Pathotypes of Bacterial Spot Pathogen Infecting Capsicum Peppers in Korea
Article information
Abstract
Sixty-seven isolates of bacterial spot pathogen (Xanthomonas spp.) collected from six provinces of Korea were tested for the identification of their pathotypes and determination of their distribution throughout Korea in an effort to genetically manage the disease. Near isogenic lines of Early Calwonder (Capsicum annuum) pepper plants carrying Bs1, Bs2 and Bs3, and PI235047 (C. pubescens) were used as differential hosts. Race P1 was found to be predominant, followed by race P7, and races P3 and P8 were also observed. This is the first report of races P7 and P8 in Korea. The races P7 and P8 were differentiated from the former races P1 and P3, respectively, on the basis of their ability to elicit hypersensitive reactions to PI235047.
Bacterial spot disease is one of the major diseases damaging pepper plants in Korea. This pathogen, formerly called Xanthomonas campestris pv. vesicatoria (Doidge) Dye, is currently subdivided into Xanthomonas euvesicatoria, X. vesicatoria, X. gardneri, and X. perforans on the basis of DNA homology and phenotypes (Hamza et al., 2010; Jones et al., 2004, 2005). However, Korean strains of the bacterial spot pathogen have not been reclassified into these four species. Although treatment of seeds with hot water or chemicals, spraying plants with chemicals including copper and antibiotics, rotation with non-host plants, and sanitation by removing volunteer plants and diseased plant residue are recommended for control of the disease (Cox, 1982; Goode and Sasser, 1980), host resistance is considered the most economically and ecologically reliable method for control of bacterial spot disease in pepper plants (Hibberd et al., 1989). Five hypersensitive resistance (HR) genes, namely Bs1, Bs2, Bs3, Bs4 and Bs7, have been reported in pepper plant accessions PI163192, PI260535, PI271322, PI235047, and UENF1556, respectively (Cook and Guevara, 1984; Cook and Stall, 1963; Kim and Hartmann, 1985; Potnis et al., 2012), and two recessive genes, bs5 and bs6, that govern non-hypersensitive resistance and act additively with each other were also screened from PI271322 and PI163192, respectively (Jones et al., 2002; Vallejos et al., 2010). One or more of the HR genes have been transferred to the Early Calwonder (ECW) strain of the pepper plant to develop near isogenic lines for the respective genes, and the varieties developed were named ECW 10R, ECW 20R, ECW 30R and ECW 70R, depending on the respective HR gene incorporated, and as ECW 12346, with Bs1, Bs2, Bs3, bs5 and bs6 incorporated (Jones et al., 2002; Stall et al., 2009; Vallejos et al., 2010). Genetic control of bacterial spot disease by hypersensitive resistance genes has not been very successful due to the occurrence of pathogenic races that can overcome the HR genes (Cook and Stall, 1969; Hibberd et al., 1987b; Kousik and Ritchie, 1995; Sahin and Miller, 1995). The hypersensitive reaction of pepper plants is known to be induced by an interaction between the Bs genes in pepper plants and the corresponding effector (avirulence, avrBs) genes in the pathogen in a gene-for-gene model (Minsavage et al., 1990; Potnis et al., 2012). Bacterial spot pathogens of peppers are classified into 11 pathotypes on the basis of interactions between the Bs1, Bs2, Bs3 and Bs4 genes that confer an HR reaction in the pepper and corresponding avirulence genes (avrBs1, avrBs2, avrBs3 and avrBs4) in the pathogen (Stall et al., 2009). ECW 10R, ECW 20R, ECW 30R, and PI235047 (Capsicum pubescens) have been used as differential hosts in the classification of pathogen races (Sahin and Miller, 1998; Stall et al., 2009). In Korea, Kim et al. (2007) transferred Bs2 and Bs3 to a local Chilseong strain of Capsicum annuum, a susceptible cultivar grown in Yeongyang in Gyeongbuk province, and produced Chilbok-2 and Chilbok-3 for use as differential hosts.
In Korea, only race P1 and race P3 had been found (Kim, et al., 1990; Lee and Cho, 1996; Pae et al., 1994) until the 1990s. These races have been subdivided into races P1 and P7, and races P3 and P8, respectively, based on their abilities to elicit HR reactions in pepper strain PI235047 in the USA (Sahin and Miller, 1998; Stall et al., 2009). The prevalence of many races makes breeding and controlling the disease by resistant cultivars complicated. Therefore, a better understanding of the pathological races distributed in a given area, as well as host-pathogen relationships, is important in planning a breeding program for pepper plants for durable resistance to the disease. Here, we report results from testing Korean strains of Xanthomonas spp. infecting peppers to identify their pathotypes according to the race identification system (Stall et al., 2009).
