Selective Interaction Between Chloroplast β-ATPase and TGB1L88 Retards Severe Symptoms Caused by Alternanthera mosaic virus Infection
Article information
Abstract
The multifunctional triple gene block protein 1 (TGB1) of the Potexvirus Alternanthera mosaic virus (AltMV) has been reported to have silencing suppressor, cell-to-cell movement, and helicase functions. Yeast two hybrid screening using an Arabidopsis thaliana cDNA library with TGB1 as bait, and co-purification with TGB1 inclusion bodies identified several host proteins which interact with AltMV TGB1. Host protein interactions with TGB1 were confirmed by biomolecular fluorescence complementation, which showed positive TGB1 interaction with mitochondrial ATP synthase delta′ chain subunit (ATP synthase delta′), light harvesting chlorophyll-protein complex I subunit A4 (LHCA4), chlorophyll a/b binding protein 1 (LHB1B2), chloroplast-localized IscA-like protein (ATCPISCA), and chloroplast β-ATPase. However, chloroplast β-ATPase interacts only with TGB1L88, and not with weak silencing suppressor TGB1P88. This selective interaction indicates that chloroplast β-ATPase is not required for AltMV movement and replication; however, TRV silencing of chloroplast β-ATPase in Nicotiana benthamiana induced severe tissue necrosis when plants were infected by AltMV TGB1L88 but not AltMV TGB1P88, suggesting that β-ATPase selectively responded to TGB1L88 to induce defense responses.
Several members of the Potexvirus group have been studied to investigate viral movement, replication, and host interaction. Recently, most of the research related to triple gene block (TGB) protein 1 (TGB1) has been focused on viral RNA-binding and NTPase/helicase activity (Verchot-Lubicz et al., 2010). Unlike studies of PVX viral movement and replication, interactions between PVX and host proteins are not well described. Among such studies the protein kinase CK2 was found to phosphorylate PVX TGB1 (Modena et al., 2008), as also reported for Tomato mosaic virus (ToMV) P30 movement protein (Lee 2008; Matsushita et al., 2003). In addition, another host protein, remorin (REM) which is located in the cytosolic leaflet of plasma membrane microdomains (lipid rafts) and in clusters at plasmodesmata (Pd), interacts with TGB1 to transport PVX to Pd (Raffaele et al., 2009). PVX TGB1 has also been reported to interact with the Argonaute1 (AGO1) effector nuclease of RNA silencing (Chiu et al., 2010) and to mediate degradation of AGO1 through the proteasome pathway. The TGB1 of Pepino mosaic virus (PepMV) has recently been reported to interact with tomato catalase 1 (CAT1) to increase CAT1 activity and enhance PepMV accumulation (Mathioudakis et al., 2013). Except for these reports, there is very limited information related to interactions between TGB1 and host proteins to date.
Alternanthera mosaic virus (AltMV) is emerging as another model potexvirus with some significant differences from PVX (Jang et al., 2013; Lim et al., 2010a), in which a single amino acid difference in TGB1 affects not only the relative efficiency of TGB1 as a suppressor of RNA silencing (Lim et al., 2010b, c), but also both the cell wall and nuclear localization of TGB1 (Lim et al., 2010b, c; Nam et al., 2013). A naturally occurring variant with proline (P) at TGB1 residue 88 has significantly reduced silencing suppression efficiency, whereas AltMV differing only by having the typical leucine (L) at this position accumulates to approximately 40-fold higher levels (Lim et al., 2010b, c, and unpublished). The N-terminal helicase domain of AltMV TGB1 is required for homologous interactions (Nam et al., 2013), as was previously reported for PVX (Leschiner et al., 2006); disruption of the TGB1 self interaction by a single residue (G31R) or double residue (GK33,34RR) mutation in helicase domain I caused both loss of detectable silencing suppression function and failure to localize at the cell wall, the nucleolus, or the nuclear periplasm (Nam et al., 2013). AltMV also has an extensive host range, including taxonomically diverse ornamental plants as well as the model species Nicotiana benthamiana and Arabidopsis thaliana, making AltMV a useful model for investigating virus:host interactions as well as viral movement and replication.
Here we report interactions of several host proteins with AltMV TGB1, identified through either yeast two-hybrid (Y2H) analysis of an Arabidopsis cDNA library, or co-purification with a TGB1 inclusion body fraction from AltMV-infected N. benthamiana. We also report a differential interaction of chloroplast β-ATPase with TGB1L88 and TGB1P88, which significantly affects the symptom response when chloroplast β-ATPase expression is reduced by virus induced gene silencing (VIGS).
