In the natural environment, plants have developed elaborate mechanisms to protect themselves from various environmental stresses. The phytohormone abscisic acid (ABA) has an important roles in serving as a signal in response to environmental stimuli and triggering changes in a number of plant physiological and developmental processes, leading to adaptation to the stress conditions (Finkelstein et al., 2002; Robert-Seilaniantz et al., 2007; Ton et al., 2009).
ABA signaling involves protein phosphorylation and deposphorylation events, which were achieved by sucrose nonfermenting1-related subfamily 2 protein kinases (SnRK2) and clade A protein phosphatase type 2Cs (PP2Cs), respectively (Geiger et al., 2009; Lee et al., 2009; Vlad et al., 2009). Genetic evidence revealed that the snrk2.6 single mutant and snrk2.2/snrk2.3 double mutant were with an ABA-insensitive phenotype (Fujii et al., 2007; Mustilli et al., 2002). A triple mutant of these kinases exhibited severe phenotypes indicative of defects in ABA signaling and water stress responses (Fujii and Zhu, 2009; Fujita et al., 2009; Nakashima et al., 2009). SnRK2s play a role of positive regulators of ABA signaling pathway. If SnRK2s function as positive regulators of ABA signaling, Clade A PP2Cs are expected to act as negative regulators in the same pathway (Lee and Luan, 2012). Earlier genetic analysis provides crucial information about function of PP2Cs (Gosti et al., 1999; Merlot et al., 2001; Schweighofer et al., 2004; Wasilewska et al., 2008). The dominant mutants of abi1-1 and abi2-1 showed an ABA-insensitive phenotype, whereas mutants in HAB1, AHG1 and PP2CA exhibited hypersensitive to ABA in seed germination (Nishimura et al., 2004; Saez et al., 2004).
To initiate ABA signaling, ABA perception by ABA receptor must be necessary in plant cells. Several different types of ABA receptors have been identified, although the function of some receptors remains to be further studied (Christmann and Grill, 2009; Ma et al., 2009; Park et al., 2009; Shen et al., 2006). Pyrabactin resistance (PYR)/PYR-like (PYL) or regulatory component of ABA receptor (RCAR) constitute a 14-member family and play a role of ABA receptor (Ma et al., 2009; Park et al., 2009). Under normal conditions, PYR/PYLs were not able to bind ABA and PP2Cs can interact with and dephosphorylate SnRKs (Lee et al., 2009). During the interaction between PP2C and SnRK2, inactivation of SnRK2 causes suppression of ABA signaling pathway (Lee and Hwang, 2009; Lee et al., 2013). When ABA level is increased, the PYR/PYLs interact with and inhibit the activity of clade A PP2Cs leading to activation of the signaling pathway (Ma et al., 2009; Park et al., 2009; Umezawa et al., 2009; Vlad et al., 2009).
Thirteen members except PYL13 are able to activate ABA-responsive gene expression by using protoplast transfection assays (Fujii et al., 2009), whereas it is unclear whether their functional differentiation is exists or not. However, it is expected that each of their functions are different according to their different expression patterns (Antoni et al., 2012; Gonzalez-Guzman et al., 2012). Recently, the function of PYL8 has been elucidated for the regulation of ABA, pyl8 mutant exhibited reduced ABA sensitivity in root and overcome inhibition of root growth by ABA (Antoni et al., 2013). In this study, we generated PYL8-overexpressing (OX) transgenic plants which were sensitive to ABA in seed germination, seedling growth and adult plant. These results suggest that PYL8 is the key protein mediating ABA signaling in seed, seedling and adult stages. Moreover, we also present data indicating that PYL8-OX transgenic plants were drought tolerance phenotype.
