Expression of Rice Chitinase Gene in Genetically Engineered Tomato Confers Enhanced Resistance to Fusarium Wilt and Early Blight
Article information
Abstract
This is the first study reporting the evaluation of transgenic lines of tomato harboring rice chitinase (RCG3) gene for resistance to two important fungal pathogens Fusarium oxysporum f. sp. lycopersici (Fol) causing fusarium wilt and Alternaria solani causing early blight (EB). In this study, three transgenic lines TL1, TL2 and TL3 of tomato Solanum lycopersicum Mill. cv. Riogrande genetically engineered with rice chitinase (RCG 3) gene and their R1 progeny was tested for resistance to Fol by root dip method and A. solani by detached leaf assay. All the R0 transgenic lines were highly resistant to these fungal pathogens compared to non-transgenic control plants. The pattern of segregation of three independent transformant for Fol and A. solani was also studied. Mendelian segregation was observed in transgenic lines 2 and 3 while it was not observed in transgenic line 1. It was concluded that introduction of chitinase gene in susceptible cultivar of tomato not only enhanced the resistance but was stably inherited in transgenic lines 2 and 3.
Fungal diseases are the major constraint to substantial yield in agronomically important crops and impacted the well-being of human’s world wide (Agrios, 1998). The fungal species Alternaria, Aspergillus, Fusarium, Verticilium causes severe damage in crops such as potato, tomato, cotton, caster and chickpea and many other monocotyledonous and dicotyledonous crops. With the advancement in biological techniques in the early 1980’s, a major area of research has been to identify, clone and characterize various genes involved in disease resistance. The identification of disease resistant genes has made it possible to evaluate their specific roles and importance in disease response pathways using transgenic plants developed with genetic engineering. From the last two decades advances have been made in utilizing a broad range of cloned genes (from plants or other organisms) to develop resistance against fungal pathogens in plants.
The pathogenesis related proteins (PR proteins) include hydrolytic enzymes (chitinases and glucanases), antimicrobial proteins (thionin, defensins, lectin), ribosome inactivating protein (RIP), phytoalexins and antifungal proteins osmotin and thumatin like. The expressions of PR are directly toxic to pathogens or reduce their growth. Chitinaseshydrolyzes poly-β-1,4-N-acetyl glucosamine known as chitin. Cell wall of most of the fungi is composed of chitin as the major component and thus in plants chitinase acts as the one of the important defenses against these fungi (Collinge et al., 1993). Plant chitinase serve as a safe and biodegradable biocontrol agent for use instead of a conventional fungicide (Karasuda et al., 2003).
Rice has a small genome and hence serving as the model for plants genetics and genomics. Several defense related important genes have been cloned from rice (Song and Goodman, 2001). These genes have been engineered into many important dicotyledonous and monocotyledonous crops such as straw berry to develop resistance against powdery mildew (Asao et al., 1997), cucumber and Chrysanthemum kitamura against Botrytis cinerea (Tabei et al., 1998; Tabaeizadeh et al., 1999), Japonica rice against sheath blight (Nishizawa et al., 1999), grape against Elisinoe ampelina (Yamamoto et al., 2000), rice against Rhizoctonia solani (Datta et al., 2000, 2001), cucumber against Phytopthora rot (Kishimoto et al., 2004), Italian ryegrass increased resistance to crown rust (Takahashi et al., 2005), peanut against Sclerotinia blight (Chenault et al., 2005), orange (Mitani et al., 2006), Indica rice against sheath blight (Nandakumar et al., 2007), banana (Maziah et al., 2007), taro against Sclerotium rolfsii (He et al., 2008) and potato against F. oxysporum (Hussain et al., 2008).
