotation related to type VI effectors (i.e., Hcp, VgrG, Rhs/YD-repeat, PAAR, phospholipase) were retained. The TMHMM server (Krogh et al., 2001) and Conserved Domain Database CDD (https://www.ncbi.nlm.nih.gov/cdd/) (Marchler-Bauer et al., 2011) were used to explore the transmembrane (TM) regions and conserved domains in potential T6SE sequences.
To reveal MIX T6SEs, we built profile HMM models for MIX-containing effectors and used these models to scan all target genomes. In total, 217 T6SS-associated effector proteins that contained MIX motifs were extracted from a recent study by Salomon et al (2014). These T6SEs were divided into five groups as MIX I, MIX II, MIX III, MIX IV, and MIX V - containing proteins (named as dataset 1 to dataset 5 in this study), including six evidential T6SEs (i.e., VP1388, VPA1263, IdsD, BTH_I2691, VCA0020, V12G01_02265). These input protein sequences were aligned using the MUSCLE 3.8 program with default parameters (Multiple Sequence Comparison by Log-Expectation) (Edgar, 2004). Next, the HMMER (http://hmmer.janelia.org/) (Finn et al., 2011), particularly the basic function of building a profile HMM from a dataset of sequences (hmmbuild), was used to establish five pHMMs for the five datasets of aligned sequences. The function of searching a profile HMM against a database (hmmsearch) in HMMER was then used to screen the trained pHMMs against protein sequences, to identify matched proteins that were considered MIX T6SE candidates. The E-value along with sequence score for whole sequence searching of matched proteins was valid based on the results of searching evidential MIX T6SEs and matched proteins with a signal peptide checked by SignalP (Petersen et al., 2011) were eliminated. To assess the ability of each pHMM to recognize potential MIX T6SEs, we performed cross-searching using five built pHMMs against all 217 collected T6SEs in the five training datasets. The detection rate of five models against non-T6SE datasets was also tested. The test datasets were extracted from two negative datasets of SecretomeP program, one from Gram-negative bacteria (196 proteins) and another from Gram-positive bacteria (1084 proteins). Next, to evaluate the ability of recognizing MIX T6SE candidates from separate bacterial genomes, we implemented a search of pHMMs against reference genomes of bacterial strains containing the known MIX T6SEs and some bacteria (e.g., Escherichia and Pseudomonas strains) with well-characterized T6SSs (Cascales and Cambillau, 2012; Cianfanelli et al., 2016; Filloux et al., 2008). Finally, we applied the pHMM-based screening approach described above to detect MIX T6SE candidates in bacterial genomes of the Burkholderia species. To explore the relationship among the revealed candidates, their amino acid sequences were aligned using the ClustalW algorithm (Thompson et al., 1994) and a phylogenetic tree was then constructed based on the Neighbor-Joining (NJ) method (Saitou and Nei, 1987) with 1000 bootstrap replicates and other default parameters in MEGA6 (http://www.megasoftware.net/) (Tamura et al., 2013). In addition, the available neighboring T6SS components of MIX T6SE candidates on the genome were checked manually. The conserved domains of the proteins were detected using the CDD program (Marchler-Bauer et al., 2011) and transmembrane (TM) regions were explored using the TMHMM server (Krogh et al., 2001).
