 |
 |
| Plant Pathol J > Volume 37(2); 2021 > Article |
|
Abstract
The present study describes the bacterial blight of walnut, caused by Xanthomonas arboricola pv. juglandis (Xaj) in the northern Gyeongbuk province, Korea. Disease symptoms that appear very similar to anthracnose symptoms were observed in walnut trees in June 2016. Pathogens were isolated from disease infected leaves, fruits, shoots, bud, flower bud of walnut, and cultured onto yeast dextrose carbonate agar plates. Isolated bacteria with bacterial blight symptoms were characterized for their nutrient utilization profiles using Biolog GN2 and Vitek 2. In addition, isolates were subjected to physiological, biochemical, and morphological characterizations. Furthermore, isolates were identified using 16S rDNA sequence analysis, and multi-locus sequence analysis using atpD, dnaK, efp, and rpoD. To confirm pathogenicity, leaves, fruits, and stems of 3-year-old walnut plants were inoculated with bacterial pathogen suspensions as a foliar spray. One week after inoculation, the gray spots on leaves and yellow halos around the spots were developed. Fruits and stems showed browning symptoms. The pathogen Xaj was re-isolated from all symptomatic tissues to fulfill Koch’s postulates, while symptoms were not appeared on control plants. On the other hand, the symptoms were very similar to the symptoms of anthracnose caused by Colletotrichum gloeosporioides. When walnut plants were inoculated with combined pathogens of Xaj and C. gloeosporioides, disease symptoms were greater in comparison with when inoculated alone. Xaj population size was more in the month of April than March due to their dormancy in March, and sensitive to antibiotics such as oxytetracycline and streptomycin, while resistant to copper sulfate.
Walnut (Juglans sinensis Dode) is one of the economically important nut-producing trees in the world, including Korea (Ozkan and Koyuncu, 2005). Walnuts are considered as one of the healthiest foods in the world, they are a rich source of many beneficial nutrients, antioxidants, and omega-3 fatty acids that are essential for well-being (Gray, 2013), especially it has substantial cardio protective effects on human beings (Djoussé et al., 2001). In Korea, the walnut tree is widely grown as ornamental plants (Korea Forest Service, 2017).
During the cultivation process, walnut trees are often subjected to various pathogens attack. Among them, Xanthomonas arboricola pv. juglandis (Xaj), which causes bacterial blight, has become an important disease for the reduction of crop produce (Lamichhane, 2014). Bacterial blight causes symptoms on leaves, twigs, immature fruits, and stems of walnut trees (Kałużna et al., 2014). Xanthomonas spp. encompass a large group of plant-associated bacteria that usually cause disease. Recently, there has been an increased focus on ecological and genomic studies of xanthomonads. Species are associated with a limited number of hosts and produce either localized or systemic infections (Maes, 1993; Roach et al., 2018). Bacterial blight of walnut can cause reductions in yield up to 70% as the pathogen attacks all parts of the plant (Lang and Evans, 2010; Mulrean and Schroth, 1982). Pathogenic bacteria can survive either on the surface of leaves or inside leaves, corresponding to an epiphytic and endophytic phase, respectively (Lindow and Brandl, 2003), and they compete for nutrients with another phyllosphere microbiome (Özaktan et al., 2010). It is well documented that Xaj has developed virulence mechanisms to inactivate the host’s defense mechanism present in the apoplast (Chisholm et al., 2006; Jones and Dangl, 2006).
In addition, X. arboricola is divided into various pathovars, some of which are classified as quarantine organisms in many countries and are responsible for diseases on nut and stone fruit trees that have emerged worldwide (Fischer-Le Saux et al., 2015). One of the earlier symptoms of leaf infection by xanthomonads is waterlogging of infected tissues, which can be relevant in humid temperate conditions (El-Banoby and Rudolph, 1979; Frutos and López, 2012). Recent studies indicate that the bacterium Xaj is most frequently associated with apical necrosis and cause infections in young nuts with visible symptoms (Moragrega and Özaktan, 2010). In the last decade, disease outbreaks caused by X. arboricola on Juglans have increased globally (Jami et al., 2005; Palacio-Bielsa et al., 2010; Shen et al., 2013). To address the consequences of such possible epidemics, it is important to understand the epidemiology of this disease, about which little is known to date. Reports of bacterial blight on walnut trees have increased but information about the causal pathogen is limited. Detection and identification of Xaj are based on isolations of the pathogen on selective medium, bioassays, and biochemical tests (Loreti et al., 2015), and DNA-based molecular techniques (Zhang et al., 2009).