For generation of transgenic PYL8 overexpression mutant, Full-length PYL8 cDNA was cloned into pENTR/D-TOPO vector (Invitrogen, Carlsbad, CA). After sequenced, the cloned genes were integrated into pK2GW7 by the LR reaction to induce constitutive expression of the PYL8 gene under the control of the CaMV 35S promoter (Karimi et al., 2002). The correct construct was introduced into the Agrobacterium tumefaciens strain GV3101 via electroporation. Arabidopsis transformation with the PYL8 gene was carried out using the floral dip method (Clough and Bent, 1998). For selection of PYL8-OX transgenic lines, seeds harvested from the putative transformed plants were plated on MS agar plates containing 50 mg/ml kanamycin. We used homozygous T3 plants for the assay.
Assays of drought tolerance were carried out using both seeding and 4-week-old plants as described before (Lee et al., 2013). To impose dehydration stress on the seedlings, watering was withheld for 2 weeks before rewatering. Water loss rates were measured to determine the drought tolerance of PYL8-OX mutant in a quantitative manner. Ten leaves were detached from each plant and placed in petridishes, and the loss of fresh weight was determined at the indicated times. Stomatal aperture bioassay was carried out as described before with the following modifications (Lee et al., 2013). In briefly, four rosette leaves of 4-week-old plants were detached and floated in stomatal opening solution (SOS: 50 mM KCl and 10 mM MES-KOH, pH 6.15, 10 μM CaCl2) in the light. After 2.5 h, buffer was replaced with SOS containing ABA of various concentrations and further incubation was carried out for 2.5 h. In each individual sample, 100 stomata were randomly observed under a Nikon eclipse 80i microscope and the width of individual stomata was recorded using Image J 1.46r (http://imagej.nih.gov/ij). Each experiment was performed in three independent repeats.
For the quantitative reverse transcription-polymerase chain reaction (qRT-PCR), total RNA was isolated from the Arabidopsis leaf tissues treated with dehydration using an RNeasy Mini kit (Qiagen, Valencia, CA, USA). After quantification using spectrophotometer, 1 μg of total RNA was applied to synthesize cDNA using a Transcript First Strand cDNA Synthesis kit (Roche, Indianapolis, USA) according to the manufacturer’s instructions. In parallel, cDNAs were synthesized without reverse transcriptase and were then subjected to qRT-PCR to reconfirm the contamination of genomic DNA in the cDNA samples. The synthesized cDNA was amplified in a CFX96 Touch™ Real-Time PCR detection system (Bio-Rad, Hercules, CA, USA) with iQTMSYBR Green Supermix and specific primers: RAB18 (forward 5′-GGAAGAAGGGAATAACA CAAAAGAT-3′ and reverse 5′-GCGTTACAAACCCTCA TTATTTTTA-3′), RD29A (forward 5′-CACA ATCACTT GGCTCCACTGTTG-3′ and reverse 5′-ACCTAGTAGCT GGTATGGAGGAACT-3′), RD29B (forward 5′-GTTGAA GAGTCTCCACAATCACTTG-3′ and reverse 5′-ATTAAC CCAATCTCTTTTTCACACA-3′), ABF2 (forward 5′-AG AGAATGACGAGTTACAACGAAAG-3′ and reverse 5′-TAAAACACCTAAGTGGGATGTCATT-3′), and Actin8 (forward 5′-CAACTATGTTCTCAGGTATTGCAGA-3′ and reverse 5′-GTCATGGAAACGATGTCTCTTTAGT-3′). The PCR was programmed as follows: 95 °C for 5 min, 45 cycles at 95 °C for 20 s, 60 °C for 20 s, and 72 °C for 20 s. The relative expression value of each gene was calculated by the Ct method (Livak and Schmittgen, 2001). The Arabidopsis actin8 gene was used for normalization.
