Histone Acetylation in Fungal Pathogens of Plants
Article information
Abstract
Acetylation of histone lysine residues occurs in different organisms ranging from yeast to plants and mammals for the regulation of diverse cellular processes. With the identification of enzymes that create or reverse this modification, our understanding on histone acetylation has expanded at an amazing pace during the last two decades. In fungal pathogens of plants, however, the importance of such modification has only just begun to be appreciated in the recent years and there is a dearth of information on how histone acetylation is implicated in fungal pathogenesis. This review covers the current status of research related to histone acetylation in plant pathogenic fungi and considers relevant findings in the interaction between fungal pathogens and host plants. We first describe the families of histone acetyltransferases and deacetylases. Then we provide the cases where histone acetylation was investigated in the context of fungal pathogenesis. Finally, future directions and perspectives in epigenetics of fungal pathogenesis are discussed.
Histone acetylation as an epigenetic regulator of gene expression
In general, epigenetics refers to changes in gene expression resulting from processes that are independent of changes in the underlying DNA sequence. These processes include DNA methylation, histone modifications, and nucleosome remodeling, all of which are related to the dynamic changes of chromatin, a highly compacted and organized complex of DNA and associated proteins. The basic structural subunit of the chromatin is a nucleosome, which consists of around 200 base pairs of DNA wrapped around an octamer of core histone proteins H2A, H2B, H3 and H4 (Kornberg and Lorch, 1999). In 1964, Allfrey and colleagues showed that histones can be modified by covalent addition of acetyl groups to lysine residues and that such modification correlates with increased transcription (Allfrey et al., 1964). Since then, numerous histone modifications and their roles in chromatin remodeling and transcriptional regulation have been discovered (Li et al., 2007; Mersfelder and Parthun, 2006). It has now become an accepted dogma that the combination of such modifications is collectively indicative of the transcriptional status of the underlying genes, leading to the concept of epigenetic code (Li et al., 2007; Turner, 2007). Histone modifications are considered to have a multitude of functions through the following mechanisms: First, histone modifications, except in the case of methylation, can cause a change in the net charge of nucleosomes, leading to alterations in DNA-histone and histone-histone interactions at the inter- or intra-nucleosomal level. Second, histone modification patterns might be read by other proteins that can influence chromatin dynamics and functions.
Among covalent modifications of histones, acetylation of lysine residues on histone 3 and histone 4 are commonly associated with active transcription and referred to as euchromatin modifications. Histone acetylation, in contrast to other modifications that are catalyzed by a specific enzyme at a specific site, occurs at multiple lysine residues and is usually carried out by protein complexes involving histone acetyltransferases utilizing acetyl CoA as a cofactor. Histone acetylation is not static but dynamically changes over time during cell development and differentiation. Such nature of histone acetylation is manifested by histone acetyltransferases (HATs) and the opposing action of histone deacetylases (HDACs). The balancing act of the two enzyme families is important for proper cellular function and development (Lee and Workman, 2007). While the relevance of HDACs in fungal plant pathogens has only just begun to be explored and communicated in literature, there is even less information available regarding HATs. Although there is not yet a great deal to address the significance of HATs in plant pathogens except speculations in relation to HDACs and knowledge from better-established systems, the importance of HATs as a group is covered for the sake of completeness and future findings to come.
Histone acetyltransferases
HATs are grouped into five families that each shares conserved structural motifs: GNAT (Gcn5-related N-acetyltransferases), MYST (MOZ, Ybf2/Sas3, Sas2, Tip60), p300/CBP, basal transcription factors (including TFIID) and nuclear receptor cofactors (Table 1: summary of search results from unpublished database, “dbHiMo: a web-based genomics platform for histone-modifying enzymes” – a database constructed based on hidden Markov model). Individual HATs can be further classified as nuclear type-A or cytoplasmic type-B HATs, regardless of the family to which they belong. While type-A HATs are largely responsible for transcriptional regulation by acetylating histones within nucleosomes, type-B HATs acetylate residues on newly synthesized histones before incorporation (Bannister and Kouzarides, 2011; Lee and Workman, 2007). The most recently proposed classification of histone modifying enzymes divided HATs further according to their catalytic activity and target histone residues and provided a new nomenclature as KATs (lysine acetyltransferases) to reflect the broad substrate specificity of the so-called HATs extending beyond histones (Allis et al., 2007) (Table 1). Below we provide short descriptions of some families with emphasis on GNAT and MYST families, and RTT109.