The present study reports for the first time on the association of Xaj with bacterial blight symptoms on various parts of walnut trees in Korea. The objective of this study was to apply different techniques to identify isolates of Xaj obtained from disease infected walnut trees in the northern Gyeongbuk province in Korea and to analyze the genetic population structure using diverse DNA typing methods: namely, repetitive sequence-based polymerase chain reaction (rep-PCR) and multi-locus sequence analysis (MLSA) if the phylogenetic diversity of the pathogen found in other countries also is present in Korea. The results of this research can help to further elucidate the disease etiology and epidemiology.
During the years from 2016 to 2018 in the month of June, the disease occurrence of bacterial brown rot on walnut trees has been investigated in the walnut orchards at two different locations (Andong and Yeongyang) in the northern Gyeongbuk province, Korea. The disease has been observed in walnut trees from the period June to October at 20 days intervals. For the disease survey analysis, 20 trees were randomly selected, and observations were made on the plants after flowering.
The disease infected samples (leaves, fruits, shoots, buds, and flowers) were collected from the walnut tree (Supplementary Table 1). The symptomatic tissues were cut into squares using a scalpel and surface disinfected by immersion in 2% sodium hypochlorite (NaOCl) solution for 1 min and rinsed 2-3 times in sterile distilled water (SDW), and then treated with 70% ethanol for 30 s and rinsed 2 or 3 times in SDW. Thus, surface-sterilized samples were macerated in 3 ml of SDW for 10 min. Subsequently, 50 μl of suspensions were plated on yeast dextrose calcium carbonate (YDC) agar plates and incubated at 28°C for 3 days. Observations were made for the development of typical yellow, convex, small bacterial colonies. The single colony was streaked on nutrient agar (NA) plates to obtain pure colonies. Bacterial suspensions were maintained at −80°C in YDC broth with sterile glycerol (20%) for long-term storage.
The identification of X. arboricola isolates was carried out based on biochemical and physiological characteristics according to Ménard et al. (2004). The following biochemical tests were performed according to the Laboratory guide for identification of plant pathogenic bacteria (Schaad et al., 2001): Gram staining, oxidative activity, starch hydrolysis, esculin hydrolysis, levan production, arginine hydrolysis, and indole production.
The morphological characterization of the bacterium was analyzed by transmission electron microscopy (TEM) according to the method described by Jeon et al. (2010). Briefly, the bacterial colony was picked with a sterile loop and placed in SDW on a Formvar-coated copper grid and stained with 2% uranyl acetate for negative staining. The preparation was examined under a JEM 1010 electron microscope (JEOL, Tokyo, Japan).
In order to analyze the carbon source utilization of the separated bacteria, a Biolog system was performed. A single colony cultured for 3 days in tryptic soy agar (TSA) medium was suspended in an inoculating fluid (IF-A) solution using a cotton swab, and a Biolog dedicated turbid meter was used. After adjusting the concentration from 90 to 95%, 100 μl was dispensed into each well of a GENIII Microplate (Biolog Inc., Hayward, CA, USA). After incubation for 24 h at 28°C, the use of carbon sources was investigated using a MicroLogTM3-Automated MicroStation System and identified by connecting to a MicroLog Gram-negative database (version 4.02).
The Vitek system is a microbial identification device that uses separated microorganisms as a biochemical analysis method. For analysis, a plastic card containing a very small amount of various drying media and biochemical reactants is used. The bacteria are multiplied and metabolized inside the card, and the results are automatically output after being analyzed on the program over time. The program used was a Vitek 2 Compact (BioMerieux, Paris, France) model. First, bacteria purely cultured in TSA medium were dispensed with 3 ml of 0.45% NaCl in a 5 cm tube to use Densicheck (bioMérieux). The turbidity was adjusted to 0.6 ± 0.05 Mcfarland, fixed in a Vitek rack, and the separated bacterial suspension was filled into a GN card, and the process after filling was performed as per the manufacturer’s instructions.