A large amount of attention has been given to studies regarding mechanisms of T6SSs and their components because they are involved in both inter-bacterial and bacterial-host interactions (Bingle et al., 2008; Cascales and Cambillau, 2012; Cianfanelli et al., 2016). In this work, the potential function T6SS clusters in representative genomes of target Burkholderia strains which are plant-associated pathogenic and beneficial bacteria, biocontrol bacteria, or isolated from environment were annotated (Table 1). In total, 86 potential T6SS clusters were found in 30 target genomes of Burkholderia strains and the components of each cluster were screened manually (Supplementary Table 1). Of those, 66 clusters (mean number of clusters per strain is 2.2) were found to contain at least 10 components out of the 13 core components (TssA - TssM) (Table 1). All these clusters were also detected by TXSScan models as single-loci with exception of one cluster in Burkholderia sp. CCGE1002 due to strict requirements regarding number of predicted core proteins by TXSScan models which requires more than 11 core proteins over 14 full componnents (TssA-TssM, and evpJ proteins) (Abby et al., 2016; Abby and Rocha, 2017).
The plant pathogens (e.g., B. glumae, B. gladioli, B. plantarri strains) generally possessed at least two or three potential functional T6SS clusters whilst most of representative plant growth-promoting bacteria (e.g., B. phytofirmans PsJN, Burkholderia sp. KJ006, B. vietnamiensis G4, B. vietnamiensis LMG 10929), symbionts (e.g., Burkholderia sp. CCGE1001, Burkholderia sp. CCGE1003) or nodule bacteria (e.g., Burkholderia sp. RPE64, Burkholderia sp. RPE67) contained around one or two clusters (Table 1). This suggested that in plant-pathogenic Burkholderia species, the type VI systems would be related to operations functioning in plant-bacterial harmful interactions. Beside encouraging the inter-bacterial competitive interactions (Bernal et al., 2017b; Ryu, 2015), the pathogens seem have evolved these multiple systems to facilitate their colonization in plant host cells. In addition, we compared the result of T6SS prediction in the collected Burkholderia strains with that in other reference Burkholderia species including human pathogens. The mean number of potential functional T6SS clusters per strain in plant-associated or environmental Burkholderia bacteria (approximately 2.2) was lower than human pathogenic Burkholderia species (approximately 4.1) (Supplementary Table 1).
Next, to assess the phylogenetic relationship among T6SS clusters, we constructed phylogenetic distributions of three proteins (i.e., TssC, TssD, and TssL) which are important components and are also found in most of the assigned T6SS clusters (Supplementary Fig. 1-3). Proteins that belong to the same annotated T6SS subtypes (i.e., i3, i4b, i2, i1, i4a) were mainly grouped together, strongly indicating that the T6SS clusters could be classified based on the phylogenetic relationship of their components. Therefore, we assigned specific subtypes for T6SS-N/A clusters, which were not annotated availably based on the groups depicted in the phylogenetic trees. Remarkably, the assigned T6SS subtypes obtained from all three phylogenetic trees of TssC, TssD, and TssL proteins were the same and are specified as an inferred T6SS subtype in Supplementary Table 1. The predicted subtype number of the collected Burkholderia strains was presented in Table 1, with the highest proportions in the T6SS-i3 (39%) and T6SS-i4b (38%) subtypes, indicating the important role of these subtypes especially in plant-pathogenic Burkholderia species.
The detection of T6SEs in bacterial species may help in developing efficient strategies to inhibit or promote their competition with other neighboring bacteria dwelling in the same niches. Here, the experimental effectors were used as query sequences to search for homologous proteins in the bacterial genomes of Burkholderia species, in order to infer potential type VI effectors. At first, hits with identity ≥ 30%, E-value < 10−6, and a length skew less than 200 amino acids were collected. The proteins revealed in hits with lengths less than 100 residues or containing a signal peptide inside were then eliminated. Next, for hits with identity ranging from 30% to lower than 50%, only proteins with at least three hits or having a