GNAT family
The members of GNAT family structurally resemble the archetypal HAT Gcn5 and have been relatively well studied. They share the highly conserved catalytic HAT domain with characteristic sequence motifs, of which motif A notably binds acetyl-coA (Lee and Workman, 2007). Some members additionally have an N-terminal PCAF homology domain for substrate selectivity (Xu et al., 1998) and a C-terminal bromodomain for binding acetylated lysine residues. As is the case with most HATs, GCN5 functions as the catalytic component in multi-subunit complexes SAGA, ADA and SLIK, playing important roles in transcriptional activation and various chromatin activities (Grant et al., 1997; Pray-Grant et al., 2002).
GCN5 has been found to be involved in induction of stress-related genes for adaptation to environmental stress in yeasts (Xue-Franzen et al., 2013). In the human pathogen Cryptococcus neoformans, Gcn5 is implicated in the adaptation to the host environment (O'Meara et al., 2010) and in the industrially important saprophyte Trichoderma reesei, induction of cellulase (Xin et al., 2013). The SAGA/ADA complex in Aspergillus nidulans is required for the induction of orsellinic acid during interaction with competing bacteria and also plays a role in the induction of various other secondary metabolite gene clusters under appropriate conditions (Nutzmann et al., 2011). Finally in Neurospora crassa, promoter acetylation by the Gcn5 ortholog NGF-1 is required in the induction of a blue light-inducible gene (Grimaldi et al., 2006).
Overall, these examples of Gcn5 orthologs in ecologically diverse fungi illustrate the process of transcriptional reprogramming at the chromatin level in response to specific environmental cues. Whether such phenomena are truly epigenetic by a definition encompassing an element of heritability, would be debatable and depend on the case. Nonetheless it is possible to expect mechanisms of epigenetic adaptation to host conditions as opposed to genetic adaptation, which will be discussed in the last section of this review.
MYST family
The MYST family, which was named after the initials of the founding members, is the largest family of acetyltransferases with diverse biological functions. In the budding yeast, Esa1 of the NuA4 complex is known to be an essential HAT required for DNA repair and cell cycle progression (Allard et al., 1999; Clarke et al., 1999). Additionally, an Esa1 ortholog in Aspergillus nidulans activates various secondary metabolite gene clusters (Soukup et al., 2012). Sas2 in yeast plays a crucial role in euchromatin/heterochromatin boundary maintenance most notably at telomeres, antagonising the Sir2 HDAC to prevent the spread of heterochromatin (Babiarz et al., 2006; Kimura et al., 2002). Sas3 is a part of the NuA3 complex and although its biological significance is unclear in yeast (Howe et al., 2001), the functions of its orthologs in animals are nevertheless substantial. In animal systems, the role of MYST family HATs in stem cells, development and cancer have been thoroughly documented (Voss and Thomas, 2009). It is of note that MYST family HAT complexes are known to have the broadest range of substrates including non-histone targets. The regulatory role and the ubiquity of protein acetylation had long been suspected to be comparable that of phosphorylation (Kouzarides, 2000). As the greater protein acetylome is being revealed, novel biological implications of HATs (or KATs) are awaiting discovery (Smith and Workman, 2009).
RTT109, a structural homolog of p300/CBP
The p300/CBP family is restricted to animals. Instead in fungi, there exists a fungal specific structural homolog, Rtt109 (Tang et al., 2008). It is responsible for the acetylation of H3K56 (Schneider et al., 2006), a histone residue at the DNA entry and exit point in the core of a nucleosome (Wang et al., 2008) as opposed to the more accessible N-terminal histone tail residues usually targeted by HATs for transcriptional regulation. Rtt109 is known to function in maintaining genome stability and participate in DNA replication (Han et al., 2007) and DNA damage repair (Chen et al., 2008). Therefore it affects cell cycle progression and sensitivity to genotoxic agents (Driscoll et al., 2007).
In the opportunistic animal pathogen Candida albicans, Rtt109 was demonstrated to be required for survival in host macrophages, where the pathogen is subjected to high oxidative stress (Lopes da Rosa et al., 2010). Plant pathogens are similarly exposed to host ROS response (Torres et al., 2006) and sometimes require internal ROS for cellular differentiation related to pathogenesis (Tudzynski et al., 2012). It is therefore possible that Rtt109 may also have implications for such processes in plant pathogenic fungi.
Histone deacetylases
HDACs reverse the effect of HATs by removing acetylation on the ε-amino group of lysine residues. The activity of HDACs thereby restores the positive charge of the lysine and has the potential to render the underlying DNA sequences relatively inaccessible to transcriptional machineries through stabilization of the local chromatin structure (Bannister and Kouzarides, 2011). This is consistent with the observation that most of HDACs are transcriptional repressors.