Materials and Methods
Virus isolates and plants
Derivatives of Alternanthera mosaic virus (AltMV) differing by a single amino acid in TGB1 have been described previously (Lim et al., 2010b, c), and are referred to here as AltMV-TGB1L88 and AltMV-TGB1P88. An isolate of Potato virus X (PVX) UK3 originally derived from infectious clone pPC2S (Baulcombe et al., 1995) was used as previously described (Lim et al., 2010b). Plants of Nicotiana benthamiana were raised from seed and typically inoculated at the four-leaf stage essentially as previously described (Lim et al., 2010b, c).
Yeast two hybrid (Y2H) analysis
In order to screen for interacting proteins by Y2H analysis, we cloned the AltMV TGB1 gene into pGBKT7 (James et al., 1996) at EcoRI and BamHI sites, in fusion with the Gal4 DNA-BD, and the resulting plasmid (pGBKT-TGB1) was used to transform yeast competent cells (strain AH109) for bait protein expression. An Arabidopsis thaliana cDNA library (CD4-30, Arabidopsis Biological Resource Center, www.abrc.osu.edu) in pAD-GAL4-2.1 (in fusion with Gal4 DNA-AD) plasmid was used to transform yeast competent cells containing pGBKT-TGB1. Transformed cells were screened on SD agar His-, Leu-, Trp- plus Aureobasidin A. Yeast colonies obtained were grown on the same media including the chromogenic substrate X-α-galactosidase; only colonies developing blue color were considered positive (James et al., 1996). A. thaliana genes encoding proteins involved in binding AltMV TGB1 were amplified from each of the selected yeast colonies using appropriate primers (Table 1), sequenced, and identified by BLAST analysis against the NCBI database.
TGB1 inclusion preparation, two-dimensional gel electrophoresis and matrix-assisted laser desorption/ionization-time of flight-mass spectrometry (MALDITOF-MS) analysis
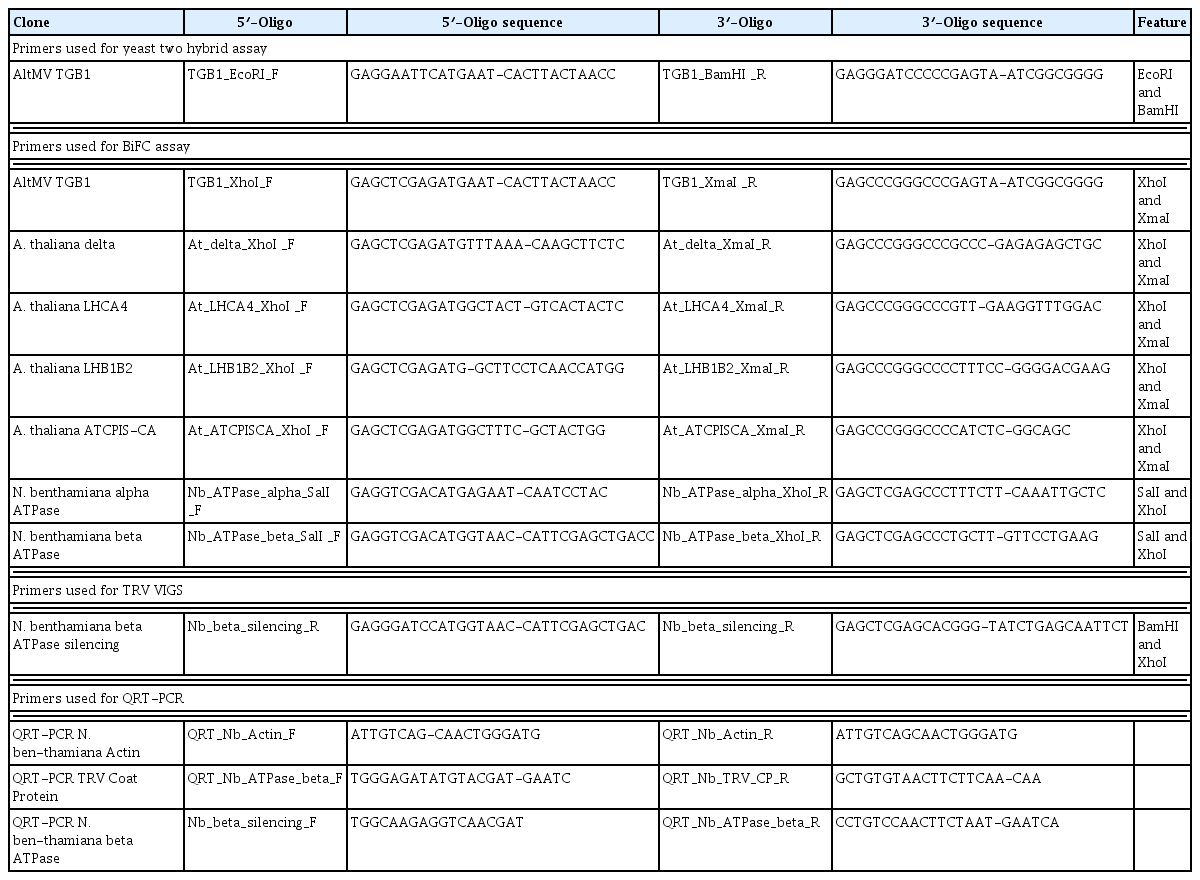
Additional host proteins were identified from an inclusion protein fraction prepared from AltMV-infected N. benthamiana; the inclusion body was partially purified from the ‘cytoplasmic inclusion pellet’ fraction according to a virus purification method originally developed for potyviruses, using a 50/60/70/80% sucrose density step gradient prepared in 0.1 M BK buffer (Hammond and Lawson, 1988). Fractions from the gradient were diluted in BK buffer, pellets collected by centrifugation (17,500 × g, 15 min), and resuspended in a small volume of BK buffer. An aliquot of the resuspended protein was stained with phosphotungstic acid (pH 7.0), and examined at a magnification of 66,000× with a JEOL 100CX II transmission electron microscope, and crystalline aggregates presumed to be TGB1 were observed most prominently in the 80% sucrose fraction (Fig. 1). This inclusion body fraction was also analyzed by 2-D gel electrophoresis; a 100 μl sample was mixed with 400 μl of ice-cold 10% TCA-acetone + 0.1% β-mercaptoethanol, incubated at −20°C for 60 min and centrifuged at 14,000 rpm for 20 min at 4°C in a microcentrifuge. The supernatant was discarded, the pellet