AltMV TGB1 Nucleolar Localization Requires Homologous Interaction and Correlates with Cell Wall Localization Associated with Cell-to-Cell Movement
Article information
Abstract
The Potexvirus Alternanthera mosaic virus (AltMV) has multifunctional triple gene block (TGB) proteins, among which our studies have focused on the properties of the TGB1 protein. The TGB1 of AltMV has functions including RNA binding, RNA silencing suppression, and cell-to-cell movement, and is known to form homologous interactions. The helicase domains of AltMV TGB1 were separately mutated to identify which regions are involved in homologous TGB1 interactions. The yeast two hybrid system and Bimolecular Fluorescence Complementation (BiFC) in planta were utilized to examine homologous interactions of the mutants. Helicase motif I of AltMV TGB1 was found to be critical to maintain homologous interactions. Mutations in the remaining helicase motifs did not inhibit TGB1 homologous interactions. In the absence of homologous interaction of TGB1, subcellular localization of helicase domain I mutants showed distinctively different patterns from that of WT TGB1. These results provide important information to study viral movement and replication of AltMV.
Among plant viruses the Potexvirus group has been a very good model for investigating viral movement and replication. The approximately 6.5 kb Potexvirus genome has five open reading frames encoding the replicase, triple gene block (TGB) proteins, and coat protein, respectively. The replicase is a single protein that contains methyltransferase, RNA helicase and RNA polymerase domains (Verchot-Lubicz et al., 2007). Potato virus X (PVX) TGB1 has been reported to be a suppressor of RNA silencing (Bayne et al., 2005; Voinnet et al., 2000), and to block systemic spread of the silencing signal. The role of the Potexvirus TGB proteins in viral movement was first elucidated for White clover mosaic virus (WClMV), in which it was determined that mutation in any of the TGB genes of an infectious clone permitted replication of the virus in protoplasts, but negated cell-to-cell spread in intact plants (Beck et al., 1991). The TGB proteins are presumed to be expressed from two 3′ co-terminal subgenomic RNAs (sgRNAs); one sgRNA for TGB1, and a di-cistronic sgRNA for TGB2 and TGB3. TGB3 may be expressed at a lower level than TGB2 as a result of leaky ribosome scanning through the TGB2 gene (Morozov and Solovyev, 2003).
The Potexvirus TGB1 serves several functions in cell to cell movement, including RNA helicase activity, silencing suppression, and mediating an increase in the plasmodesmata (PD) size exclusion limit (Verchot-Lubicz et al., 2007). TGB1 of PVX has been reported to be associated with both cytoplasmic and nuclear curved lamellar inclusions in the form of proteinaceous sheets with attached electron dense beads; these structures were not associated with plasmodesmata (PD), and were not labeled by antibodies raised against virions (Davies et al., 1993); fibrillar TGB1 inclusions of Foxtail mosaic virus (FMV) were localized only to the cytoplasm, and were frequently associated with viral aggregates, and occasionally with the cell wall (Rouleau et al., 1994). In contrast, Bamboo mosaic virus (BaMV) TGB1 forms angular electron dense crystalline bodies in both the cytoplasm and the nucleus, that were labeled by virus-specific antibodies early in infection in cells where virions were also visible, but not later in the infection cycle (Lin and Chen, 1991).
The recently isolated Alternanthera mosaic virus (AltMV) has several distinctive properties compared to other potexviruses. In particular, the TGB proteins of AltMV provide more information to study Potexvirus movement, replication, and gene silencing suppressor function. The AltMV TGB1 protein has a size (26 kDa) similar to those of other potexviruses (Verchot-Lubicz et al., 2007), and has also been demonstrated to function as a silencing suppressor (Lim et al., 2010b; Lim et al., 2010c). The TGB2 (12 kDa), TGB3 (7 kDa), and coat protein (22 kDa) are also similar in size to those of other potexviruses (Hammond et al., 2006; Verchot-Lubicz et al., 2007). However, TGB1 and TGB3 of AltMV have unique functions for replication and movement. First, AltMV TGB1 has strong or weak silencing suppressor function depending on whether the 88th amino acid is leucine or proline (Lim et al., 2010b; Lim et al., 2010c). Among naturally existing AltMV quasispecies, a TGB188P with a weak silencing suppressor was selected and evaluated, which revealed that silencing suppressor function efficiency was dependent on localization to the nucleolus (Lim et al., 2010b). Secondly, viral movement from the epidermal layer to the mesophyll required an additional function with assistance of AltMV TGB3 (Lim et al., 2010a).