PYR/PYL belongs to the family of the star-related lipid transfer (START) protein and constitutes a 14-member family as well as is homologous to the Bet v 1 superfamily (Ma et al., 2009; Park et al., 2009; Radauer et al., 2008). Although Bet v 1 superfamily has a common structure, their function has evolved diversely (Radauer et al., 2008). Bet V 1 is a member of the pathogenesis-related protein-10 (PR-10), which functions as pollen allergen (Radauer et al., 2008). The CaAMP1, which is orthologue of PYR/PYL in Capsicum annuum, has antimicrobial activity and involved in in broad spectrum resistance to pathogen infection (Lee et al., 2008). In addition, the members of superfamily are also related to binding and metabolism of hydrophobic compounds such as lipids, hormones, and antibiotics (Radauer et al., 2008). However, PYR/PYL family has a similar structure and plays a major role of ABA receptors (Fujii et al., 2009), even if different members of PYR/PYL family were differentially expressed in different tissues and at different growth stages (Santiago et al., 2009).
PYR/PYL family has 14 members, therefore they have functional redundancy and the phenotypes of each mutant have been masked (Gonzalez-Guzman et al., 2012; Park et al., 2009). To analyze the in vivo effect of overexpression of the PYL8, we generated Arabidopsis transgenic plants overexpressing the entire PYL8 precursor. The vegetative and reproductive development of the PYL8-OX transgenic plants was not significant difference from that of wild-type plants (data not shown). The level of ABA sensitivity was estimated by measuring the seed germination, seedling and adult plant phenotypes during exposure to various ABA concentrations (Fig. 1). To determine whether PYL8-OX plants exhibited enhanced ABA sensitivity during germination, the seeds of the wild-type and transgenic plants were germinated on MS media containing 0, 0.3, 0.5 and 1.0 μM ABA (Fig. 1A). No significant differences in seed germination were observed between wild-type and PYL8-OX plants on the basal MS medium without ABA. In the presence of 0.5 and 1.0 μM ABA, PYL8-OX plant seeds showed more sensitive than wild-type. We further examined the ABA sensitivity of transgenic plants at seedling stage (Fig. 1B, 1C). When seeds of wild-type and the transgenic plants were germinated on MS medium containing 0.5 μM ABA, PYL8-OX plants were highly sensitive to ABA at the seedling stage (Fig. 1B). We analyzed the root lengths of the wild-type and the transgenic plants 7 days after exposure to 0.3 μM ABA (Fig. 1C). The root lengths of PYL8-OX plants were 1.7-6.0 folds shorter than those of wild-types. The root growth revealed that transgenic plants were more sensitive to ABA-mediated inhibition of root growth compared to that in wild-type plants. Furthermore, treatment with 50 μM ABA induced a whitish etiolation phenotype in the PYL8-OX leaves (Fig. 1D). The chlorophyll contents of transgenic leaves were significantly lower than those of wild-type plants (Fig. 1D). Together, these results demonstrate that PYL8-OX is more sensitive than the wild-type to ABA during seed germination and post-germinative growth, suggesting that PYL8 serves as positive regulator of ABA.
Previous genetic study showed that pyl8 single mutant exhibited reduced ABA sensitivity in root (Antoni et al., 2013) and the overexpression of PYL8 has seed specific phenotype to ABA (Saavedar et al., 2010). In this study, the PYL8-OX plants exhibited completely sensitive in seeds and vegetative tissues because of globally enhanced ABA response, which indicates that PYL8 may function as a positive regulator of ABA signaling. Interestingly, PYL8 interacted with HAB1 (Santiago et al., 2009) and the HAB1-OX plants may reduce the ABA sensitive phenotype, which is in opposite with the PYL8-OX plants (Saez et al., 2004). These suggested that PYL8 and HAB1 share same pathway, and function oppositely each other in ABA signal transduction.