functional annotation related to type VI effectors were retained. Overall, a total of 322 potential T6SE proteins were inferred based on the homology search of the 30 genomes of Burkholderia species. Of these, 215 (66.8%) proteins were predicted as non-classically secreted proteins (NCSPs) by the SecretomeP program (Bendtsen et al., 2004) and only 17 proteins (5.3%) were found to contain at least one TM domain based on the TMHMM program (Krogh et al., 2001). The number distribution of the T6SE candidates along with the T6SS clusters in the analyzed Burkholderia bacterial strains are presented in Fig. 1. The plant-pathogenic Burkholderia strains obtained the highest average number 14.5 of putative T6SEs, followed by the group of environmental bacteria or biocontrol agent with 11.6, and the group of plant-associated beneficial bacteria got around 7.1. Clearly, the type VI effectors in Burkholderia plant pathogens would be associated with the ability of toxic infecting on their plant host cells. However, there seems to be no clear relationship between the number of T6SS clusters and number of T6SE candidates when considering them in all bacterial strains. The GenBank accession numbers of all induced T6SE proteins and related annotations can be found in Supplementary Table 2.
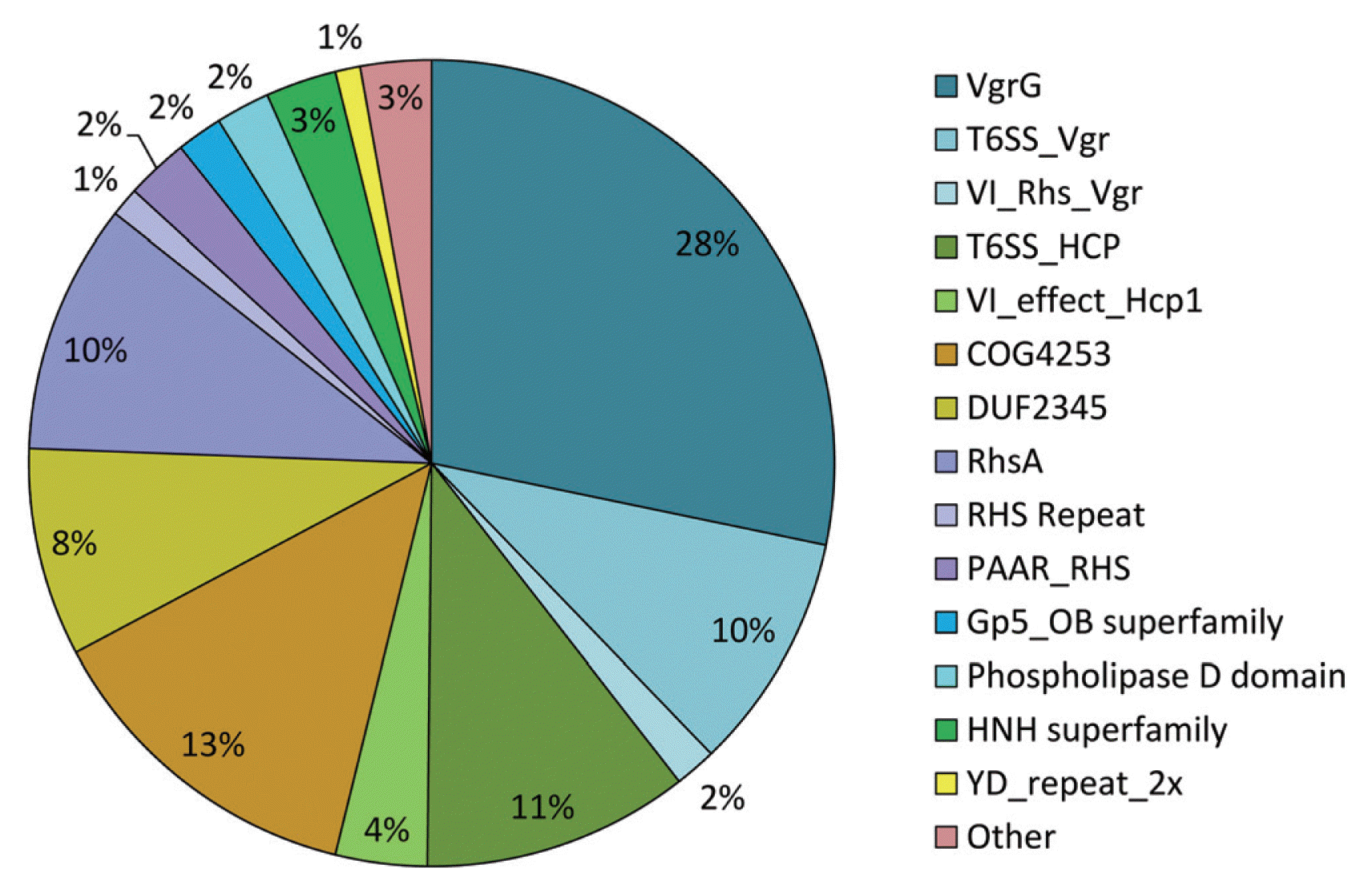
Domains or motifs may provide biologically meaningful features in the characterization of putative type 6 effectors and could be related to processes that connect the cargo effectors with the component effectors (Cianfanelli et al., 2016; Russell et al., 2014a). Based on the CDD program with concise results, conserved domains in the putative T6SEs induced in Burkholderia species were assigned with an E-value < 10−4. The distribution of domain types is depicted in Fig. 2, showing the highest fractions of VgrG-related domains (i.e., VgrG, T6SS_Vgr, VI_Rhs_Vgr) and Hcp-related domains (i.e., T6SS_HCP, VI_effect_Hcp1) with 40% and 15%, respectively. Lower fractions were observed for COG4253 (13%) and DUF2345 (8%), which are unknown functional domains usually found in type VI associated proteins that could be involved in the transport of putative effector islands (Ma et al., 2013). The Rhs - related domain type (i.e., RhsA, RHS, PAAR_RHS) also showed the fraction of up to 13% in total. Important domains like the Phospholipase D domain, included in the type VI secretion phospholipase D effectors targeting both eukaryotes and prokaryotes (Jiang et al., 2014), and especially the HNH super family comprising the conserved AHH, WHH, HNHc, and/or Tox-SHH domains that play an important role in the toxicity of proteins (Veluchamy et al., 2009) were also observed. The remaining domains (i.e., DUF3274, Cls, DUF2235, Ntox8 superfamily, PAAR, Pput2613-deam superfamily, SCP1201-deam, TVP38, and YadA_anchor superfamily) were pooled with others and hence were gathered together in term of other domain. The results suggested that these domains of type VI effectors in Burkholderia species are important in the secretion process or function in plant host cells, being consistent with findings from recent studies regarding the type VI effector - related domains (Flaugnatti et al., 2016; Jiang et al., 2014).
Five profile hidden Markov models (termed as pHMM1 to pHMM5) were generated separately for the five datasets of T6SE candidates (Supplementary Table 3). The number of T6SEs in the training datasets recognized by pHMMs through cross-searching are presented in Supplementary Table 4, showing the ability of each pHMM to reveal potential MIX T6SEs. As expected, all five pHMMs could recognize most of the proteins in their own training datasets (diagonal values, from 96.7% to 100%). The pHMM5 seemed to be the most unique model, mainly recognizing proteins in dataset 5. The pHMM3 was found to be the most effective model in the recognition of candidates