In this paper, we have investigated the properties of AltMV TGB1 protein and also examined the subcellular localization patterns of TGB1 fusion proteins expressed in Nicotiana benthamiana leaf cells by agroinfiltration, and identified some differences between AltMV and other characterized potexviruses. AltMV TGB1 did not localize to the cell wall in the absence of TGB1 homologous interaction, and the helicase I of AltMV TGB1 related to homologous interaction was found to be required for silencing suppressor activity. This report provides a detailed analysis of the TGB1 depending on specific amino acids which affect silencing suppressor function while maintaining cell to cell movement and homologous interactions. Through our experiments, we found that there were relationships between nuclear localization, cell wall localization and gene silencing suppressor function which correlated with TGB1 homologous interactions.
In the genus Potexvirus the triple gene block proteins have been targets for investigating self-interaction and host protein interactions. To identify the crucial region for homologous interaction of AltMV TGB1, the amino acid sequence of AltMV TGB1 (Hammond et al., 2006) was compared to the homologous sequence of PVX TGB1 by the CLUSTALW program within the Biology Workbench package (http://workbench.sdsu.edu). As indicated in the PVX TGB1 sequence, seven conserved motifs of the helicase domain were identified in AltMV TGB1 (Fig. 1).

Alternanthera mosaic virus (AltMV) TGB1 helicase domain alignment. CLUSTALW alignment of AltMV TGB1 and PVX TGB1 showing helicase domains identified in PVX (Morosov et al., 1999).
It has been previously reported that there is homologous interaction in AltMV TGB1 (Lim et al., 2010c), and here we have tested how gene silencing suppressor function is related to homologous TGB1 interactions. A previous report (Leshchiner et al., 2006) indicated that helicase domain I was critical for PVX TGB1 homologous interactions, so we designed mutant derivatives from domain I, II, and III of the AltMV TGB1, and we cloned these in pGAD and pGDBU. Yeast two-hybrid vectors (James et al., 1996), were designed to produce fusions of either the Gal4 activation domain (AD) or the Gal4 binding domain (Abdel-Ghany et al., 2005) to the N-terminus of the CP and TGB proteins to be tested for interactions. To construct fusions to the AD (pGAD-TGB1) and the BD (pGBDU-TGB1, pGBDU-TGB1G31R, pGBDU-TGB1L88P, pGBDU-TGB1D81R) were amplified using the TGB1 specific primers (TGB1 F; 5′GAG GGA TCC ATG AAT CAC TTT ACT AAC C-3′, TGB1 R; 5′-GAG CTG CAG CCC GAG TAA TCG GCG GGG-3′, restriction sites are underlined). The TGB1 PCR products were introduced separately between the BamHI and PstI sites of both plasmid pGAD, and of plasmid pGBDU. Yeast two hybrid assay with AD:TGB1 and BD: TGB1G31R or BD:TGB1GK33/34RR exhibited no growth within 40 hrs, indicating no interactions between helicase domain I mutant derivatives of TGB1. None of the derivatives of TGB1 helicase domain II and III showed failure to interact, indicating that A1tMV TGB1 also required helicase domain I for homologous interactions (Fig. 2A).

Helicase domain I is a crucial region for self-interaction and maintenance of silencing suppression activity. (A) Yeast two hybrid assay with AltMV TGB1 mutant derivatives, demonstrating loss of interaction between helicase I mutants. (B) Co-infiltration with smGFP to assay silencing suppressor function of TGB1 and its derivatives. TGB1L88, TGB1P88, TGB1G31R, TGB1D81R, TGB1Q101R, TGB1GK33/34RR, and TGB1G88 cloned in pGD binary vector were coinfiltrated with pGD:smGFP. Fluorescence was captured at 2 days post infiltration.
At the same time all derivatives mutated in helicase domain I, II, and III were cloned into pGD vector for agroinfiltration with smGFP to be analyzed for silencing suppressor function. A smGFP silencing suppression assay was used as previously described (Bragg and Jackson, 2004; Lim et al., 2010c). Because 35S promoter-controlled GFP induces silencing of GFP expression (Shiboleth et al., 2007; Silhavy and Burgyan, 2004), we used only pGD: smGFP co-infiltrated with the protein being assayed for silencing suppressor function. As shown in Figure 2B, in the absence of homologous interaction, the silencing suppressor function was diminished.