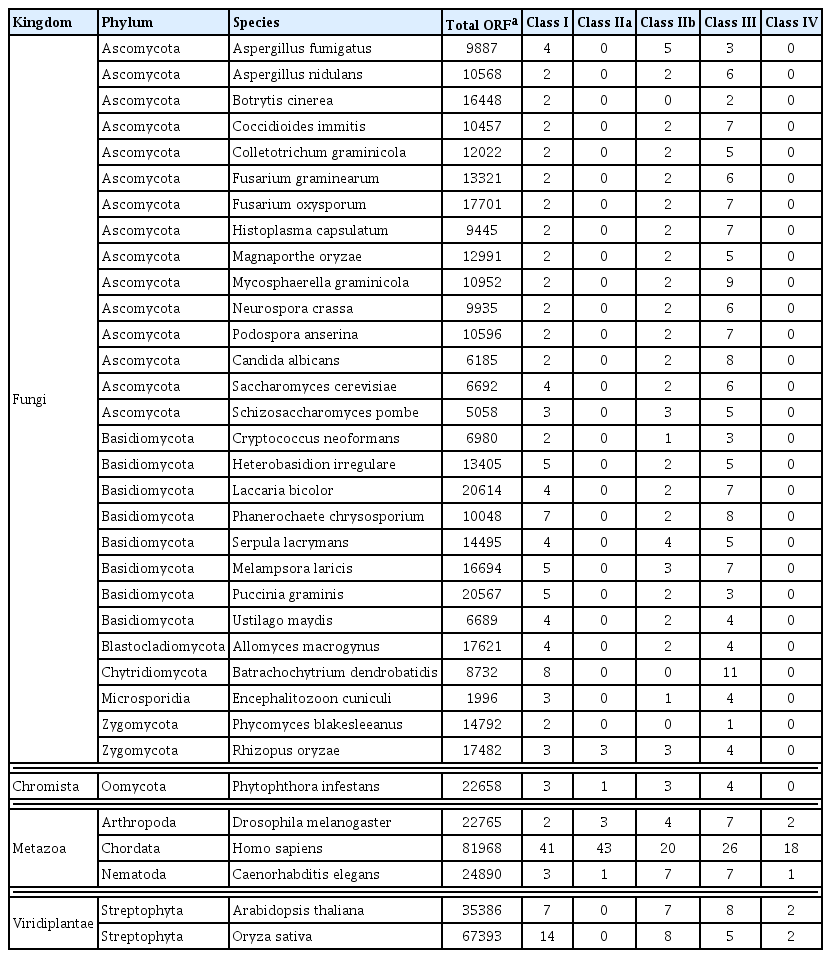
HDACs are grouped into four classes according to phylogenetic analysis and sequence homology (Gregoretti et al., 2004) (Table 2: summary of search results from the unpublished database, “dbHiMo: a web-based genomics platform for histone-modifying enzymes”). Class I and II contain enzymes that are closely related to Rpd3 and Hda1, respectively, in Saccharomyces cerevisiae with class II being further divided into subclasses IIa and IIb. Class III consists of Sir2 (silent information regulator-2) or sirtuin (Sir2-like protein) family, while class IV has only a single member, HDAC11. Unlike class I, II, and IV enzymes, the Sir2 family is NAD+-dependent. Although little is known about the function or regulation of HDAC11, class IV enzymes are highly conserved in diverse organisms. Below is the brief description of each class except class IV.
Class I HDACs
Rpd3 from the budding yeast, S. cerevisiae is a founding member of class I HDACs and contains an N-terminal deacetylase domain and a C-terminal tail region (Vidal and Gaber, 1991). In mammalian orthologs including HDAC1, HDAC2 and HDAC3, the C-terminal tail harbors one or two casein kinase-2 (CK2) phosphorylation sites and a sumoylation motif (Sengupta and Seto, 2004; Taunton et al., 1996). In yeast, Rpd3 functions as a catalytic subunit in a multi-protein complex called Sin3. In mammals, HDAC1 and HDAC2 form the catalytic core of the mammalian Sin3, NuRD (Nucleosome Remodeling deacetylase) and CoREST (Co-repressor of Re1-silencing transcription factor) complexes (Yang and Seto, 2008). These complexes are known to interact with sequence-specific transcription factors for transcriptional repression.