of all three datasets 2-4, in which it could recognize proteins in dataset 2 and 4 with high fractions of 96.7% and 97.1%, respectively. Despite this, we used all three models of pHMM2, pHMM3, pHMM4 along with pHMM1 and pHMM5 for further detection of potential MIX T6SEs to obtain comprehensive results. In addition, in the tests to assess the false positive rates of pHMMs against negative datasets, only one protein (Q81J59) out of 1280 non-T6SE proteins was carried out by pHMM2 with E-value < 0.001 (Supplementary Table 4). Particularly, it got a quite low sequence-score of 15.5, suggesting that the proteins detected with higher cores would be more confident candidates of MIX T6SEs. Next, matched proteins obtained in the process of searching pHMMs against the reference genomes were determined (Supplementary Table 5), comprising all known MIX T6SEs along with four candidates included in the training datasets. Remarkably, two of the inferred MIX T6SE candidates, which were not included in the training datasets, were recently discovered to be experimental type VI effectors (EC042_4534 and BTH_I2698) (Flaugnatti et al., 2016; Russell et al., 2013) and two others (V12G01_01565 and BTH_I2701) shared significant similarity with known effectors (VP1388 and BTH_I2698) when checked by a BLASTP search (E-value < 10−6 and identity ≥ 70%). EC042_4534 and BTH_I2698 were explored only by the pHMM5 model, and have been indicated as belonging to the Tle1 (type VI lipase effector) family, one of the five divergent families (Tle1-5), suggesting that the T6SEs recognized by this model (pHMM5) are highly related to the Tle1 effector family (Russell et al., 2013). These results demonstrated that the pHMM-based screening approach could be useful to reveal potential MIX T6SEs from separate bacterial genomes and was therefore applied to the bacterial genomes of Burkholderia species.
MIX T6SE candidates inferred in the plant-associated or enviromental Burkholderia bacterial strains were then obtained using the pHMM-based screening approach described above, comprising 66 proteins in total (Table 2). Impressively, 20 of those proteins (~30.3%) were recognized by only pHMM5 and thus these proteins may belong to the Tle1 effector family, the anti-effectors aforementioned. It is also notable that we found no any MIX T6SE candidate inferred from genomes of some plant-associated beneficial bacteria including Burkholderia sp. CCGE1001, Burkholderia sp. CCGE1002, Burkholderia sp. CCGE1003, Burkholderia sp. RPE64, and Burkholderia sp. RPE67 in the screening process. Of the novel candidates, 3 proteins that were not included in the training datasets, shared significantly high similarity with at least one of the evidential T6SEs when checked by BLASTP search (E-value < 10−6 and identity ≥ 70%) (Table 2). Additionally, a larger number were found to have T6SS component proteins located in their proximity (Supplementary Table 5). This corresponded with the discovery that MIX-containing type VI effectors usually reside near components of T6SSs on the genome map (Hachani et al., 2016; Russell et al., 2014a). The phylogenetic tree of all 66 candidates clearly manifested two main groups of which group I covered most MIX T6SE candidates detected by one or more models from pHMM1-4 and group II covered all 20 MIX T6SE candidates that were recognized by pHMM5 alone (Fig. 3). The relationship presented in the phylogenetic distribution could provide hinders of translocating ability among close T6SEs in different Burkholderia bacterial strains. The conserved domains were founded in only 27 out of 66 candidates in which the fraction of DUF2235 domains (uncharacterized alpha/beta hydrolase domain) was the highest (~69%), followed by lower fraction of UhpC superfamily domain (~10%) (Supplementary Fig. 4). A considerable fraction (~62%) of potential T6SE sequences was found to contain at least one transmembrane region inside based on the TMHMM program results, in which the number of proteins with 3 TM domains received the highest proportion (~36.4%) (Supplementary Table 5). The results are in agreement with recent findings about domains associated with MIX motif - containing type 6 effector candidates (Cianfanelli et al., 2016; Flaugnatti et al., 2016; Salomon et al., 2014).