In order to confirm the homologous interactions, bimolecular fluorescence complementation (BiFC) assays were performed using pSPYCE(MR):TGB1 infiltrated with pSPYNE(R)173:TGB1 mutant derivatives or pSPYNE(R)173: TGB1 (wild type; WT). Constructs were agroinfiltrated essentially as previously described, in the presence of pGD: p19 (Lim et al., 2008), and enhanced yellow fluorescent protein (eYFP) fluorescence was observed at 2 days post agroinfiltration (dpa) by laser-scanning confocal microscopy (LSCM). The BiFC results confirmed that the helicase domain is necessary for TGB1 homologous interactions. The constructs of TGB1L88, TGB1P88, TGB1D81R, and TGB1Q101R yielded positive interaction, and TGB1GK33/34RR did not interact (Figure 3 and data not shown).
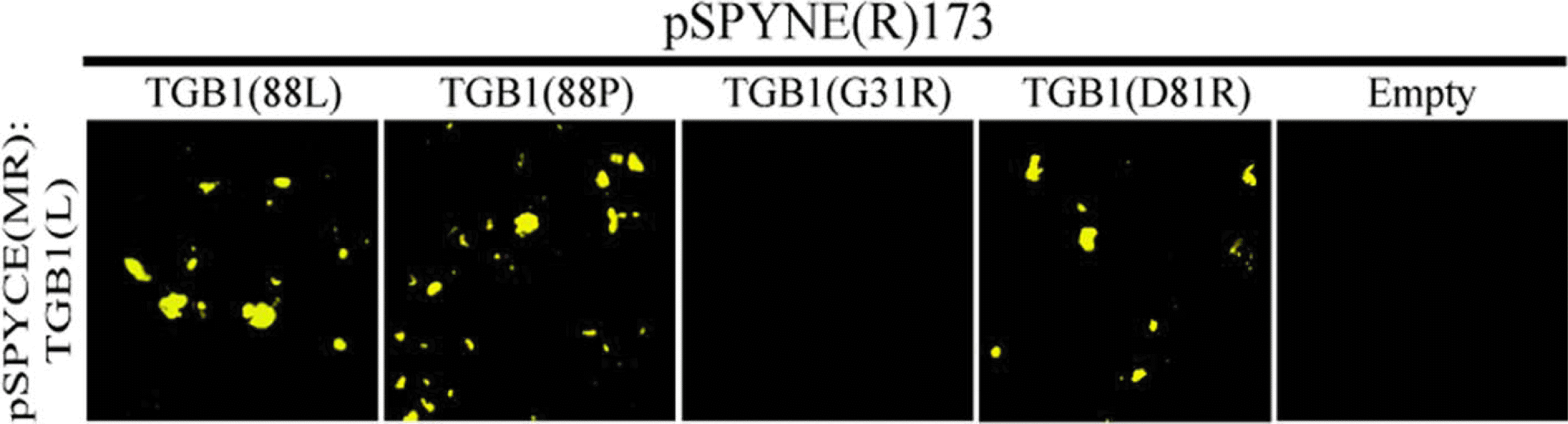
Homologous interaction of AltMV TGB1 confirmed by Bimolecular fluorescence complementation (BiFC) assay. SPYNE(R)173:AltMV TGB1(L) was co-infiltrated with SPYCE(MR)::TGB1(L), TGB1(P), TGB1(G31R), TGB1(GK33/34RR), TGB1(D81R), TGB1(Q101R) and empty SPYCE(MR) respectively through Agrobacterium. The image was captured by LSCM at 2 days post-infiltration (TGB1(GK33/34) lack of interaction and TGB1(Q101R) positive interaction, results not shown).
The TGB1 protein of potexviruses has the functions of ATPase activity, RNA binding, and cell to cell movement (Lin et al., 2004; Liou et al., 2000; Wung et al., 1999). The helicase domains of TGB1 have also been reported to be multifunctional for homologous interaction, RNA helicase activity, ATP and Mg2+ binding, and NTPase activities (Verchot-Lubicz et al., 2010). We have previously demonstrated that leucine residues between helicase domains II and III of AltMV, PVX, and the NMV isolate of Plantago asiatica mosaic virus affect the efficiency of RNA silencing suppression activity (Lim et al., 2010b, c). Results presented here show that AltMV TGB1 self-interaction is also important to maintain silencing suppression function, in addition to confirming reports with other potexviruses that helicase domain I is required for TGB1 homologous interaction.