Tomato is the most occurring vegetable in the world (Bose and Som, 1986). This crop is affected by over 200 diseases worldwide (Watterson, 1986). A wide range of fungal pathogens is reported to infect this crop (Tabaizadeh et al., 1999) and results in the loss of millions of dollars. Among fungal diseases the major diseases are early blight (Alternaria solani), late blight (Phytophthora infestans) and wilt (F. oxysporum). All commercially cultivated tomato lacking Immunity (I) genes are susceptible to Fol. Huang and Lindout (1997) tested 17 accessions of six Lycopersicon species for resistance to Fol race 1 and race 2. Most of the accessions were highly susceptible, some were intermediate resistance and three accessions were highly resistant.
Several important genes isolated from other organisms have been successfully engineered into tomato (Punja et al., 1998). Resistance of tomato against fungal pathogen has been enhanced by using antifungal genes from many other crops (Jongedijk et al., 1995; Lin et al., 2004; Parashina et al., 2000; Thomzik et al., 1997; Tabaizadeh et al., 1999; Van-den-Elzen et al., 1995) but still there is no single report of expression of rice chitinase in tomato to develop resistance against fungal pathogens. There is a need to bring strategies to develop resistant varieties by using some important genes like rice chitinase in under developed countries including Pakistan. Once the presence of transgene is confirmed it is important to evaluate the resistance of transgenic lines by pathogenicity test. Root dip method of artificial inoculation for screening of tomato cultivar (pathogenecity tests) to study resistance/susceptibility to Fol under green house conditions have been widely used in number of studies (Gale et al., 2003; Osuinde et al., 2002; Shishido et al., 2005). A wide genetic variation for wilt resistance has been observed in tomato. Similarly the assessment of tomato cultivars for resistance/susceptibility to EB has been reported earlier by using number of ways. The disease severity was determined by percentage of defoliation, rate of lesion expansion (Foolad et al., 2000) and lesion diameter (Gardner, 1988). The present study is designed to evaluate the resistance of genetically modified tomato prior to field trials by pathogenicity test.
Materials and Methods
Evaluation of the transgenic plants for their resistance to fungal pathogens
Three transgenic lines of tomato (Jabeen et al., 2009) Solanum lycopersicum Mill cv. Riogrande harboring rice chitinase (RCG3) were evaluated for resistance to fusarium wilt caused by F. oxysporum and early blight caused by A. solani.
Fusarium wilt test
Six weeks old non-transgenic and transgenic plants were evaluated to compare the level of tolerance to F. oxysporum. For this purpose plants were dipped in the freshly prepared inoculum of F. oxysporum for one minute. Plant treated with water was used as control. Plants were incubated at 28°C for three weeks.
Early blight test
Six weeksold non-transgenic and transgenic plants were evaluated for their resistance to A. solani by detached leaflet assay.
Detached leaflet assay
Three leaves from the upper leaflets were detached from each transgenic and non-transgenic plant. These leaves were placed in petri plates containing 0.1% agar. A 50 μl drop of the spore suspension (containing 2,000 conidia) was deposited on the upper surface of each leaf using micropipette (Foolad and Ntahimpera, 2000). Leaves treated with water were used as control. The inoculated leaves were incubated in the dark for 24 h at 22°C then maintained under cool white florescent diurnal light with a 12 h photoperiod. Lesion size (length × perpendicular width) was measured on the 7th day. EB severity on each leaf was recorded on a scale of 0 to 5 (Chaerani et al., 2007; Vakalounakis, 1983) where
0 = No visible lesion on leaf
1 = Up to 10% leaf area affected
2 = 11–25% leaf area affected
3 = 26–50% leaf area affected
4 = 51–75% leaf area affected
5 = More than 75% leaf area affected or leaf abscised
The disease scales were converted into percentage of EB index (PEBI) for each plant using the following formula (Pandey et al., 2003).
Determination of inheritance of chitinase gene in T1 seedling
In order to determine the inheritance of chitinase gene in T1 progeny six weeks old seedling was tested for resistance to F. oxysporum and A. solani by simple inoculation method. The Mendelian segregation in the seedlings was determined to check the inheritance of chitinase gene.