In conclusion, we focused on the detection and characterization of T6SS clusters and potential T6SEs at genome-scale in plant-associated pathogenic and beneficial Burkholderia bacteria based on computational analyses. 66 potential functional type VI systems were found in 30 collected Burkholderia strains, showing the higher number of the systems for plant pathogens compared with that of plant beneficial bacteria. Furthermore, the phylogenetic trees of important components (i.e., TssC, TssD, TssL) showed the close relationship of species that have close lifestyles, besides the distribution based on specific subtypes of T6SS clusters. Also 322 potential T6SEs in the analyzed genomes based on the homology search method were identified and their important conserved domains were explored. In addition, using the screening approach based on profile hidden Markov models of such MIX T6SEs, 57 proteins that were not included in training datasets were recognized as novel MIX T6SE candidates in Burkholderia species. Our findings of potential T6SS along with its effectors in Burkholderia species could provide useful hinders contributing to managing the inter-bacterial interactions of this genus, in order to repress the infection of phytopathogens on their host plants through encouraging the competitive ability of beneficial bacterial strains in the same niches.
Acknowledgments
This work was supported by grants from the Strategic Initiative for Microbiomes in Agriculture and Food, Ministry of Agriculture, Food and Rural Affairs, Republic of Korea (No. 916009021SB010), and from the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2016R1D1A1B03930678).
Fig. 1
Distribution of putative T6SEs and annotated T6SS clusters. The number of T6SS loci ranges from 1 to 5, existing in all 30 analyzed genomes, and the number of T6SE candidates ranges from 4 to 20 in which the mean numbers of type VI effectors for three groups of plant-associated pathogenic bacteria, plant-associated beneficial bacteria, environmental bacteria and/or biocontrol agent were 14.5, 7.1, and 11.6, respectively.
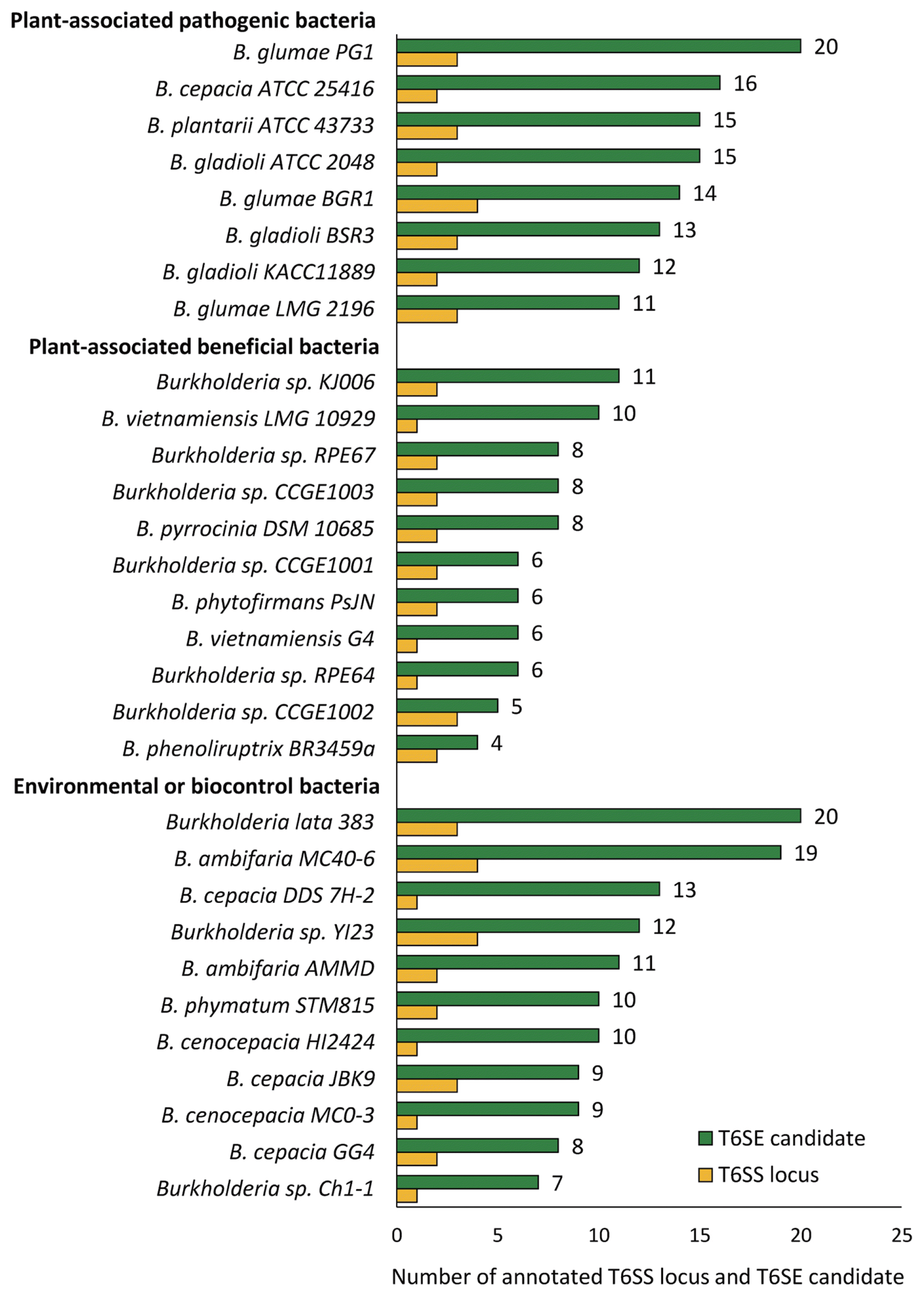
Fig. 3
Phylogenic tree of 66 MIX T6SE candidates detected in Burkholderia bacterial species. The tree was constructed in MEGA6 using the Neighbor-Joining method with 1000 bootstrap replicates. Branches corresponding to partitions reproduced in less than 50% bootstrap replicates were collapsed. All positions with less than 95% site coverage were eliminated. The evolutionary distances were computed using the JTT matrix-based method and are in the units of the number of amino acid substitutions per site. Two main groups were observed on the inferred tree; group I covered most proteins inferred by one or more models of pHMM1-4 and group II covered 20 proteins that were all recognized by only pHMM5.
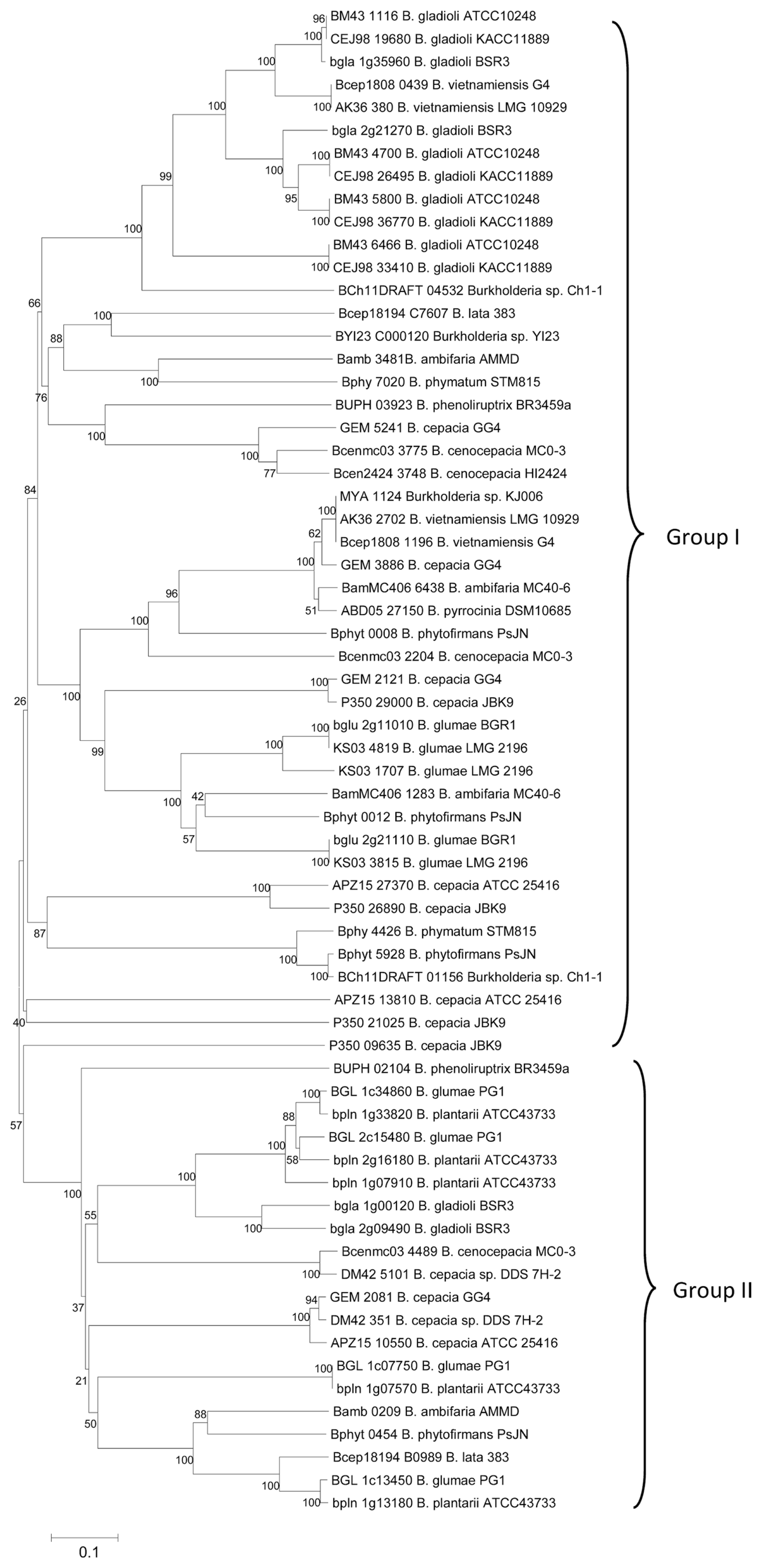
Table 1
Distribution of T6SS loci with at least 10 components identified in Burkholderia species genomes
Table 2
Putative MIX T6SE proteins detected from the genomes of Burkholderia species
| Bacterial strain | Locus_tag | Detection model | Specify |
|---|---|---|---|
| B. ambifaria AMMD | Bamb_0209 | pHMM5 | Non-training |
| B. ambifaria AMMD | Bamb_3481 | pHMM1-4 | Non-training |
| B. ambifaria MC40-6 | BamMC406_1283 | pHMM1-4 | Non-training |
| B. ambifaria MC40-6 | BamMC406_6438 | pHMM2-4 | Non-training |
| B. cenocepacia HI2424 | Bcen2424_3748 | pHMM1,2,3 | Non-training |