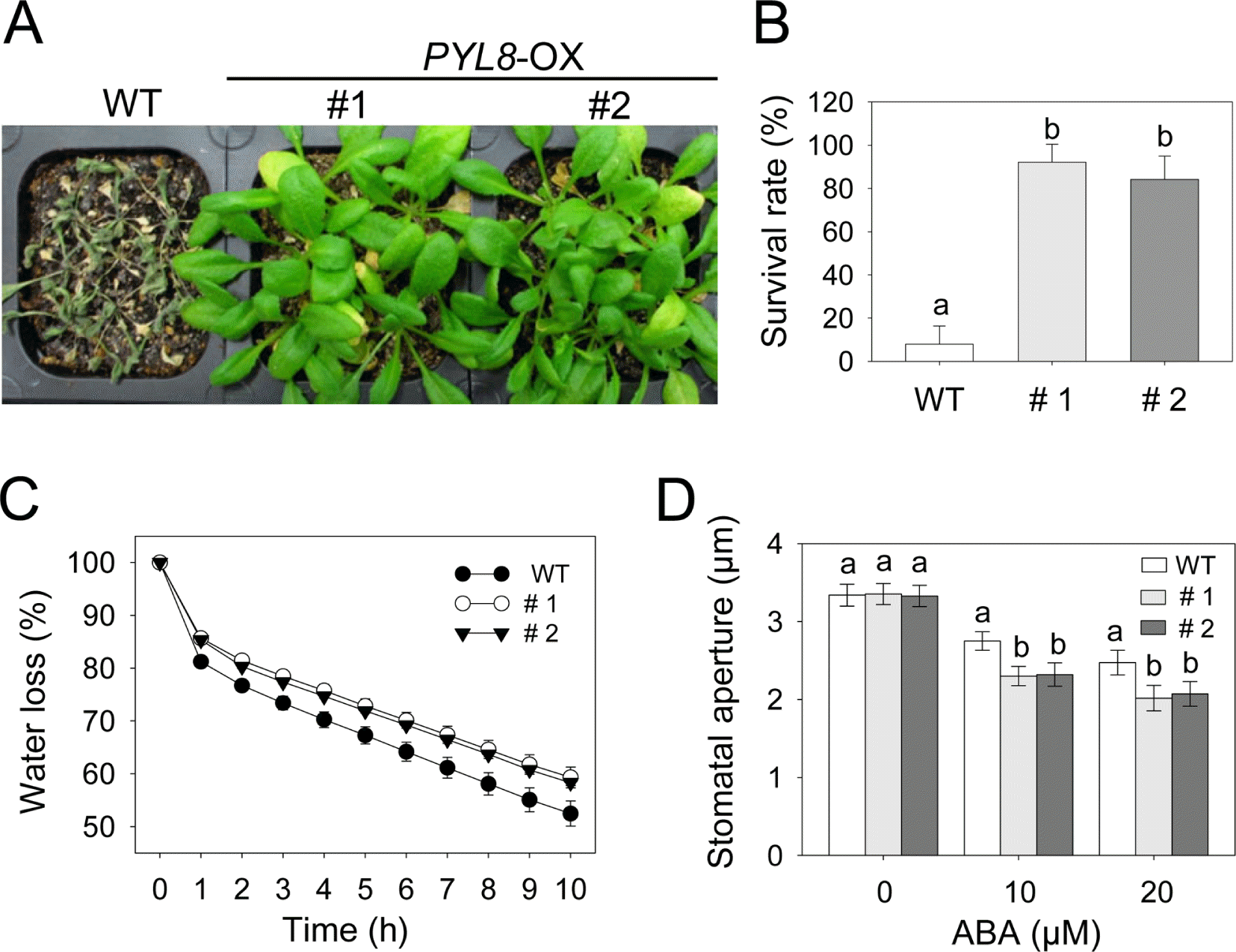
To determine whether sustained transcriptional upregulation of PYL8 affects drought tolerance, we compared phenotype and survival rates of PYL8-OX and wild-type plants under dehydration condition (Fig. 2A and 2B). When grown under well-watered conditions, PYL8-OX plants did not displayed any altered phenotype (data not shown). However, after drought treatment by withholding watering for 14 days, the transgenic lines exhibited enhanced drought tolerance compared to that in wild-type plants (Fig. 2A). After rewatering, PYL8-OX plants showed a high survival rate (approximaterly 83-89%), whereas wild-type plants had a much lower survival rate (approximaterly 6%) (Fig. 2B). In line with drought tolerant phenotype, the fresh weight loss of leaf tissues was lower in transgenic plants compared to that in the wild-type plants (Fig. 2C).