The selective pathogenic isolates (ANU-XAJ025 and ANU-XAJ035) were subjected to molecular identification based on sequence homology of its 16S rDNA gene (Weisburg et al., 1991). Genomic DNA was isolated using a kit (Genomic DNA Extraction Kit for bacteria, Biofact Co., Seoul, Korea) following the manufacturer’s instructions. The 16S rDNA gene was amplified using polymerase chain reaction (PCR) with Taq DNA polymerase, and primers 27F (5′-AGA GTT TGA TCM TGG CTC AG-3′) and 1492R (5′-GGY TAC CTT GTT ACG ACT T-3′) were used for amplification. The conditions for thermal cycling were as follows: denaturation at 94°C for 5 min followed by 35 cycles at 94°C for 1 min, annealing at 56°C for 30 s and extension at 72°C for 1 min. At the end of the cycle, the reaction mixture was held at 72°C for 10 min and then cooled to 4°C. The electrophoresed PCR product was purified using a PCR purification kit (Biofact Co.). The PCR product obtained was sequenced by an automated sequencer (Genetic Analyzer 3130, Applied Biosystems, Carlsbad, CA, USA). The above primers were used for this purpose. The sequences were compared with the homologs of the strains contained in a genomic database using an NCBI-BLAST tool. Sequence alignment and phylogenetic tree construction were performed using a DNA star program (version 5.02, DNA Star Inc., Madison, WI, USA) and MEGA 4.0 (Biodesign Institute, Tempe, AZ, USA).
Molecular identification of putative pathovars of X. arboricola was performed using the universal primer pair XajF/XajR, whereas Xaj was identified by specific primer set Xaj-neF/Xaj-neR, developed by Gironde et al. (2009) (Supplementary Table 2). X. arboricola. pv. fragariae was used as a negative control using the same set of primers. PCR was carried out in a 25-μl volume of Taq reaction buffer containing 1.0 μl of template DNA, 100 pmol each primer, 200 μM each deoxynucleotide triphosphate, 2.0 mM MgCl2, and 1.0 U of Taq DNA polymerase (Bioneer Co., Daejeon, Korea). For negative controls, the DNA template was replaced with SDW. Amplifications using TXajF/XajR and XajF/XajR primer sets were programmed for 1 cycle of 4 min at 94°C, 30 cycles of 1 min at 94°C, and 1 min at 72°C. PCR amplifications were performed using a thermal cycler machine (MyGenie96/384 Thermal Block, Bioneer Co.).
Partial coding sequences of the housekeeping genes atpD, dnaK, efp, and rpoD were amplified for the isolated bacterium using the primers listed in Supplementary Table 3 (Boudon et al., 2005; Fargier et al., 2011). PCR was carried out in a 20-μl volume of Taq reaction buffer containing 1.0 μl of template DNA, 100 pmol each primer, 200 μM each deoxynucleotide triphosphate, 2.0 mM MgCl2, and 1.0 U of Taq DNA polymerase (Bioneer Co.). PCR amplifications were performed with an initial denaturation at 94°C for 5 min; 35 cycles of denaturation at 94°C for 30 s, annealing at 54°C for 30 s, and extension at 72°C for 1 min; and a final extension at 72°C for 10 min. PCR products were purified and then sequenced. The distance dendrogram of the atpD, dnaK, efp, and rpoD gene sequences determined in this study was generated by the Mega 4.0 package (Biodesign Institute) according to the neighbor-joining method.