TGB1 helicase domain I is required for TGB1 self-interaction. Therefore, we wanted to know how TGB1 subcellular localization was affected in the absence of homologous interaction. In order to demonstrate change of localization of TGB1 according to site of the mutations of helicase domains of TGB1, we cloned TGB1 derivatives into pGDG vector to examine sub-cellular localization using agroinfiltration, as previously examined with AltMV TGB3 (Lim et al., 2010a). The naturally-occurring variant TGB1L88G was also cloned into pGDG. Variants TGB1L88G, TGB1G31R, TGB1GK33/34RR, TGB1D81R, and TGB1Q101R were separately cloned into the binary vector pGDG for agroinfiltration, and subcellular localization at the nucleus and cell wall was visualized by laser scanning confocal microscopy (LSCM) within 48 h of infiltration. According to our previous investigations related to TGB1 nuclear localization, TGB1 amino acid 88 is very important for nucleolar localization of TGB1 (Lim et al., 2010b). Mutant TGB1L88G showed nuclear but not nucleolar localization, which further confirmed the importance of leucine at position 88 for nucleolar localization (Figure 4A). Depending on the presence of L or P at position 88, TGB1 localized at the nucleolus and/or in the nuclear periplasm (Lim et al., 2010b), however in the absence of homologous interaction of TGB1, different patterns of nuclear localization were observed which were not found with WT TGB1 or other mutant derivatives (Figure 4A). There was no nucleolar or nuclear periplasm localization of TGB1 derivatives lacking homologous interaction, and TGB1 fluorescence was instead distributed uniformly throughout the nucleus. In addition, the cell wall of plasmolyzed cells stained with calcofluor blue (Sigma-Aldrich Chemical, st, Louis, MO) was visualized together with GFP:TGB1 and its derivatives (Figure 4A). WT AltMV TGB1 localized at the cell wall in the absence of TGB2 and TGB3, however TGB1 cell wall localization was not detected for TGB1 variants lacking self-nteraction (Figure 4A). Interestingly, nucleolar localization of TGB1L88 correlated with enhanced TGB1 cell wall localization (Figure 4A, B). TGB1 fusion protein expression from the binary vector was confirmed by western blot using a GFP-specific antibody, and detection of an about 50 kDa GFP-reactive band in all of the constructs confirmed expression of the various GFP:TGB1 mutant fusions (Figure 4C).
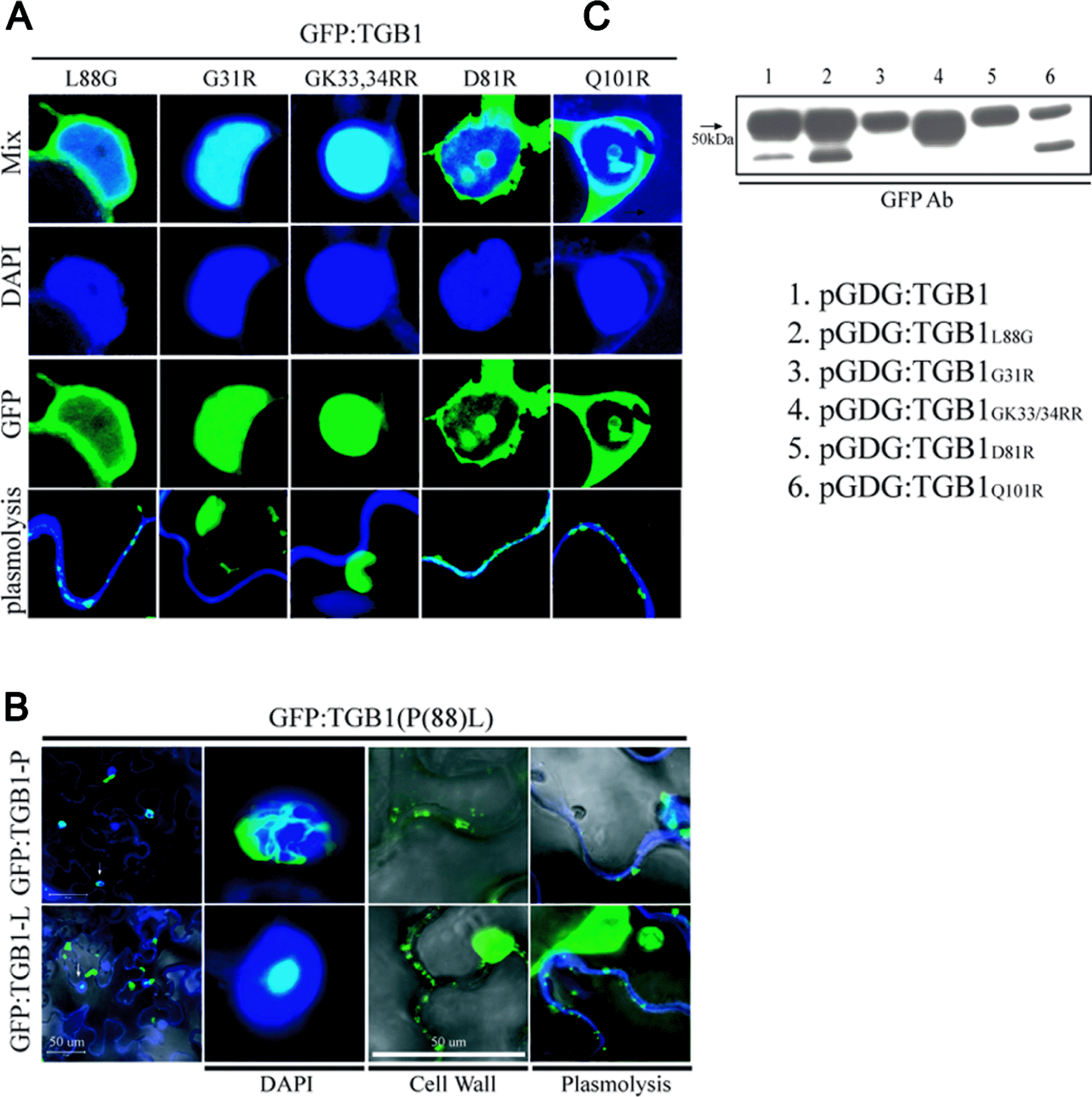
AltMV TGB1 derivatives showed different patterns of nuclear and cell wall localization. (A) GFP:TGB1L88G, GFP:TGB1G31R, GFP:TGB1GK33/34RR, GFP:TGB1D81R, and GFP:TGB1Q101R were expressed in N. benthamiana leaves via agroinfiltration, and laser-scanning confocal microscopy images of epidermal cells were captured at 2−3 days post infiltration. To stain the nucleus DAPI solution (blue) was used. Cells were treated with 0.7 M sucrose for 24 h to induce plasmolysis and cell walls were stained with calcofluor blue. (B) GFP:TGB1L88 and GFP:TGB1P88 were compared for cell wall localization. The fluorescent signal is overlaid onto differential interference contrast (DIC) images. Bars = 50 μm. (C) Purified protein from Agroinfiltrated region in N. benthamiana were separated by electrophoresis in 12% SDS-PAGE gel, blotted to PVDF membrane, and reacted with GFP antibody.