| B. cenocepacia MC0-3 | Bcenmc03_3775 | pHMM1-4 | Non-training |
| B. cenocepacia MC0-3 | Bcenmc03_4489 | pHMM5 | Non-training |
| B. cenocepacia MC0-3 | Bcenmc03_2204 | pHMM2-4 | Training |
| B. cepacia ATCC 25416 | APZ15_13810 | pHMM1,3 | Non-training |
| B. cepacia ATCC 25416 | APZ15_27370 | pHMM2-4 | Non-training |
| B. cepacia ATCC 25416 | APZ15_10550 | pHMM5 | Non-trainingH |
| B. cepacia GG4 | GEM_2121 | pHMM2-4 | Non-training |
| B. cepacia GG4 | GEM_3886 | pHMM2-4 | Non-training |
| B. cepacia GG4 | GEM_5241 | pHMM1-4 | Non-training |
| B. cepacia GG4 | GEM_2081 | pHMM5 | Non-trainingH |
| B. cepacia JBK9 | P350_09635 | pHMM1,3,4 | Non-training |
| B. cepacia JBK9 | P350_21025 | pHMM2-4 | Non-training |
| B. cepacia JBK9 | P350_26890 | pHMM2-4 | Non-training |
| B. cepacia JBK9 | P350_29000 | pHMM2-4 | Non-training |
| B. cepacia sp. DDS 7H-2 | DM42_5101 | pHMM5 | Non-training |
| B. cepacia sp. DDS 7H-2 | DM42_351 | pHMM5 | Non-trainingH |
| B. gladioli ATCC10248 | BM43_1116 | pHMM2-4 | Non-training |
| B. gladioli ATCC10248 | BM43_4700 | pHMM2-4 | Non-training |
| B. gladioli ATCC10248 | BM43_5800 | pHMM2-4 | Non-training |
| B. gladioli ATCC10248 | BM43_6466 | pHMM2-4 | Non-training |
| B. gladioli BSR3 | bgla_1g00120 | pHMM5 | Non-training |
| B. gladioli BSR3 | bgla_1g35960 | pHMM2-4 | Non-training |
| B. gladioli BSR3 | bgla_2g09490 | pHMM5 | Non-training |
| B. gladioli BSR3 | bgla_2g21270 | pHMM2-4 | Training |
| B. gladioli KACC11889 | CEJ98_19680 | pHMM2-4 | Non-training |
| B. gladioli KACC11889 | CEJ98_33410 | pHMM2-4 | Non-training |
| B. gladioli KACC11889 | CEJ98_36770 | pHMM2-4 | Non-training |
| B. gladioli KACC11889 | CEJ98_26495 | pHMM2-4 | Non-training |
| B. glumae BGR1 | bglu_2g11010 | pHMM2-4 | Non-training |
| B. glumae BGR1 | bglu_2g21110 | pHMM1-4 | Training |
| B. glumae LMG 2196 | KS03_3815 | pHMM1,3,4 | Non-training |
| B. glumae LMG 2196 | KS03_4819 | pHMM2-4 | Non-training |
| B. glumae LMG 2196 | KS03_1707 | pHMM1,3 | Non-training |
| B. glumae PG1 | BGL_1c07750 | pHMM5 | Non-training |
| B. glumae PG1 | BGL_1c13450 | pHMM5 | Non-training |
| B. glumae PG1 | BGL_1c34860 | pHMM5 | Non-training |
| B. glumae PG1 | BGL_2c15480 | pHMM5 | Non-training |
| B. lata 383 | Bcep18194_B0989 | pHMM5 | Non-training |
| B. lata 383 | Bcep18194_C7607 | pHMM1-4 | Training |
| B. phenoliruptrix BR3459a | BUPH_02104 | pHMM5 | Non-training |
| B. phenoliruptrix BR3459a | BUPH_03923 | pHMM2-4 | Training |
| B. phymatum STM815 | Bphy_4426 | pHMM1 | Non-training |
| B. phymatum STM815 | Bphy_7020 | pHMM1-4 | Training |
| B. phytofirmans PsJN | Bphyt_0008 | pHMM2-4 | Non-training |
| B. phytofirmans PsJN | Bphyt_0012 | pHMM2-4 | Non-training |
| B. phytofirmans PsJN | Bphyt_0454 | pHMM5 | Non-training |
| B. phytofirmans PsJN | Bphyt_5928 | pHMM1 | Non-training |
| B. plantarii ATCC43733 | bpln_1g07570 | pHMM5 | Non-training |
| B. plantarii ATCC43733 | bpln_1g07910 | pHMM5 | Non-training |
| B. plantarii ATCC43733 | bpln_1g13180 | pHMM5 | Non-training |
| B. plantarii ATCC43733 | bpln_1g33820 | pHMM5 | Non-training |
| B. plantarii ATCC43733 | bpln_2g16180 | pHMM5 | Non-training |
| B. pyrrocinia DSM10685 | ABD05_27150 | pHMM2-4 | Non-training |
| B. vietnamiensis G4 | Bcep1808_0439 | pHMM2-4 | Training |
| B. vietnamiensis G4 | Bcep1808_1196 | pHMM2-4 | Training |
| B. vietnamiensis LMG 10929 | AK36_2702 | pHMM2-4 | Non-training |
| B. vietnamiensis LMG 10929 | AK36_380 | pHMM2-4 | Non-training |
| Burkholderia sp. Ch1-1 | BCh11DRAFT_01156 | pHMM1 | Non-training |
| Burkholderia sp. Ch1-1 | BCh11DRAFT_04532 | pHMM2-4 | Training |
| Burkholderia sp. KJ006 | MYA_1124 | pHMM2-4 | Non-training |
| Burkholderia sp. YI23 | BYI23_C000120 | pHMM2,3,4 | Non-training |
References
Abby, SS, Cury, J, Guglielmini, J, Néron, B, Touchon, M and Rocha, EPC 2016. Identification of protein secretion systems in bacterial genomes. Sci Rep. 6:23080