Pathogenicity tests were carried out on disease-free walnut plants under artificial inoculation conditions. For the pathogenicity test, the bacterial suspensions cultured at 28°C for 3 days in YNA medium were suspended with SDW and adjusted to a concentration of 108 colony-forming unit (cfu)/ml. Bacterial suspensions were inoculated onto detached leaves, branches, and fruits of 3-year-old healthy walnut plants. For the pathogenicity test on leaves, the leaves were wounded with a sterile needle, followed by inoculation with bacterial suspensions (108 cfu/ml) of Xaj using a foliar spray method, and dried for 1 h. For the pathogenicity test on branches, the selected healthy branches were inoculated with bacterial suspensions using a foliar spray method. The disease symptoms were observed one week after incubation at 28°C. For the pathogenicity test on fruits, the healthy fruits were surface sterilized with 5% sodium hypochlorite (NaOCl) for 5 min and rinsed with SDW. Each fruit was made with 6 wounds on the surface about 2 mm in diameter and inoculated with 1 ml of bacterial suspensions into the wound, dried at room temperature for 1 h, and incubated for one week at 28°C. To compare the differences between the disease symptoms caused by Xaj with the disease symptoms of walnut anthracnose caused by Colletotrichum gloeosporioides, one set of walnut fruits were inoculated with conidia suspensions (105 spores/ml) of C. gloeosporioides, while another set of walnut fruits were inoculated with both bacterial and fungal pathogen suspensions. One set of fruits treated with SDW severed as control. Disease symptoms were observed at 5, 7, 11, and 13 days after incubation at 28°C. To fulfill Koch’s postulates, the pathogen Xaj was re-isolated from all symptomatic tissues to determine morphologically identical to the original isolates.
Flower and leaf buds were randomly collected from the walnut orchard during months of March and April of the following year in Andong, Gyeongsangbuk Province, Korea, where bacterial blight occurred. The collected buds were surface disinfected as above. The surface sterilized samples were ground well using mortar and pestle and diluted to 10-fold serially, and spread onto YDC medium. Yellow colonies were selected from the generated colonies, and direct colony PCR was performed using Xaj specific primer set, Xaj-neF/Xaj-neR (Schaad et al., 2001).
To assess the antibiotic sensitivity test, a total of seven commercially available antibiotics were used against Xaj. About 100 μl bacteria suspensions (107 cfu/ml) cultured for 48-h were spread onto the freshly prepared NA medium. The sterile paper disks (6 mm in diameter) were impregnated with antibiotics at different concentrations (0.5×, 1.0×, and 2.0×) after proper dilution as per the manufacturer’s instructions and placed onto agar plates. The discs loaded with SDW served as a negative control. The clear zone of the inhibition was recorded 48 h after incubation at 28°C and the sensitivity to antibiotics was observed.
Bacterial blight on walnut plants affects all parts including leaf, stem, and fruits (Fig. 1). The symptoms of bacterial blight of walnut appeared as yellow halos around the brown irregular spots on leaves, and later the lesions spread on the entire leaf (Fig. 1A and B). On the fruits, the lesions become sunken as they enlarge (Fig. 1C). Then the lesions tissue become soft in dark brown to black color, and when they become wet, they expand and become burst, resulting in oozing out the bacterial infection from the fruits (Fig. 1D). In addition to the usual cankers on walnut, X. arboricola also causes a newly reported vertical oozing canker on the stem regions or on the main trunk, which often lead to trunk deformities (Fig. 1E). The kernel inside the fruit becomes fully damaged (shown in cross-section) by the pathogen which leads to contamination of other fruits (Fig. 1F).
Typical Xanthomonas colonies were isolated from the diseased leaves, stem, flower buds, and fruits of a 3-year-old walnut tree in the month of June. A total of 50 isolates of Xanthomonads were collected from two different areas (Andong and Yeongyang) in the northern Gyeongbuk province, Korea. Of which, only 14 isolates were identified as Xaj species obtained from two different locations (Andong and Yeongyang) in the northern Gyeongbuk province, Korea (Supplementary Table 1). Among these, only one isolate (ANU-XAJ025) was used for further studies identified as Xaj. The colonies on YDC agar plates appeared as pale yellow, slimy, circular, mucoid round (with a diameter of 2 to 4 mm) after 3 days of incubation at 28°C. When single colonies cultured on NA plates, they appeared yellow and round. During this process, bacteria such as Pseudomonas aeruginosa and P. syringae, and fungi such as C. gloeosporioides and Botryosphaeria dothidea were also detected.