Previously, Lim et al. (Lim et al., 2009) reported that TGB1 of a hordeivirus could localize at the cell wall in the absence of self-interaction. However, in contrast to hordeiviruses, TGB1 of the potexvirus AltMV needs self-interaction for cell wall localization.
Through subcellular localization of TGB1 and its derivatives, including helicase domain I mutants, we determined that TGB1 nucleolar localization may also be correlated to cell wall localization. In comparison, the ORF3 protein of Groundnut rosette virus (GRV) localized at the nucleolus, and a mutant failing to enter the nucleolus resulted in failure of long distance movement (Kim et al., 2007). Similarly, nucleolar localization of AltMV TGB1 was correlated with increased TGB1 binding at the cell wall. In the absence of TGB1 self-interaction, nuclear localization of mutant TGB1 showed quite a different pattern compared to the wild type, and cell wall localization was not detected at all. This suggests that the nucleolar localization utilizes the same signals as for targeting to the cell wall, or at least that localization to both requires the formation of dimers. In case of TGB1P88, nuclear but not nucleolar localization is observed; however AltMV expressing TGB1P88 still moves systemically (Lim et al., 2010a), which is not equivalent to the result from GRV, in which systemic movement was abolished when nucleolar localization was lost (Kim et al., 2007). Without nucleolar localization, TGB1P88 is still detected at the cell wall but at significantly lower proportion than for TGB1L88. It is assumed that AltMV TGB1P88 moves less efficiently from cell to cell due to the weak gene silencing suppressor function, and that nucleolar localization is somehow related to TGB1 binding at the cell wall. In most potexviruses, TGB1 is detected as paracrystalline inclusion bodies in the nucleus (Verchot-Lubicz et al., 2010) (Hammond et al unpublished), but it is not clear why TGB1 and inclusion bodies occur in the nucleus. Partial information as to why plant RNA virus specific protein enters the nucleolus is provided by the case of the ORF3 protein of GRV; ORF3 nucleolar import via Cajal bodies accompanies fibrillarin and RNP particles that are required for the long-distance spread of the virus and systemic infection (Canetta et al., 2008). Based on their findings, we need to test whether AltMV TGB1 self-interaction is necessary for further interaction with a nucleolar import protein like fibrillarin. With these data we could anticipate the relationship between TGB1 cell wall localization and nucleolar localization, and that self-interaction is required for binding a nucleolar import protein.
Acknowledgements
This work was supported by a grant from the NextGeneration BioGreen 21 program (No. PJ008063), Rural Development Administration, Republic of Korea.