washed twice by vortexing with 500 μl of ice-cold acetone containing 0.1% β-mercaptoethanol, and collected by incubation and centrifugation as before. The washed pellet was dried under vacuum at 4°C for 10 min and dissolved in 50 μl solubilization buffer (9 M urea, 1% 3-[{3- cholamidopropyl} dimethylammonio]-1-propane sulfonate) (CHAPS), 1% (v/v) immobilized pH gradient (IPG) buffer 4–10 (Amersham Biosciences, Piscataway, NJ, USA) and 1% dithiothreitol (DTT)), by ultrasonication in an ice-cold water bath. Undissolved particles were removed by centrifugation at 14,000 rpm for 10 min at 4°C.
Electron micrographs of an inclusion preparation from AltMV-infected N. benthamiana plants. Left, a paracrystalline array thought to be a TGB1 inclusion, together with loose aggregates that may also represent TGB1; Right, loose aggregates that may be accumulations of TGB1. Both show material purified through a sucrose step gradient, and stained with potassium phosphotungstate.
First dimension iso-electric focusing (IEF) was performed in an IPGphor system (GE Healthcare, Wauwatosa, WI, USA) with immobilized pH gradient gels (Immobiline pH 4–10, 13 cm, GE Healthcare). The conditions for isoelectric focusing followed by 12.5% SDS-polyacrylamide gel electrophoresis (Laemmli, 1970), protein staining, and protein spot analysis with MALDI-TOF-MS spectrometer (Applied Biosystems, Framingham, MS, USA) were as described in Lakshman et al. (2008). The peptide mass fingerprinting data were used to identify proteins by searching the non-redundant protein database of the National Center for Biotechnology Information (NCBI) with the Mascot search engine (Perkins et al., 1999, http://www.matrixscience.com) and taxonomy limited to plants and viruses. Mascot uses a probability based scoring system (Perkins et al., 1999). The parameters used for database searches with MALDI-TOF-MS peptide mass fingerprinting data were as described in Lakshman et al. (2008).
Bimolecular fluorescence complementation (BiFC) assay
The full-length genes encoding proteins identified from the Y2H and MALDI-TOF-MS assays were amplified from oligo(dT)-primed cDNA prepared from A. thaliana and N. benthamiana total RNA, respectively, using primers based on the appropriate GenBank sequences (primers listed in Table 1). Amplified PCR products were cloned into BiFC vectors (Waadt et al., 2008) in order to confirm interactions with TGB1. The AltMV TGB1L88, TGB1P88, and TGB1G31R (Nam et al., 2013) genes were subcloned into both pSPYCE(M) and pSPYNE173 to confirm homologous interactions; the host protein genes were subcloned into pSPYNE(R)173 and paired with AltMV TGB1 variants inserted to pSPYCE(MR) (Waadt et al., 2008). Constructs were agroinfiltrated into N. benthamiana essentially as previously described, in the presence of pGD:p19 (Lim et al., 2008) and enhanced yellow fluorescent protein (eYFP) fluorescence was observed at 3 days post agro-infiltration (dpa) by laser-scanning confocal microscopy (LSCM) as described (Jang et al., 2013).