Abby, SS and Rocha, EPC 2017. Identification of protein secretion systems in bacterial genomes using MacSyFinder. Methods Mol Biol. 1615:1-21.


Alfano, JR and Collmer, A 2004. Type III secretion system effector proteins: double agents in bacterial disease and plant defense. Annu Rev Phytopathol. 42:385-414.


Altschul, SF, Gish, W, Miller, W, Myers, EW and Lipman, DJ 1990. Basic local alignment search tool. J Mol Biol. 215:403-410.


Barret, M, Egan, F, Fargier, E, Morrissey, JP and O’Gara, F 2011. Genomic analysis of the type VI secretion systems in Pseudomonas spp.: novel clusters and putative effectors uncovered. Microbiology. 157:1726-1739.


Bendtsen, JD, Jensen, LJ, Blom, N, Von Heijne, G and Brunak, S 2004. Feature-based prediction of non-classical and leaderless protein secretion. Protein Eng Des Sel. 17:349-356.


Bernal, P, Allsopp, LP, Filloux, A and Llamas, MA 2017a. The Pseudomonas putida T6SS is a plant warden against phytopathogens. ISME J. 11:972-987.


Bernal, P, Llamas, MA and Filloux, A 2017b Type VI secretion systems in plant-associated bacteria. Environ Microbiol. (in press).

Bernard, CS, Brunet, YR, Gueguen, E and Cascales, E 2010. Nooks and crannies in Type VI secretion regulation. J Bacteriol. 192:3850-3860.



Bingle, LE, Bailey, CM and Pallen, MJ 2008. Type VI secretion: a beginner’s guide. Curr Opin Microbiol. 11:3-8.


Bladergroen, MR, Badelt, K and Spaink, HP 2003. Infection-blocking genes of a symbiotic Rhizobium leguminosarum strain that are involved in temperature-dependent protein secretion. Mol Plant-Microbe Interact. 16:53-64.


Broms, JE, Meyer, L, Sun, K, Lavander, M and Sjostedt, A 2012. Unique substrates secreted by the type VI secretion system of Francisella tularensis during intramacrophage infection. PLoS One. 7:20

Burtnick, MN, Brett, PJ, Harding, SV, Ngugi, SA, Ribot, WJ, Chantratita, N, Scorpio, A, Milne, TS, Dean, RE, Fritz, DL, Peacock, SJ, Prior, JL, Atkins, TP and DeShazer, D 2011. The cluster 1 Type VI secretion system is a major virulence determinant in Burkholderia pseudomallei. Infect Immun. 79:1512-1525.



Cascales, E and Cambillau, C 2012. Structural biology of type VI secretion systems. Philos Trans R Soc Lond B Biol Sci. 367:1102-1111.



Cianfanelli, FR, Monlezun, L and Coulthurst, SJ 2016. Aim, load, fire: the type VI secretion system, a bacterial nanoweapon. Trends Microbiol. 24:51-62.


Coenye, T and Vandamme, P 2003. Diversity and significance of Burkholderia species occupying diverse ecological niches. Environ Microbiol. 5:719-729.


Compant, S, Nowak, J, Coenye, T, Clement, C and Ait Barka, E 2008. Diversity and occurrence of Burkholderia spp. in the natural environment. FEMS Microbiol Rev. 32:607-626.


de Bruin, OM, Duplantis, BN, Ludu, JS, Hare, RF, Nix, EB, Schmerk, CL, Robb, CS, Boraston, AB, Hueffer, K and Nano, FE 2011. The biochemical properties of the Francisella pathogenicity island (FPI)-encoded proteins IglA, IglB, IglC, PdpB and DotU suggest roles in type VI secretion. Microbiology. 157:3483-3491.



De Maayer, P, Venter, SN, Kamber, T, Duffy, B, Coutinho, TA and Smits, THM 2011. Comparative genomics of the type VI secretion systems of Pantoea and Erwinia species reveals the presence of putative effector islands that may be translocated by the VgrG and Hcp proteins. BMC Genomics. 12:576-576.



Edgar, RC 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792-1797.



Santos, P, Vinuesa, P, Martinez-Aguilar, L, Hirsch, AM and Caballero-Mellado, J 2013. Phylogenetic analysis of burkholderia species by multilocus sequence analysis. Curr Microbiol. 67:51-60.


Filloux, A, Hachani, A and Bleves, S 2008. The bacterial type VI secretion machine: yet another player for protein transport across membranes. Microbiology. 154:1570-1583.


Finn, RD, Clements, J and Eddy, SR 2011. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39:W29-W37.



Flaugnatti, N, Le, TT, Canaan, S, Aschtgen, MS, Nguyen, VS, Blangy, S, Kellenberger, C, Roussel, A, Cambillau, C, Cascales, E and Journet, L 2016. A phospholipase A1 antibacterial Type VI secretion effector interacts directly with the C-terminal domain of the VgrG spike protein for delivery. Mol Microbiol. 99:1099-1118.


Hachani, A, Wood, TE and Filloux, A 2016. Type VI secretion and anti-host effectors. Curr Opin Microbiol. 29:81-93.


Ham, JH, Melanson, RA and Rush, MC 2011. Burkholderia glumae: next major pathogen of rice? Mol Plant Pathol. 12:329-339.


Jeong, Y, Kim, J, Kim, S, Kang, Y, Nagamatsu, T and Hwang, I 2003. Toxoflavin produced by Burkholderia glumae causing rice grain rot is responsible for inducing bacterial wilt in many field crops. Plant Dis. 87:890-895.


Jiang, F, Waterfield, N, Yang, J, Yang, G and Jin, Q 2014. A Pseudomonas aeruginosa type VI secretion phospholipase D effector targets both prokaryotic and eukaryotic cells. Cell Host Microbe. 15:600-610.


Jones, DT, Taylor, WR and Thornton, JM 1992. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 8:275-282.