Statistical analysis
All field experiments were carried out in randomized completely block design (RCBD) and lab experiments were carried out in completely randomized design (CRD). Each experiment was replicated thrice. Data was analysed by using Minitab 13, Statistica and MSTAT-C softwares. Significance of data was checked by using least significance (LSD) test.
Results
Resistance of transgenic plants and non-transgenic control plants against fungal pathogens was evaluated by pathogenesity test.
Assay with Fusarium oxysporum
Transgenic and non transformed control plants after 4 weeks of their establishment in soil were incubated with F. oxysporum f. sp. lycopersici. After two days of infection at lower leaves turned golden yellow in non-transformed control plants while leaves remained fresh and green in transgenic plants. On the fifth day leaves of the whole non-transgenic plant became yellow and finally plant wilted on 7th day of infection while transgenic plant stayed alive in the infected soil for three weeks (Fig. 1).
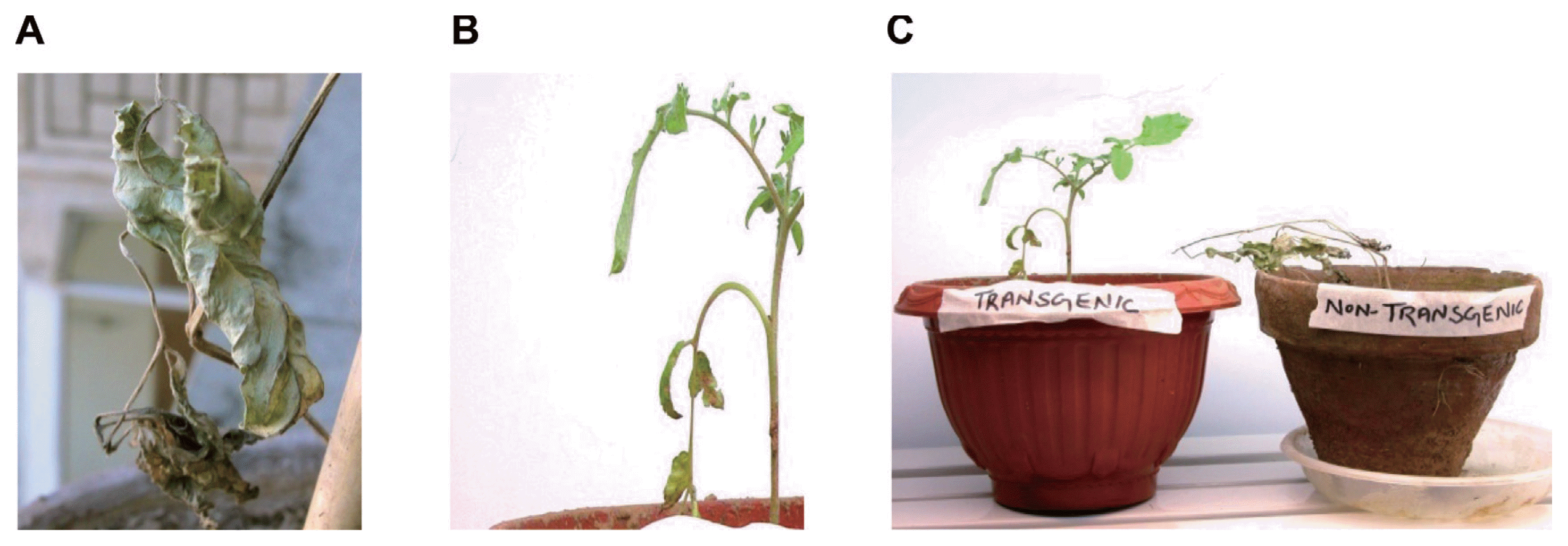
Photographic presentation of assay with Fusarium oxysporum f. sp. lycopersici. (A) Foliage of the non-transgenic control plant after 5 days of infection. (B) Foliage of the transgenic plant after 5 days of infection. (C) Non-transgenic plant completely wilted while transgenic plant survived after 7 days of infection.