Virus-induced gene silencing (VIGS)
A 300 bp product from the 5’ portion of the chloroplast β-ATPase gene was amplified from N. benthamiana cDNA using primers based on the GenBank sequence (Z00044) (Table 1), and inserted into TRV RNA2 (Dinesh-Kumar et al., 2003). This β-ATPase silencing VIGS construct was transformed into Agrobacterium tumefaciens GV3101. Plants of N. benthamiana at the four leaf stage (two weeks old) were infiltrated with A. tumefaciens containing wild-type TRV or TRV VIGS constructs respectively (Dinesh-Kumar et al., 2003).
Quantitative real-time reverse transcription polymerase chain reaction (Q-RT-PCR) assay
At 14 days after potexvirus inoculation, β-ATPase mRNA expression level was measured in sets of β-ATPase-silenced and TRV control plants, each separately infected with AltMV-TGB1L88, AltMV-TGB1P88, or PVX respectively. The levels of N. benthamiana Actin, TRV coat protein (CP), and N. benthamiana β-ATPase RNAs were measured by Q-RT-PCR, using the primers shown in Table 1, and normalized with respect to actin. Levels of β-ATPase expression were compared as ΔCt value (actin CT – β-ATPase CT) relative to Actin expression.
Results and Discussion
Y2H identification of A. thaliana proteins interacting with TGB1
Four host proteins were identified from the Y2H assay as interacting with AltMV TGB1 bait protein, by bioinformatic analysis of the sequence of the reactive cDNA clones. These proteins were identified as chloroplast-localized IscA-like protein (ATCPISCA), chlorophyll binding protein (LHB1B2), mitochondrial ATP synthase delta′ chain subunit (ATP synthase Delta′), and light harvesting chlorophyll-protein complex I subunit A4 (LHCA4) (Table 2); all are nuclear-encoded proteins.
MALDI-TOF-MS identification of N. benthamiana proteins interacting with TGB1
Of the 20 protein spots (Fig. 2) isolated from a 2-D gel and subjected to MALDITOF-MS analysis, seven spots were positively identified with proteins from the protein data bank (Table 3). Spots 1 and 2 are isomers significantly matched with ATP synthase CF1 α subunit of N. tabacum (chloroplast α-ATPase), whereas spots 3 and 4 are isomers related to ATP synthase CF1 β subunit of N. sylvestris (chloroplast β-ATPase); the respective isomers have similar molecular weights but migrated differently in IEF and are probably distinct phosphorylated forms. Both chloroplast ATPase genes are encoded in the chloroplast genome (Shinozaki et al., 1986). Spot 11 is related to the catalase isozyme 1 of Cucurbita pepo. Spots 16 and 17 have similar molecular weights but different charges and matched with the TGB1 protein of AltMV and most likely represent different phosphorylated isomers.

2-D gel from purified TGB1 inclusion body preparation. The 20 spots indicated were collected and analyzed by MALDI-TOF Mass Spectrometry.
Selection of chloroplast-associated genes for further examination
Previous reports indicated that protein kinase CK2, REM, and AGO1 interacted with PVX TGB1 (Chiu et al., 2010; Módena et al., 2008; Raffaele et al., 2009), and that tomato CAT1 interacts with PepMV TGB1 (Mathioudakis et al., 2013). Of these proteins, AGO1 and CAT1 each localize to both the cytoplasm and the nucleus (Mathioudakis et al., 2013; Zhang et al., 2006), as does TGB1 of PVX (Samuels et al., 2007) and AltMV (Lim et al., 2010c; Nam et al., 2013). Unlike PVX, AltMV infects Arabidopsis thaliana and therefore we could initiate screening of an Arabidopsis cDNA library using the Y2H system with TGB1 as bait. One initial goal of this screening was to identify host proteins which might transport TGB1 to the nucleus. In parallel, an inclusion body purification was tested to screen for co-purified host proteins; interestingly, the proteins identified from these different methods were not duplicated. The differences may be due in part to the distinct host systems, A. thaliana and N. benthamiana used for the Y2H analysis and inclusion purification, or because the inclusion bodies represent a different stage of the viral life cycle with a possibly inactive aggregated form of TGB1 compared to a monomeric or dimeric active form represented in the Y2H interactions. However, it is notable that both assays identified chloroplast-related proteins, as TGB1 has not previously been observed to associate with the chloroplasts. This is in contrast to the obvious chloroplast targeting to AltMV TGB3, and the apparent utilization of the chloroplast external membrane as the viral replication site (Jang et al., 2013; Lim et al., 2010a); AltMV accumulates primarily in the mesophyll layer, with in situ hybridization to AltMV RNA concentrated around the chloroplasts (Lim et al., 2010a). In this report we therefore present an examination of the chloroplast-related genes and mitochondrial ATP synthase Delta′ gene; the interactions with catalase and possible relationship to the nuclear and cytoplasmic distribution of TGB1 will be examined and reported separately.