On NA solid medium, morphological characteristics of the Xaj from walnut were light yellow, mucoid, raised colonies, and sticky in nature (Fig. 2A), an enlarged picture of single bacterium colonies shown in Fig. 2B. When viewed by a TEM, the bacteria were found to be rod-shaped (Fig. 2C), approximately 1.0 μm (diameter) × 2.0 μm (length). The enlarged image of an individual bacterium (Fig. 2D), while a single bacterium with one flagellum is shown in Fig. 2E, and the dividing bacterium without flagellum is shown in Fig. 2F. Physiological and biochemical characteristics of Xaj were compared with a reference isolate, Xanthomonas pv. oryzae (Table 1). The bacteria were negative for Gram staining and oxidase activity. It is positive for starch, esculin hydrolyses, and levan production; while negative for arginine hydrolysis and indole production. The Biolog results for Xaj showed that it could be classified as Xanthomonas arboricola, because its closest match with the reference results was shown in the utilization of various carbohydrates examined (Table 2). Comparing these traits to the Biolog database revealed that this strain had a match probability of 94% to X. arboricola. Based on the results of biochemical reactions using the Vitex 2 analysis (Supplementary Table 4), the ANU-Y25 strain was identified as Xaj.
The ANU-XAJY25 isolate was further characterized by 16S rDNA gene sequencing. The 16S rDNA gene sequences from the two isolates ANU-XAJY25 (accession no. MT573402) and ANU-XAJA35 (accession no. MT573403) suggested that ANU-XAJY25 and ANU-XAJA35 isolates belonged to the Xanthomonas genus and had the highest homology to X. arboricola pv. juglandis (99%). Thus, both morphological and molecular characteristics confirmed this species as Xaj. In the phylogenetic tree, two isolates were clustered with other Xanthomonas spp., that are closely related to X. arboricola (boot strap support of 63%) (Fig. 3).
In order to confirm pathovar identity, PCR amplification was performed and obtained a product of 481-bp with the pathovar-specific primer pair XajF/XajR from all the tested isolates (Fig. 4A). PCR products of 181-bp size were obtained from the pathovar Xaj with the specific primer set Xaj-ne-F/Xaj-ne-R (Fig. 4B), whereas the negative control pv. fragariae did not generate a visible product upon ethidium bromide staining of agarose gel (Fig. 4). For further genetic determination of Xaj, concatenated housekeeping data of atpD, dnaK, efp, and rpoD gene loci were analyzed. The dendrogram constructed on the basis of the concatenated gene sequences of domestic isolates and compared with overseas strains (Fig. 5A). The strains (ANU-A35, ANU-A36, and ANU-A37) isolated from young walnut trees of the Andong region are grouped with those strains of Xaj from New Zealand and Italy; while the strains (ANU-Y12, ANU-Y13, and ANU-Y15) isolated from the old walnut trees of Yeongyang have high similarity to those of Chinese isolate. The amplified housekeeping gene products were shown in Fig. 5B. The two Xaj domestic strains (ANU-XAJY25 and ANU-XAJA35) have been published in the NCBI database with accession numbers MT573402 and MT573403, respectively, and then grouped in a monophyletic cluster. The sequence analysis of the concatenated data of all the loci of Xaj strains showed clear genetic differences with other overseas Xanthomonas pathovars.