Leaf assay with Alternaria solani
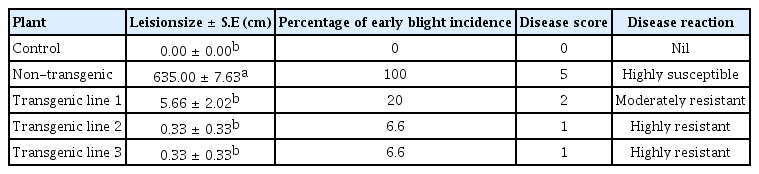
Leaves of the transgenic and non transgenic control plants were also tested for their resistance against A. solani. After 14 days of incubation with A. solani complete necrosis and chlorosis of the non-transgenic plant was observed because fungus covered the whole lamina while in case of transgenic leaves infection spreaded in media (Fig. 1A,1B) and fungus could not grow on the leaf lamina. Susceptibility/resistance of the control (treated with water) and transgenic and normal leaves (infected with A. solani) was determined in terms of the lesion size and percentage of early blight incidence (PEBI). Comparison of the lesion size of three transgenic lines and non transgenic leaves is summarized in Table 1 and Table 2. Results show (Table 1) that lesion size and PEBI of non-transgenic (treated with A. solani) was high compared to transgenic leaves (transgenic infected with A. solani). In case of non transgenic PEBI was 100 (HS, score = 5) while in case of transgenic lines it ranged from 6.6 to 20. All the transgenic lines were resistant (moderate or high). Transgenic line 1 was moderately resistant (PEBI = 20, score = 2) and transgenic lines 2 and 3 were resistant (PEBI = 6.66, score = 1). It was concluded that chitinase enhanced the level of resistance in all the transgenic lines.
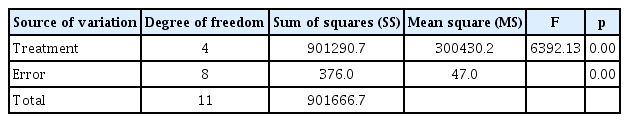
Analysis of the variance of the mean leision size in the leaves of the control (treated with water), non-transgenic and transgenic plants (treated with A. solani)
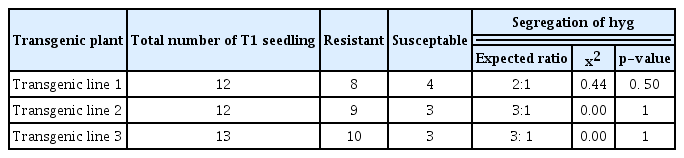
Transgene inheritance for hygromycin resistance
Fruits from the R0 transgenic plants were harvested and seeds were isolated. These seeds were subjected for germination on medium containing 50 mg/l hygromycin to get the hygromycin resistant R1 progeny. The genetic segregation to hygromycin resistance was observed in R1 progeny seedling. Expected Mendelian 3:1 ratio (Table 3) was observed in transgenic lines 2 and 3 while transgenic line 1 had shown no expected segregation.
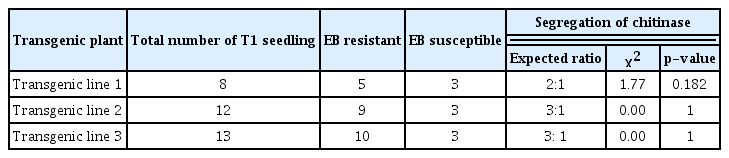
Transgene inheritance for resistance to fungal pathogens
The resistance of T1 six weeks old hygromycin resistant seedling to early blight and wilt was evaluated by simple inoculations. The pattern of segregation of three independent transformant is listed in Table 4 for Fol and Table 5 for A. solani. Mendelian segregation was observed in transgenic line 2 and 3 while it was not observed in transgenic line 1. It was concluded that chitinase gene was stably inherited in transgenic lines 2 and 3 only.