Confirmation of TGB1 interactions by BiFC assay
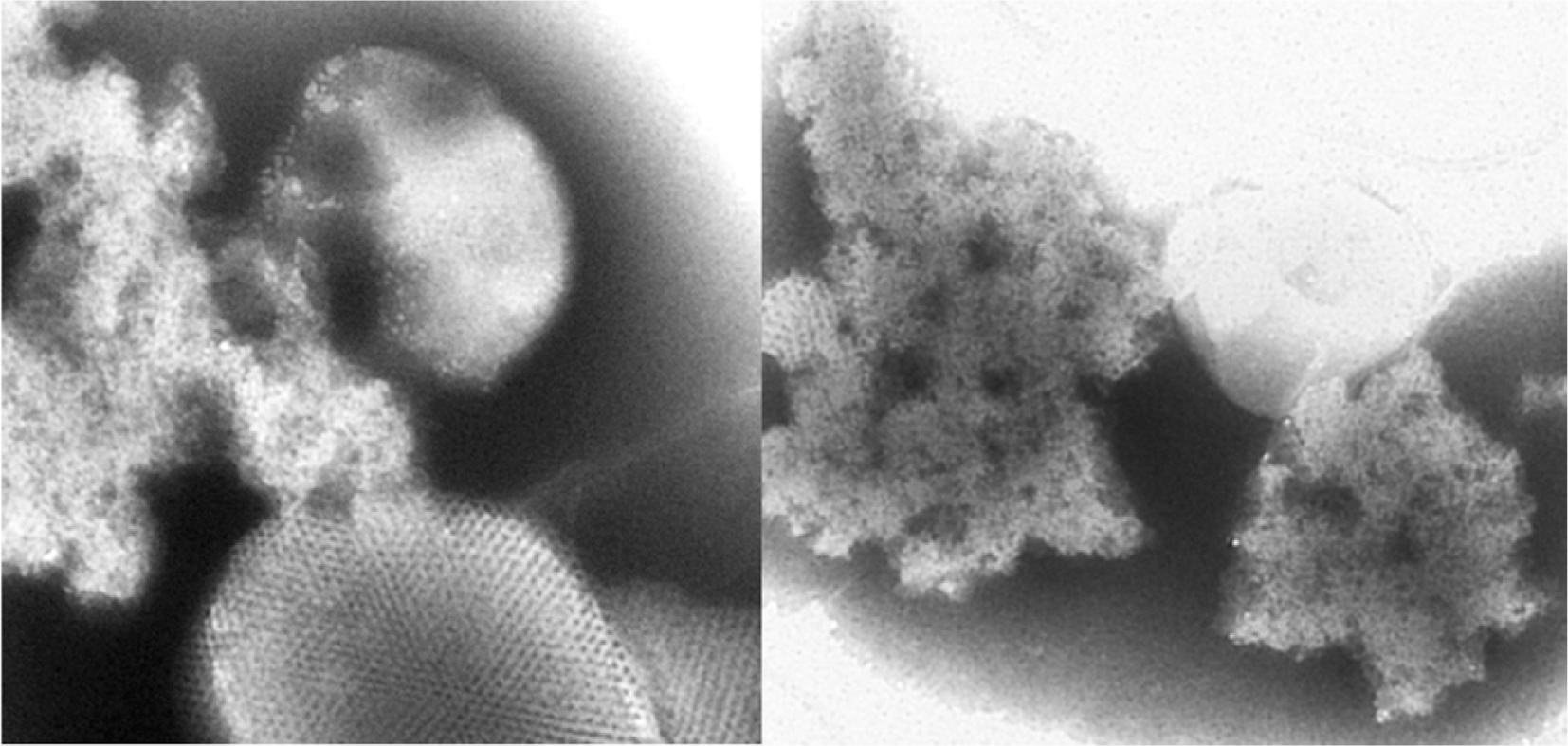
The chloroplast ATP synthase genes identified by MALDI-TOF-MS, and the four genes identified by Y2H were further examined by BiFC assay (Fig. 3). The four host proteins selected from Y2H (ATPISCA, LHB1B2, Delta, and LHCA4) all showed similar positive interactions with TGB1L88, TGB1P88, and helicase I domain mutant TGB1G31R. Chloroplast α-ATPase did not interact with either WT TGB1L88 or any derivatives in BiFC assays, suggesting that the α-ATPase subunit co-purified with TGB1 by association with the ß-ATPase subunit rather than interacting directly with TGB1. There were differential results between TGB1L88 and TGB1P88 or TGB1G31R in interactions with chloroplast β-ATPase by BiFC (Fig. 3). The differential interaction of β-ATPase to TGB1L88 and TGB1P88 is of considerable interest because the typical TGB1L88 has strong silencing suppression function, and confers higher accumulation to AltMV than TGB1P88, which has far weaker silencing suppression activity (Lim et al., 2010b, c).