The pathogenicity of isolated pathovar Xaj was tested on detached leaves, stems, and fruits of 3-year-old walnut plants (Figs. 6 and 7). The leaves from 3-year-old walnut trees were inoculated with 1 ml bacterial suspensions at the concentration of 108 cfu/ml. Xaj started to induce blight symptoms on the leaves appearing with black spots, which later developed into yellow halos 1 week after inoculation (Fig. 6A), whereas no symptoms were developed in non-inoculated fruits (Fig. 6B). In the case of the stem, the leaf veins showed sunken symptoms in inoculated stems, turned totally black one-week after inoculation (Fig. 6C), while no symptoms were observed in non-inoculated stems (Fig. 6D). In the case of a pathogenicity test on fruits, disease symptoms started to develop on fruits inoculated with bacterial suspensions or fungal spores or combined inoculation (1:1 ratio) 5 days after inoculation (DAI) (Fig. 7). Black lesions on fruits were detected 7 DAI, and the entire fruit was developed with black lesions and some fruits showed rotting symptoms from 13 DAI (Fig. 7A). Fruits inoculated with spore suspensions of C. gloeosporioides, the onset of disease symptoms began to appear on fruits 5 DAI, and later lesions spread rapidly from 7 DAI, and the entire fruit was developed with anthracnose symptoms from 13 DAI (Fig. 7B). Symptoms with this bacterial pathogen were similar to those of walnut anthracnose. When the fruits inoculated with a combination of both bacterial and fungal pathogen suspensions, similar symptoms began to appear 5 DAI, but the expansion of the lesions was found to be faster than the individual inoculation. White hyphae began to grow on the surface of fruits, and some fruits began to crack 11 DAI, while there was the formation of mycelium and spores on 13 DAI (Fig. 7C). The disease severity was higher when fruits inoculated with combined inoculation than inoculation with either alone, whereas non-inoculated fruits remained symptomless (Fig. 7D). The pathogen was re-isolated from the symptomatic tissues and cultured on YDS medium at 28°C for 3 days, to fulfill Kock’s postulates.
Xaj population was detected both in flower and leaf buds (Fig. 8). However, the abundance of Xaj population is varied from both flower buds and leaf buds based on the time gap of collection. The average population size of Xaj cells was detected as 220.6 cells/bud in March, which was increased in April by 5 times. In over all, Xaj population size was detected more in flower buds than in leaf buds, and the proportion of Xaj colonies recovered from total bacterial colonies isolated from both flower and leaf buds increased to a greater level in the month of April than March (Fig. 8). The proportion of Xaj colonies obtained from leaf buds were 36.8% and 97.4% in March and April, respectively, while the colonies obtained from flower buds were detected as 87.8% and 97.4% in March and April, respectively (Fig. 8 line graph). These results suggest that the Xaj pathogen survives in the flower and leaf buds at a dormant stage during winter season, but starts to multiply when the temperature rises in the month of April.
In the antibiotic sensitivity test, the pathogen Xaj strain in our study showed high sensitivity to oxytetracycline and streptomycin with inhibition zones of 18.3 mm and 16.6 mm, respectively at 2.0× concentration, while the zone of inhibition was 15.3 mm when oxytetracylcine and streptomycin applied in combination (Table 3). The pathogen Xaj was very less sensitive to copper hydroxide, and combined application of copper oxychloride and kasugamycin as well. Whereas, the pathogen was strongly resistant to copper sulfate basic and dithianon, for which no inhibition zone was observed.
Bacterial blight of walnut is one of the most severe economic diseases of Juglans sinensis Dode and the disease has been known for the past century from all over the world, where walnut orchards cultivation is being practiced (Barionovi and Scortichini, 2008; Giovanardi et al., 2016; Webber et al., 2020). In this study, we isolated and characterized the pathogen Xaj from the orchards of walnut that causes bacterial blight in the northern Gyeongbuk province, Korea, on various parts, including leaves, fruits, and stems, resulting in severe damage to the crop. When the disease is under severe conditions, many nuts may fall prematurely, while others reach full size, but the kernel becomes blackened, leading to becoming wrinkled (Galán, 2009).
Various methods were used to characterize Xaj isolates from walnuts in our study and found the biochemical and physiological characteristics, and molecular analysis was effective for identification of isolates to the species X. arboricola, as supported by previous studies (Garita-Cambronero et al., 2018; Hajri et al., 2010). Morphophysiological features of the pathogenic Xaj isolates were studied by culturing and observing colonies on NA medium; on the other hand, a previous study by Giovanardi et al. (2016) reported that the similar colonies were observed in GYCA medium. Xaj is one of the most disease-causing pathovars within the X. arboricola complex that cause severe economic damage in various fruits and nuts in the world (Timilsina et al., 2020). The other pathovar such as X. arboricola pv. pruni causes bacterial spots on prunus (Roselló et al., 2012). Recently, Giovanardi et al. (2016) investigated the morphological and genotypic features of Xaj isolates and compared the assessment of different virulence on walnut fruits, resulting in showing a clear intra-pathovar variation by the presence of different haplotypes.