The degree of drought tolerance is determined by at least two cellular and molecular parameters. The ABA responsiveness of stomatal movement, which determines transpiration rates, can modify drought tolerance (Cheong et al., 2007; Lee and Hwang, 2009). Other studies have correlated drought tolerance with the level of ABA- or drought-related gene expression, which may lead to increased drought tolerance (Gonzalez-Guzman et al., 2012; Li et al., 2011). Previous microarray data and GUS expression studies have shown that PYL8 were expressed in guard cells (Gonzalez-Guzman et al., 2012; Yang et al., 2008). To examine whether lower levels of transpiration rate resulted from ABA hypersensitivity in the stomatal closure of the transgenic plants and PYL8 contribute to regulate stomatal aperture, we measured size of stomatal pores in the absence or presence of ABA (Fig. 2D). When wild-type and transgenic plants were not treated with ABA, no significant difference was detected in the size of stomatal pores. However, stomata of wild-type plants showed more open than in the transgenic plants upon both 10 and 20 mM ABA treatments. The size of stomatal pores in PYL8-OX was reduced to 60-62%, whereas they were only reduced to 74% in the wild-type by treatment with 20 μM ABA. These results suggest that PYL8 overexpression increases ABA sensitivity in guard cells, leading to reduced water loss under drought stress condition.
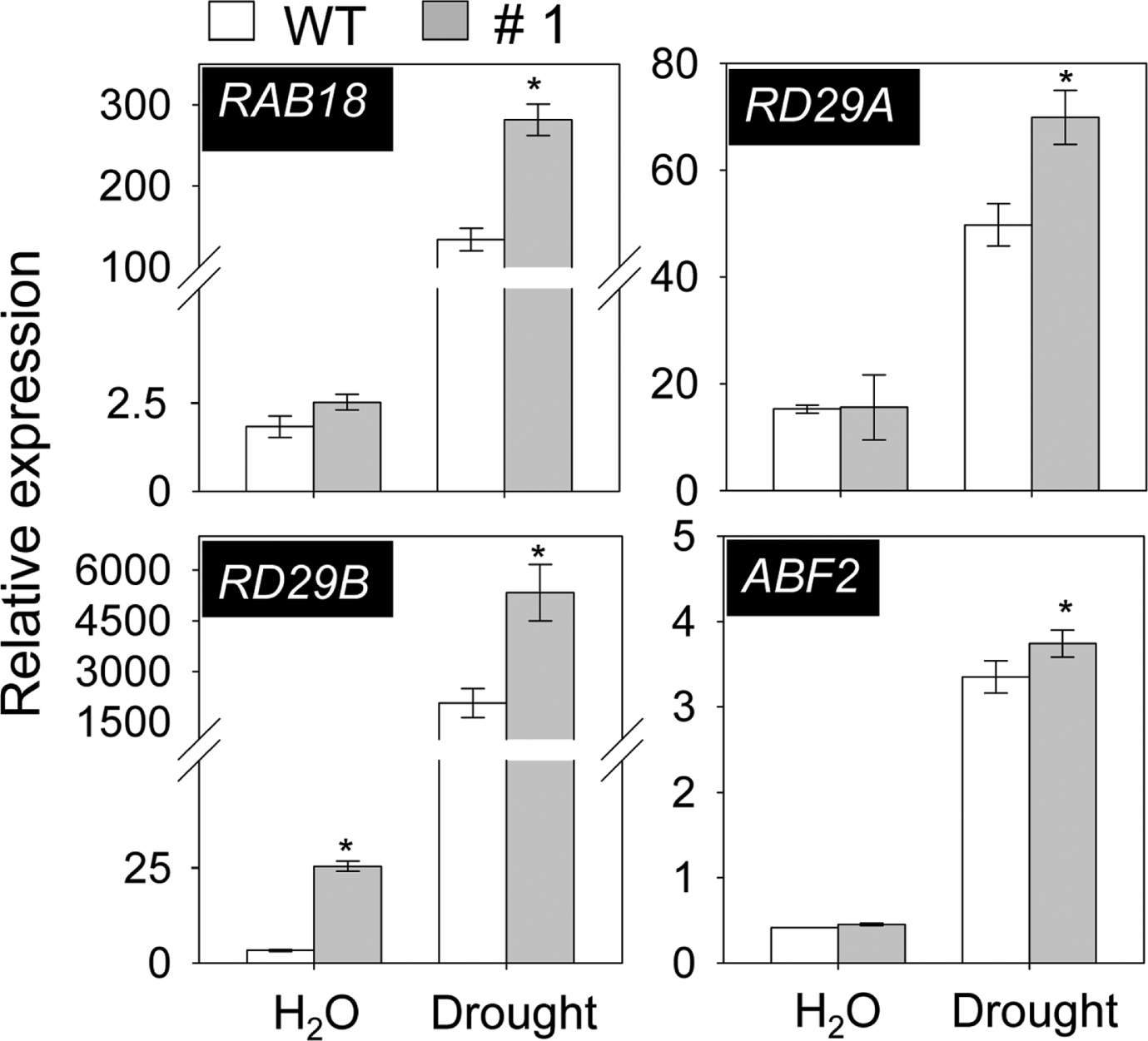
Previous studies reported that ABA treatment has led to increased expression levels of ABA marker genes, including RAB18, RD29A, RD29B and ABF2 (Abe et al., 2003; Lang and Palva, 1992; Li et al., 2011). To determine whether change in expression levels of these genes were contributed to drought tolerance in PYL8-OX plants, qRT-RCR analysis was performed. In line with ABA sensitive and drought tolerant phenotype of PYL8-OX plants, the expression levels of marker genes significantly increased in response to drought conditions (Fig. 3), indicating that alteration of ABA maker genes expression may be the one of reason for altered drought tolerance phenotype in PYL8-OX plants.