Bimolecular fluorescence complementation assay resulting from protein:protein interactions reconstituting functional enhanced yellow fluorescent protein (eYFP), Host protein genes from Arabidopsis thaliana and Nicotiana benthamiana cloned in binary vector pSPYNE(R)173 [= eYFP(N-term)::Host protein] were co-infiltrated into leaves of N. benthamiana with TGB1 derivatives TGB1(L), (P), or (G31R) in pSPYCE(MR) [= eYFP(C-term)::TGB1]. All binary vectors were transformed in Agrobacteria. At 48 hours post infiltration, laser scanning confocal microscopy was used to detect eYFP protein expression. The A. thaliana genes were: ATCPISCA (chloroplast-localized IscA-like protein), structural molecule (ATCPISCA); LHB1B2, chlorophyll binding (LHB1B2); mitochondrial ATP synthase delta′ chain (Delta); and LHCA4 (Light-Harvesting Chlorophyll-Protein Complex I Subunit A4). The N. benthamiana genes were chloroplast alpha (α-ATPase) and beta (β-ATPase) ATPase. Bar = 50 μm.
Effect of VIGS reduction of β-ATPase expression on AltMV-TGB1L88 symptom expression
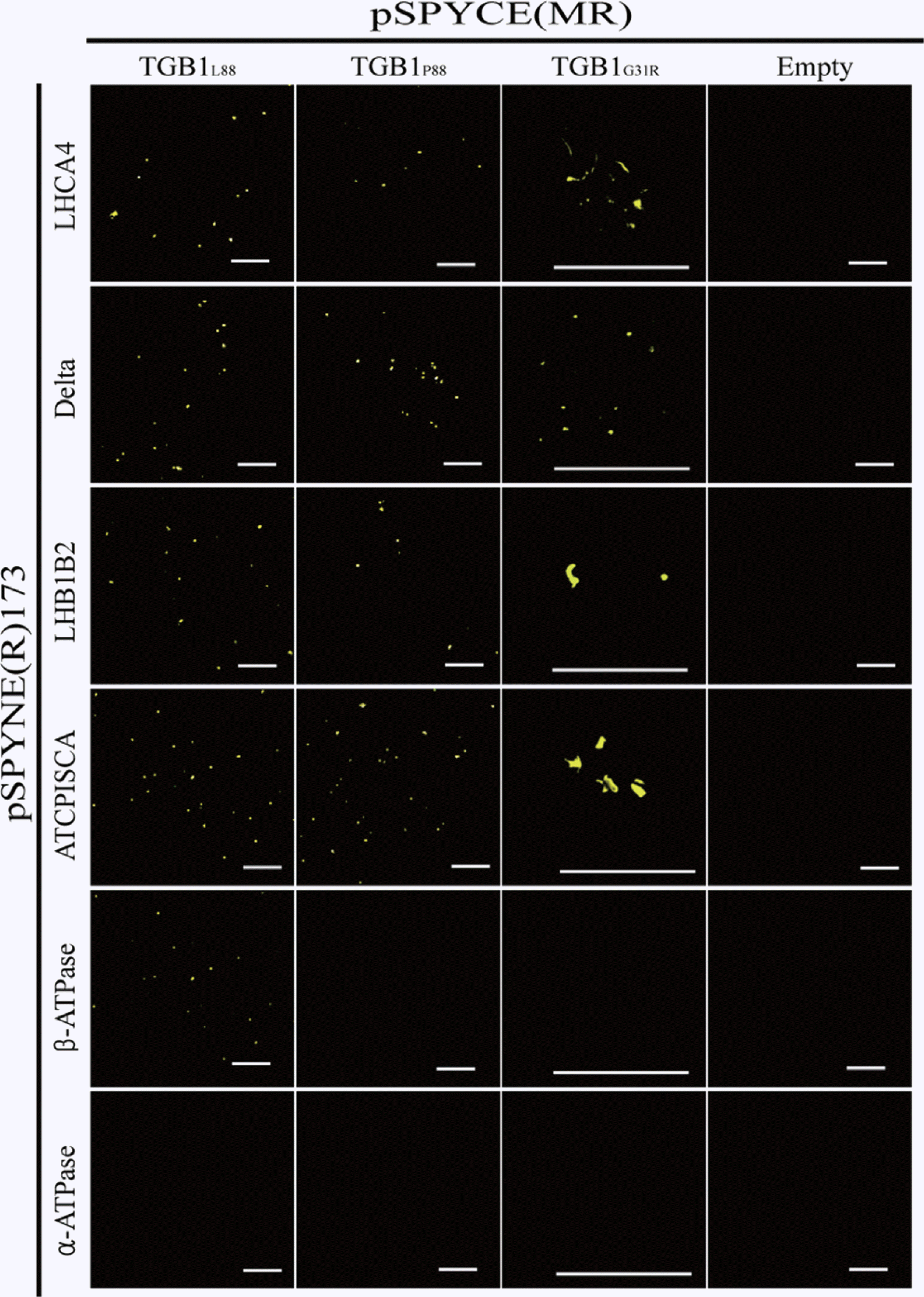
In order to investigate the effects of minimizing expression of β-ATPase in N. benthamiana on AltMV symptom expression, we induced β-ATPase silencing using a TRV VIGS vector (Dinesh-Kumar et al., 2003). Plants of N. benthamiana in which β-ATPase was silenced by VIGS showed a 6.9 fold reduction in β-ATPase mRNA expression, without significant phenotypic difference compared to control plants infected with WT TRV (Fig. 4). There were no distinct symptom differences between WT-TRV and β-ATPase-silenced plants infected with either PVX or AltMV-TGB1P88, but plants infected with AltMV-TGB1L88 showed more severe symptoms and stunting, and necrotic collapse of lower leaves at 20 days after potexvirus challenge (Fig. 4), and were eventually killed (data not shown). However, there was no significant change of AltMV-TGB1L88 or AltMV-TGB1P88 RNA copy number in silenced plants (data not shown). It was unexpected that the normal interaction between TGB1L88 and β-ATPase appears to retard severe symptom development and protect plants from severe necrotic reaction and death.