The differential host sets consisting of Early Calwonder (ECW) and its isogenic lines ECW10R, ECW20R, and ECW30R, carrying Bs1, Bs2 and Bs3, respectively, and PI235047 with Bs4 gene, were kindly provided by USDA while Chilseong NILs, Chilbok-2, and Chilbok-3 were bred by us. All materials were sown in 200-cell trays with Wonjo mix (Nongkyung Agroindustrial Co., Ltd., Korea) and 16 plants of each line were transferred to 32 cell-trays one month after sowing in July 2012. In September 2012, the plants were transferred to pots 13 cm in diameter, again filled with Wonjo Mix, to grow more leaves.
Isolates of Xanthomonas spp. have been collected from various locations of Korea since the 1990s. These cultures have been maintained in water and on Yeast Extract Dextrose Calcium Carbonate (YDC) slants with routine subculture. Dry specimens of the collected leaves have also been stored in the refrigerator. In 2012, additional isolates were collected in the Chungnam, Jeonbuk and Jeonnam area of Korea. Overall, 67 isolates, 42, 3, 6, 3, 8 and 5 of which were originally collected from Daegu and Gyeongbuk, Chungnam, Chungbuk, Gangwon, Jeonnam and Jeonbuk province, respectively, were tested to determine their pathological races in September 2012. The confirmation study was conducted only with ECW NILs and PI235047 after transplanting the differential host plants to 25 cm pots in January 2013.
Isolates of pathogenic Xanthomonas spp., stored as stocks strains, were subcultured on yeast extract dextrose calcium carbonate (YDC) agar plates and incubated for 2–3 days at 28–29ºC to obtain pure cultures. Two-day-old, typical round, convex, mucoid, and yellow colonies were suspended in distilled water with a sterile cotton swab to obtain a suspension. The bacterial concentration was adjusted to approximately 108 cfu/ml (0.2 OD at 470 nm) using a spectrophotometer (Kim, 1983). The bacterial suspension was infiltrated into the abaxial surface of the leaves on each side of the midrib using a syringe with the needle removed. Two leaves were used as replication for each isolate.
Races were determined on the basis of hypersensitive (HR) and susceptible (Sus) reactions in response to infiltration of the bacterial suspension into leaf tissues, according to the race identification system reported by Stall et al. (2009). A hypersensitive reaction was recognized by tissue collapse at the infiltration site, with the tissue turning from dark green to light black within 24–48 h post inoculation, while a susceptible or non-hypersensitive reaction by water-soaked lesions appearing 3 or 4 days post infiltration. HR lesions turned tan to papery white with light twisting while non-HR lesions spread with yellow margins with time. Infiltration sites on tender leaves of susceptible plants often turned wet clear with water-soaking and yellowing margins (Fig. 1).

Hypersensitive (HR) and susceptible (Sus) reaction to infiltration of bacterial spot pathogen. Hypersensitive reaction 24 h (A) and 48 h, (B) post infiltration; susceptible reaction 48 h, (C) and 72 h, (D) post infiltration showing wet clear lesion with water-soaked margin. HR lesions on PI235047 carrying Bs4 gene formed in reaction to race P1 and P3, (E) and non-hypersensitive reaction to race P7 and P8, (F) 1 week post infiltration.
The reactions of the differential hosts to the 67 isolates collected from the six provinces of Korea are given in Table 1. Chilbok-2 and Chilobok-3 carrying Bs2 and Bs3, respectively, were included in the test but their reactions were the same as ECW 20R and ECW 30R, respectively, therefore, not listed here. Races P1, P3, P7, and P8 were found in the six provinces of Korea. Among the 67 isolates, 29 (43.3.0%) were race P1, 3 (4.5%) were race P3, 23 (34.3%) were race P7, and 12 (17.9%) were race P8. The races appeared to be evenly distributed in the six provinces although the number of isolates collected was limited and concentrated in Daegu and Gyeongbuk province. Races P7 and P8 have been subdivided from races P1 and P3, respectively, on the basis of their abilities to elicit an HR reaction in PI235047 carrying the Bs4 gene (Sahin and Miller, 1998; Stall et al., 2009). Thus, race P1 was found to be predominant in Korea, followed by race P7, and races P3 and P8 were also identified (Table 1). To date, races P4, P5, P6, P9 and P10 that can overcome the Bs2 gene have not been