Plant cell can keep ABA level from the nanomolar to the micromolar range depending on water potential (Christmann et al., 2007). ABA level keeps very high and PYL8 expression is downregulated in the dehydration condition (Christmann et al., 2007; Szostkiewicz et al., 2010). However, PYL8-OX plants had resistant phenotype to drought stress (Fig. 2) and the more expression level of ABA marker gene was detected in PYL8-OX plants. These data suggested two explanations that can account for the imbalance between expression level and phenotype of PYL8-OX plants. One explanation is the concentration of ABA receptor is not main concern to develop resistance. The other is the best working condition of PYL8 is not high but certain concentration. The orthologue of PYR/PYL, CaAMP1-OX plants showed pleiotropic effects, including tolerance to high salt and drought during all growth stages. These results also indicated that PYR/PYLs function as ABA receptors in various plant species.
Future studies on PYR/PYLs function should lead to a greater understanding of the ABA signal transduction in various stress conditions. The PYR/PYL family consists of 14 members and more than 2 subfamilies, and PYR/PYLs may produce different responses to various concentrations of ABA or different cell types. Therefore, it will be important to determine which PYR/PYL is working in regards to the condition and concentration dependence of ABA in different tissues and at different growth stages. Further genetic and biochemical analysis should be conducted to assess the exact function of ABA receptors under certain stress conditions or in a particular cell type of plants. Such studies would lead to a better understanding of the ABA signal transduction pathway.




 PDF Links
PDF Links PubReader
PubReader Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Print
Print