Symptom differences in plants infected with AltMV-TGB1L88 and AltMV-TGB1P88. Plants of N. benthamiana agro-inoculated with wild-type TRV (upper row) or TRV carrying a β-ATPase silencing insert (lower row), and either unchallenged or challenged 20 days later with PVX, AltMV-TGB1P88 or AltMV-TGB1L88, respectively (left to right). Plants in which β-ATPase was silenced using TRV:β-ATPase showed hypersensitive reaction to AltMV-TGB1L88 infection. All images were captured at 20 days after potexvirus challenge. β-ATPase mRNA expression level was measured by real time qPCR and fold difference was shown above (WT TRV infected; upper row), or below (TRV:β-ATPase infected; lower row) each plant image.
Potential involvement of chloroplast proteins in host defense
Chloroplast-related genes have been implicated in interactions with a number of viral proteins. Examples include interactions of the Rubisco large subunit with Potato virus Y (PVY) CP (Feki et al., 2005), and chloroplast division-related protein MinD with PVY HC-Pro (Jin et al., 2007). Other potyviral interactions include sugarcane ferredoxin-5 with Sugarcane mosaic virus HC-Pro (Cheng et al., 2008), and Rieske Fe/S protein of several hosts with the P1 protein of Soybean mosaic virus (Shi et al., 2007). The N. benthamiana photosystem I protein PSI-K interacts with Plum pox virus (PPV) CI protein, and VIGS of PsiK increased PPV accumulation, while PPV infections lowers PSI-K levels (Jiménez et al., 2006). The thylakoid membrane protein IP-L interacts with Tomato mosaic virus (ToMV; tobamovirus) CP (Zhang et al., 2008). Interactions specifically with potexviral proteins include that of PepMV TGB1 with tomato CAT1 (Mathioudakis et al., 2013) noted above, and plastocyanin of N. benthamiana, which interacts with PVX CP; VIGS of plastocyanin reduced both the severity of PVX symptoms, and accumulation of CP (Qiao et al., 2009). N. tabacum protochlorophyllide reductase was shown to interact with PVX TGB3 (Fridborg et al., 2003).