Class II HDACs
Hda1, Hos1 and Hos2 (Hda one similar) in the budding yeast contain an N-terminal deacetylase domain and a long C-terminal extension of unknown function (Yang and Seto, 2008). In mammalian orthologs (especially IIa members), only the deacetylase domain is conserved in the extended N-terminal region harboring binding sites for myocyte enhancer factor-2 (MEF2) and 14-3-3 proteins, which play a role in subcellular trafficking of class IIa HDACs (Verdin et al., 2003). Hda1 of S. cerevisiae and its ortholog Clr3 of Schizosacchromyces pombe are found in multi-proteins complexes. However, non-catalytic subunits of these complexes are not conserved in class IIa HDAC complexes of metazoans, indicating that the complexes are specific to fungal species (Yang and Seto, 2008).
Class III HDACs
Class III HDACs are sirtuin family enzymes. This class of proteins is related to the transcriptional repressor Sir2 (Silent information regulator 2) in the budding yeast. Yeast has five sirtuins including Sir2 and four homologs, Hst1-4 (Homolog of Sir two) (Grozinger and Schreiber, 2002). The sirtuin deacetylases contain a conserved catalytic domain that is not related to deacetylase domain of other HDAC classes. Thus, the sirtuins catalyze deacetylation by a different mechanism that requires NAD+ as a cofactor. The best-characterized sirtuin protein is the yeast Sir2 that is found in two major complexes: one responsible for silencing telomeric and HM loci and the other for suppressing genetic recombination at the rDNA loci (Gartenberg, 2000). Notably, Sir2 has been linked to aging in yeast by its dependence on NAD+ and roles in repression of rDNA recombination. Such role of Sir2 greatly influenced studies of aging in human and related model systems (Guarente, 2013; Park et al., 2013; Smith et al., 2007; Suvarna, 2012).
Histone acetylation in fungal pathogenesis on host plants
A growing body of evidence suggests epigenetic mechanisms including histone acetylation is pertinent to interactions between hosts and pathogens (Gomez-Diaz et al., 2012). Previous studies on epigenetic mechanisms as regulators of pathogenesis are centered mainly on human and animal pathogens such as Plasmodium falciparum, Mycobacterium tuberculosis, and influenza virus (Chookajorn et al., 2007; Croken et al., 2012; Hamon and Cossart, 2008; Marazzi et al., 2012; Pennini et al., 2006). Recently, plants challenged with a phytopathogenic bacterium, Pseudomonas syringae, was reported to be primed for defense against biotic stresses in the next generation without any additional treatment, suggesting that host plants also take advantage of epigenetic mechanism for protection from pathogen attack (Luna et al., 2012; Slaughter et al., 2012).
Despite emerging importance of epigenetic components in host-pathogen interactions, the role of such mechanisms regarding plant pathogens has received comparatively little attention. Yet there are a few lines of evidence that support the implication of epigenetic mechanisms in fungal pathogenesis on plants. Here we provide examples where histone acetylation/deacetylation plays a role in fungal pathogenesis. The examples can be broadly classified into two categories: (1) cases of epigenetic processes governing pathogen responses and (2) cases of pathogen-induced alteration of host cells.
Epigenetic regulation of pathogen traits
In Fusarium graminearum, the causal agent of Fusarium head blight in wheat and barley, it was shown that FTL1 is required for conidiation and pathogenesis (Ding et al., 2009). FTL1 is homologous to Sif2, which is a component of the Set3 complex in Saccharomyces cerevisiae. Set3 complex contains two histone deacetylases, Hos2 and NAD+-dependent Hst1, other than Sif2, Snt1, and YIL112w (Pijnappel et al., 2001). Deletion of HDF1, an ortholog of S. cerevisiae Hos2 in F. graminearum also resulted in significant reduction in virulence as well as defects in deoxynivalenol production, asexual and sexual reproduction (Li et al., 2011). Similarly, in Fusarium fujikuroi, the causal agent of bakanae disease on rice, deletion of two genes encoding HDACs, FfHDA1 (Class II HDAC) and FfHDA2 (Class I HDAC) showed that HDAC activities are crucial for secondary metabolite biosynthesis and virulence on rice. Direct mechanistic link between histone acetylation and expression of gene clusters involved in secondary metabolite biosynthesis of other mycotoxogenic fungi suggests that HDAC-mediated regulatory pathways are evolutionarily conserved and co-opted to invade host plants in pathogenic fungal species (Roze et al., 2007; Shwab et al., 2007).
Recently, Ding et al. reported that in the rice blast fungus, Magnaporthe oryzae, Tig1, which is an ortholog of F. graminearum FTL1, is important for conidiogenesis and growth inside host cells (Ding et al., 2010). The authors also showed that other components of Set3 complex such as Snt1, Hos2, and Set3 are required for full HDAC activity and virulence in the fungus. Interestingly, tig1 mutant was unable to differentiate bulbous invasive hyphae and induced high concentration of reactive oxygen species within plant cells. Hypersensitivity of the mutant to H2O2 and osmotin (a plant protein toxic to fungal pathogens) suggests that Set3 complex containing Tig1 participates in activating pathways that enable the fungus to cope with plant defense responses. Although we now have only a handful of studies aimed at understanding epigenetic control of fungal pathogenesis, current data suggest that histone acetylation/deacetylation is a crucial regulatory mechanism during plant infection.