Krogh, A, Larsson, B, von Heijne, G and Sonnhammer, EL 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 305:567-580.


Li, J, Yao, Y, Xu, HH, Hao, L, Deng, Z, Rajakumar, K and Ou, H-Y 2015. SecReT6: a web-based resource for type VI secretion systems found in bacteria. Environ Microbiol. 17:2196-2202.


Ma, J, Sun, M, Bao, Y, Pan, Z, Zhang, W, Lu, C and Yao, H 2013. Genetic diversity and features analysis of type VI secretion systems loci in avian pathogenic Escherichia coli by wide genomic scanning. Infect Genet Evol. 20:454-464.


Marchler-Bauer, A, Lu, S, Anderson, JB, Chitsaz, F, Derbyshire, MK, DeWeese-Scott, C, Fong, JH, Geer, LY, Geer, RC, Gonzales, NR, Gwadz, M, Hurwitz, DI, Jackson, JD, Ke, Z, Lanczycki, CJ, Lu, F, Marchler, GH, Mullokandov, M, Omelchenko, MV, Robertson, CL, Song, JS, Thanki, N, Yamashita, RA, Zhang, D, Zhang, N, Zheng, C and Bryant, SH 2011. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 39:D225-D229.


Mitter, B, Petric, A, Shin, MW, Chain, PS, Hauberg-Lotte, L, Reinhold-Hurek, B, Nowak, J and Sessitsch, A 2013. Comparative genome analysis of Burkholderia phytofirmans PsJN reveals a wide spectrum of endophytic lifestyles based on interaction strategies with host plants. Front Plant Sci. 4:120



Mougous, JD, Cuff, ME, Raunser, S, Shen, A, Zhou, M, Gifford, CA, Goodman, AL, Joachimiak, G, Ordonez, CL, Lory, S, Walz, T, Joachimiak, A and Mekalanos, JJ 2006. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science. 312:1526-1530.



Petersen, TN, Brunak, S, von Heijne, G and Nielsen, H 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 8:785-786.


Pukatzki, S, Ma, AT, Sturtevant, D, Krastins, B, Sarracino, D, Nelson, WC, Heidelberg, JF and Mekalanos, JJ 2006. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc Natl Acad Sci USA. 103:1528-1533.



Rao, PS, Yamada, Y, Tan, YP and Leung, KY 2004. Use of proteomics to identify novel virulence determinants that are required for Edwardsiella tarda pathogenesis. Mol Microbiol. 53:573-586.


Russell, AB, LeRoux, M, Hathazi, K, Agnello, DM, Ishikawa, T, Wiggins, PA, Wai, SN and Mougous, JD 2013. Diverse type VI secretion phospholipases are functionally plastic antibacterial effectors. Nature. 496:508-512.



Russell, AB, Peterson, SB and Mougous, JD 2014a. Type VI secretion system effectors: poisons with a purpose. Nat Rev Microbiol. 12:137-148.


Russell, AB, Singh, P, Brittnacher, M, Bui, NK, Hood, RD, Carl, MA, Agnello, DM, Schwarz, S, Goodlett, DR, Vollmer, W and Mougous, JD 2012. A widespread bacterial type VI secretion effector superfamily identified using a heuristic approach. Cell Host Microbe. 11:538-549.



Russell, AB, Wexler, AG, Harding, BN, Whitney, JC, Bohn, AJ, Goo, YA, Tran, BQ, Barry, NA, Zheng, H, Peterson, SB, Chou, S, Gonen, T, Goodlett, DR, Goodman, AL and Mougous, JD 2014b. A type VI secretion-related pathway in Bacteroidetes mediates interbacterial antagonism. Cell Host Microbe. 16:227-236.


Ryu, C-M 2015. Against friend and foe: type 6 effectors in plant-associated bacteria. J Microbiol. 53:201-208.


Saitou, N and Nei, M 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 4:406-425.

Salomon, D, Kinch, LN, Trudgian, DC, Guo, X, Klimko, JA, Grishin, NV, Mirzaei, H and Orth, K 2014. Marker for type VI secretion system effectors. Proc Natl Acad Sci USA. 111:9271-9276.



Schell, MA, Ulrich, RL, Ribot, WJ, Brueggemann, EE, Hines, HB, Chen, D, Lipscomb, L, Kim, HS, Mrazek, J, Nierman, WC and Deshazer, D 2007. Type VI secretion is a major virulence determinant in Burkholderia mallei. Mol Microbiol. 64:1466-1485.


Schwarz, S, West, TE, Boyer, F, Chiang, W-C, Carl, MA, Hood, RD, Rohmer, L, Tolker-Nielsen, T, Skerrett, SJ and Mougous, JD 2010. Burkholderia type VI secretion systems have distinct roles in eukaryotic and bacterial cell interactions. PLoS Pathog. 6:e1001068



Silverman, JM, Austin, LS, Hsu, F, Hicks, KG, Hood, RD and Mougous, JD 2011. Separate inputs modulate phosphorylation-dependent and -independent type VI secretion activation. Mol Microbiol. 82:1277-1290.



Solis, R, Bertani, I, Degrassi, G, Devescovi, G and Venturi, V 2006. Involvement of quorum sensing and RpoS in rice seedling blight caused by Burkholderia plantarii. FEMS Microbiol Lett. 259:106-112.


Tamura, K, Stecher, G, Peterson, D, Filipski, A and Kumar, S 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725-2729.



Thompson, JD, Higgins, DG and Gibson, TJ 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680.



Tseng, T-T, Tyler, BM and Setubal, JC 2009. Protein secretion systems in bacterial-host associations, and their description in the Gene Ontology. BMC Microbiol. 9:S2



- TOOLS
-
METRICS

- Related articles
-
RNAseq-based Transcriptome Analysis of
Burkholderia glumae Quorum Sensing2013 September;29(3)



 PDF Links
PDF Links PubReader
PubReader Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Supplement
Supplement Print
Print