found in Korea. ECW 20R, ECW 12346, and Chilbok-2 carrying Bs2 gene exhibited hypersensitive resistance to all strains collected in Korea. However, ECW 10R, carrying Bs1, was susceptible to all isolates tested, indicating that races P0, P2 and P5 are not present in Korea, and ECW 30R and Chilbok-3 plants, carrying the Bs3 gene only, were resistant to races P1 and P7 only. In the 1990s, only races P1 and P3 were reported in Korea (Kim et al., 1990; Lee and Cho, 1996; Pae et al., 1996). Thus, the pathotype distribution in Korea has been essentially unchanged for the last 15 years, although races P7 and P8 could be differentiated from race P1 and P3, respectively, according to their ability to induce HR reactions in PI235047 carrying the Bs4 gene. Regarding the relative frequency of pathotypes, Kim et al. (1990) and Lee and Cho (1996) reported that race P3 was detected more frequently than race P1, while Pae et al. (1994) reported that races 1 and 3 were found at a similar frequency. In the present study, however, the majority of the 67 isolates identified were race P1 (43.3%), followed by race P7 (34.3%). Race P1 and race P7 are formerly race 1. Thus, the relative frequencies of pathotypes found in this study differed from the previously reported results. The discrepancy in the frequency of pathotypes may be due to the different collection sites of the studies, since races P1 and P3 can be observed in the same field and even in the same leaf (Kim et al., 1990). Pohronezny et al. (1992) reported a sudden shift in the predominant race from P2 to P1 in Southern Florida, and Kousik and Ritchie (1996) also reported a rapid shift from race P1 to race P2 to race P3 in North Carolina, due to the inactivation of effector genes (avrBs1, avrBs2 and avrBs3) in the pathogen by various mutations, including insertion elements. Likewise, a shift from race P3 to race P1 could have happened in Korea as reported by Oh et al. (2011). However, resistant varieties of pepper plants carrying any of the HR genes have not been distributed for cultivation in Korea. Selection pressure for a race shift, therefore, has not been present in Korea. The difference in the frequency of the races is more likely due to a difference in collection sites, since all strains that were newly isolated for this study from the Chungnam, Jeonbuk, and Jeonnam areas in 2012 were either P1 or P7. Thus, the hypersensitive resistance gene, Bs2, is still functioning in Korea and may be used in breeding for resistance. The validity of Bs1 and Bs3, for resistance, however, is over in Korea (Kim et al., 2009). In breeding for resistance, the exploitation of quantitative resistance not specific to races or the integration of the major HR genes with quantitative resistance would be desirable for more durable resistance (Hibberd, 1989; Jones et al., 2002; Stall et al., 2009; Vallejos et al., 2010). Substantial genetic diversity has been found among Korean strains of the bacterial spot pathogen (Chung et al., 1997). Abundant sources of non-hypersensitive resistance have also been reported (Kim et al., 2009; Sowell, 1960; Sowell and Dempsey, 1977; Tran and Kim, 2007) and are available at hand. Most of the sources of resistance are in hot chili type peppers in horticultural characters including fruit shape. Therefore, these sources of resistance may be more readily available for breeding Korean chili peppers, the most common type of pepper grown in Korea, than for breeding bell type peppers. In the current classification system of bacterial spot pathogen affecting peppers, bacterial spot disease may be caused by one of four species, including X. euvesicatoria, X. vesicatoria, X. gardneri, and X. perforans, or by any mixture of these variants (Jones et al., 2004, 2005; Stall et al., 2009). The association of pathotypes with the four species of Xanthomonas has not yet been reported, but this information would be useful to breed peppers resistant to bacterial spot disease.
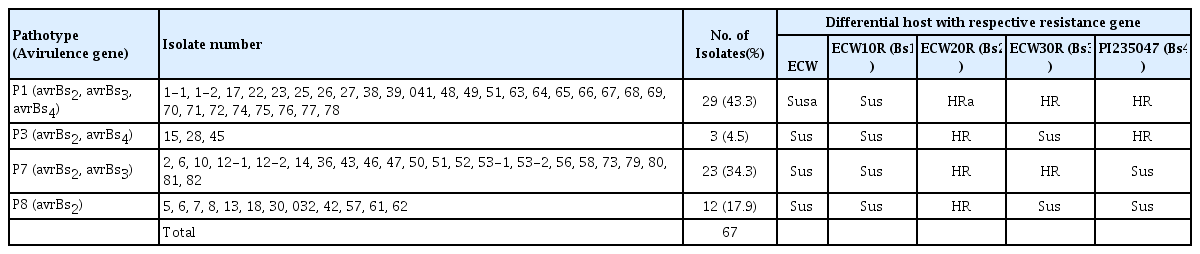
Reaction of differential hosts for identification of pathotypes to isolates of bacterial spot pathogen infecting pepper plants in Korea
Acknowledgment
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2013R1A1A2008630), and by a grant (710001-07-06) from the Agriculture, Food and Rural Affairs Research Center Support Program, Ministry of Agriculture, Food and Rural Affairs.