Pathogen-induced epigenetic changes in hosts
The HC-toxin is a host-selective toxin produced by Cochliobolus carbonum. Only the maize plants that carry functional carbonyl reductase (HC-toxin reductase) gene(s) can detoxify HC-toxin (Meeley and Walton, 1991). Therefore, it is a determinant of specificity and virulence in interaction between C. carbonum and the maize plant. HC-toxin is not cytotoxic but cytostatic. Plant cell protoplasts treated with HC-toxin survive longer than do control protoplasts. Such nature of HC-toxin left open the question of how HC-toxin contributes to pathogenesis of C. carbonum on sensitive maize plants, given the necrotrophic lifestyle of the pathogen. Later, it was shown that HC toxin specifically inhibits maize HDACs, leading to proposition of a model where HC-toxin helps the pathogen to establish compatibility with maize via interference of histone acetylation/deacetylation (Brosch et al., 1995).
Depudecin is another HDAC inhibitor produced by a pathogenic fungus, Alternaria brassicicola (Kwon et al., 1998; Matsumoto et al., 1992). Depudecin is chemically unrelated to HC-toxin but, like HC-toxin, it can cause detransformation of oncogene-transformed mammalian cells. Genetic analysis of depudecin biosynthesis genes showed that depudecin is also a virulence factor but its effect is not as dramatic as that of HC-toxin (Wight et al., 2009). Zhou et al. reported that HDAC19 expression is induced in Arabidopsis thaliana challenged with Alternaria brassicicola and correlates with changes in expression of jasmonic acid and ethylene-regulated genes (Zhou et al., 2005). In conjunction with the role of fungal toxins as HDAC inhibitors, it is likely that plant HDACs are conserved regulators of defense-related genes in both monocot and dicot plants and are targeted by pathogens to cause alterations in transcriptional programs of host plants in favor of pathogens.
Future directions and perspectives
In this review, we attempted to summarize the wealth of information on histone acetylation/deacetylation in model systems including S. cerevisiae and relate that information to our current understanding of how fungal pathogens cause diseases on their host plants. Two things have been clarified from our efforts.
Firstly, we know surprisingly little about epigenetic mechanisms responsible for fungal pathogenesis. Considering the emerging significance of epigenetic mechanisms in understanding host-pathogen interactions, a comprehensive survey of epigenetic factors regulating pathogenesis will be required in the future. For histone acetylation/deacetylation, it is conceivable that HATs and HDACs in fungal pathogens would play a role in the coordinate expression of a plethora of genes during infection processes. Of particular interest would be gene sets that are induced and regulated by histone acetylation/deacetylation specifically during interaction with the host plant. These genes could include effectors, cell wall degrading enzymes, pathogenicity factors, metabolic enzymes and others involved in infectious development and adaptation to the host environment. Such endeavors would reveal many traits of both pathogens and hosts that are under epigenetic control and therefore provide potential targets to which improved disease control strategies can be devised and applied.
Secondly, fungal pathogens have epigenetic mechanisms for regulation of developmental processes of their own as well as for manipulation or subversion of host species. This suggests that both genetic and epigenetic variations should be considered to explain host-pathogen co-evolution. Current paradigm resides in the gene-for-gene theory proposing sequence alterations or loss of effector genes to account for the breakdown of R gene-mediated resistance. A recent study on the Oomycete, Phytophthora sojae, the causative agent of root and stem rot in soybean plants, presents a thought-provoking example on the issue. The authors of the work showed that P. sojae uses an epigenetic silencing mechanism to evade effector-mediated immune response in host plants carrying a cognate R gene over multiple generations while the sequence of the effector gene remains unchanged (Qutob et al., 2013). This observation suggests that epigenetic control of gene expression is a flexible means of generating phenotypic diversity in a population without committed changes in the DNA sequence. However, we lack evidence for the presence and/or prevalence of such mechanisms in fungal pathogens of plants. We believe that a paradigm shift from a gene-centric view to an epigenetic outlook, with the aid of cutting-edge epigenomics technologies, would promise unprecedented insights into host-pathogen interactions in the years to come.
Acknowledgements
This work was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2013-056580). S. Kwon is grateful for a graduate fellowship through the Brain Korea 21 Plus Program.