Discussion
Three transgenic lines of (TL1, TL2 and TL3) and their R1 progeny harboring rice chitinase gene were evaluated for resistance to fusarium wilt caused by F. oxysporum lycopersicii and early blight caused by A. solani.
Assay with Fusarium oxysporum
Three PCR positive R0 transgenic plants were tested for resistance to F. oxysporum by root dip method. Results indicated that all transgenic plants were highly resistant to F. oxysporum and no symptoms of wilt were observed. It was concluded that chitinase gene (RCG3) expressed its antifungal activity in all the transgenic plants tested. (Kumar et al., 2004) produced transgenic pigeonpea plant with rice chitinase gene and induced resistance against wilt caused by Fusarium spp. (Hussain et al., 2008) produced transgenic plants harboring rice chitinase gene in potato. These plants were highly resistant to local Pakistani isolate of F. oxysporum.
Assay with Alternaria solani
These PCR positive T0 transgenic plants were also examined for resistance to early blight (EB) caused by A. solani by detached leaf assay. Detached leaf assay method has already been reported for the evaluation of resistance in transgenic lines against different fungal pathogens in number of crops like cucumber (Tabei et al., 1998) and rice (Nishizawa et al., 1999). Results indicated that chitinase gene induced resistance to A. solani in all the transgenic plants (PEBI was less than 20) compared to the control (PEBI was 100).
Rice chitinase gene (RCC2 and RCG3) has already been widely reported for its broad spectrum antifungal activity against number of fungal species in many monocots and dicots (Nishizawa et al., 1999) observed resistance in transgenic rice plants harboring rice chitinase gene against blast (Magnaporthe grisea). (Tabei et al., 1998) and (Kishimoto et al., 2002) produced cucumber transgenic lines highly resistant to gray mold caused by fungal pathogen B. cinerea (Yamamoto et al., 2000) produced transgenic grapevine plants expressing rice chitinase gene resistant to powdery mildew caused by fungus Uncinula necator and anthracnose caused by Elisinoe ampelina. Maziah et al. (2007) reported resistance against fungal pathogens in banana plants harboring rice chitinase gene. Results of the present study and earlier reports revealed that rice chitinase could be utilized to enhance the resistant against number of fungal pathogens in agronomically important crops.
Transgene inheritance for hygromycin resistance
In order to determine the transgene inheritance Seeds were isolated from the three R0 transgenic lines and hygromycin resistance of R1 progeny was determined on the MS- medium supplemented with 50 mg/l hygromycin. Hygromycin resistant R1 seedlings were also evaluated for resistance to Fol and A. solani by inoculation method. Mendelien segregation ratio 3:1 (resistant: susceptible) was observed in R1 offsprings of the two resistant transgenic lines. (Ignacimuthu and Ceasar, 2012) reported that expression of rice chitinase (RCC2) gene in finger millet resulted in the development of resistance against leaf blast disease. Inheritance of RCC2 was also confirmed in R1 progenies. The segregation of disease resistance among the progenies was according to predicted Mendalien ratio of 3:1. Parasad et al. (2013) reported the enhanced resistance against soil borne and foliar fungal pathogens in transgenic peanut plants harboring rice chitinase gene. They observed that transgene inheritance was on the pattern of Mendelian inheritance in all R1 progenies. In our study Mendelian segregation was not followed by the R1 progenies of the transgenic line 1 although it was moderately resistant. This contradictions in our study and earlier report is may be due to the transient integration of rice chitinase gene in transgenic line 1 developed during the present study. This difference is may be the difference in the varietal response to transgene integration among difference species.
It was concluded that expression of rice chitinase gene in tomato confers enhanced resistance againsttwo major fungal diseases of tomato. If it is stably inherited the offspring can be as strong as their parents. Resistance of these transgenic lines against many other fungal pathogens can also be tested.
Acknowledgments
The plasmid of RCG3 was kindly provided by Dr. Yuko Nishizawa, National Institute of Agro-biological Resources, Tsukuba, Japan.