It has recently been shown that one nuclear-encoded chloroplast protein, NRIP1, is important in plant viral defense through interactions with both the tobacco N gene receptor and the TMV 126 kDa replicase protein (Caplan et al., 2008), while silencing of a second, the PsbO subunit of photosystem II oxygen-evolving complex, increased accumulation of TMV, but not of either PVX or Alfalfa mosaic virus (Abbink et al., 2002). A third nuclear-encoded chloroplast protein, the Rubisco small subunit, RbcS, interacts with the ToMV movement protein, and is involved in both tobamovirus movement and Tm-22-mediated extreme resistance (Zhao et al., 2013). Interestingly, AltMV TGB3 interacts with PsbO, and over-expression of TGB3 is associated with chloroplast vesiculation and veinal necrosis, suggesting that the interaction may interfere with basal host defenses (Jang et al., 2013).
Chloroplast ATPase consists of a large multisubunit complex, CF1, attached on the stromal side of the thylakoid membrane to an integral membrane complex, known as CF0 (Strotmann and Bickel-Sandkotter, 1984). CF1 consists of five different polypeptides, with a stoichiometry of α3, β3, γ, δ, ε, while CF0 contains probably four different polypeptides, with a stoichiometry of a, b, b’, c12 (Strotmann, 1984). The membrane-embedded proton translocating ATP synthase of chloroplasts, CF1-CF0, catalyzes ATP synthesis and ATP hydrolysis balanced to effect proton translocation across the thylakoid membrane (Groth and Strotmann, 1999). There has been a lot of research to pursue the structure of chloroplast ATPase, but there is little information on how chloroplast ATPase might be related to virus infection. Very recently, Bhat et al. (2013) reported that TMV 183 kDa and 126 kDa replication-association protein complexes were enriched with chloroplast γ-ATPase, which apparently worked to induce resistance against TMV infection, as γ-ATPase silenced plants had higher TMV titer and rapid cell to cell and systemic movement. Interestingly, proteolytic fragments of γ-ATPase functioned to induce salicylic acid (SA), jasmonic acid (JA) and ethylene causing defense response against insect attack (Schmelz et al., 2006, 2007). It is also notable that SA and JA are formed from amino acid biosynthetic precursors generated primarily in the chloroplast (Mauch et al., 2001; Schmid and Amrhein, 1995; Wasternack et al., 2006), which may suggest that disabling chloroplast function impairs host defense signaling (Dardick, 2007). Except for these recent reports and our own data we were unable to find information linking chloroplast ATPase and virus infection. One of the subunits, β-ATPase, reacted differentially to TGB1L88 and TGB1P88, which play different roles in replication and host response to AltMV. Our work using β-ATPase silencing introduces the possibility of a β-ATPase-mediated host defense response between TGB1 and N. benthamiana in the same way as γ-ATPase functions to elicit SA, JA and ethylene induced defense responses against insect attack (Schmelz et al., 2007).
More work is necessary to clarify this relationship, especially as chloroplast β-ATPase is encoded in the chloroplast genome, as are most of the other chloroplast ATPase subunits (Shinozaki et al., 1986), while the γ-ATPase subunit (AtpC) interacting with TMV 183 kDa and 126 kDa replication-associated proteins is nuclear-encoded (Bhat et al., 2013; Groth and Strotmann, 1999). It is surprising that there would be a cytoplasmic interaction between a chloroplast encoded protein and TGB1, as TGB1 is only known to accumulate in the nucleus and the cytoplasm; we have never observed TGB1 accumulation within the chloroplasts by either electron microscopy or confocal microscopy of TGB1 fusion proteins (Lim et al., 2010b; Nam et al., 2013; and data not shown). However, both chloroplast-encoded α-ATPase and β-ATPase co-purified with TGB1 aggregates, which have been observed only in the cytoplasm and the nucleus. Thus amino acid position 88 of TGB1 appears to have multiple roles not only in gene silencing suppressor activity, but also in host defense response through β-ATPase interaction. Therefore, AltMV TGB1 will clearly be a good target for further investigations of viral pathogenesis and host protein interactions.
Acknowledgements
This work was supported by a grant from the Next-Generation Biogreen 21 program (Plant Molecular Breeding Center No. PJ009033), Rural Development Administration, Republic of Korea. The authors would like to thank Drs. Savithiry Natarajan (USDA-ARS, SGIL), Wesley Garrett (USDA-ARS, ABBL), and Sukla Lakshman (USDA-ARS, DGIL), Beltsville, MD for help with 2-D proteomic analysis.