Identification of Xaj isolates was further confirmed by PCR, using the specific primer pairs XajF/XajR. This primer pair proved to be very specific in the study of Kałuzna et al. (2014) when isolates of Xanthomonas species were analyzed with the Xaj-ne-F/Xaj-ne-R primer pair. All isolates generated the 181 bp amplification product, but our results are in contradiction to a previous study by Balaž et al. (2016), where all isolates of X. hortorum did not generate the same size of amplicons. These results demonstrated the importance of PCR method for the identification of pathovars for fast diagnostic analysis. The present work constitutes the first step towards a better understanding of the etiology of the disease. On the basis of biochemical tests, Biolog analysis, pathogenicity tests, and analysis performed using rep-PCR, this study ascertained that Xaj is the causal agent of the disease. Interestingly, when the pathogen Xaj is associated with the fungal pathogen C. gloeosporioides, the symptoms were more severe than when inoculated with bacterial or fungal pathogens individually. These results are corroborative with a previous report by Belisario et al. (2002), where the bacterial pathogen X. arboricola was associated with pathogenic fungi such as Alternaria sp., and Fusarium sp. to cause brown apical necrosis of walnut fruits at an increased level. Similarly, Ma et al. (2013) reported that an increased level of root rot of ginseng was caused by a combination of fungal pathogens such as Alternaria panax or Cylindrocarpon destructans and bacteria Pseudomonas sp. or Ralstonia sp. However, the mechanism of synergism of pathogens in the disease complex is still unclear.
MLSA analysis identified all of our isolates as Xaj and placed them in one cluster, clearly differentiated from other X. arboricola pathovars. The sequence analysis of atpD, dnaK, efp, and rpoD genes of Xanthomonas isolates from disease infected walnut trees in Korea showed phylogenetic dendrogram of the concatenated data analysis. These housekeeping genes have become the standard today to investigate intraspecific variability for phylogenetic analyses of bacterial species and were proposed as an alternative to DNA-DNA hybridizations for species delineation (Fischer-Le Saux et al., 2015; Gevers et al., 2005). MLSA allows strain assignment at the species level (Pérez-Losada et al., 2013; Young et al., 2008), and represents an easy, reproducible, and portable approach for pathogen identification at the intra-species level (Maiden, 2006). In our study, the genetic relationship among Xaj isolates in Korea was found to be similar to the strains from New Zealand, Italy, and China based on the use of conserved housekeeping genes.
It is interesting to note that, in our pathogenicity test, results showed that detached parts such as leaves, stems, and fruits of walnut tree are susceptible to Xaj, hence the pathogen is considered highly virulent for walnut. To our knowledge, this is the first report on the identification and characterization of Xaj strain that causes bacterial blight on walnut trees in the northern Gyeongbuk province, Korea, which encompasses representative strains of all pathovars classified in the species. Pathogenicity tests of most of the strains studied were previously reported in pathovar descriptions (Hajri et al., 2010; Yirgou, 1964). Furthermore, an antibiotic sensitivity test revealed that the pathogen Xaj was sensitive to a few antibiotics, while resistant to copper sulfate basic. These results are in contradiction with a recent study by Webber et al. (2020), where they demonstrated that X. arboricola pv. corylina was sensitive to copper. Results obtained in this study indicate that some pathogenic and genetic variation exists in the Xaj pathogen population in Korea when isolated from leaves, stems, fruits, and flower and leaf buds. On the other hand, population study of Xaj in flower and leaf buds determines the survival of Xaj pathogen in walnut tree during winter season without exhibiting any visual symptoms, while its population increased to five times greater in April than in March. Therefore, it is desirable to apply an appropriate antimicrobial agent in March before the number of the population increases. This finding could provide a basis for further research and understanding of the variation among Xaj populations and their epidemiology, and a deeper insight into the occurrence of blight infections on leaf, stem, and fruits of walnut in Korea.
Acknowledgments
This work was supported by grants from the National Institute of Forest Science and Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2018R1A6A1A03024862).
Electronic Supplementary Material
Supplementary materials are available at The Plant Pathology Journal website (http://www.ppjonline.org/).
Supplementary Table 1.
Isolation of Xanthomonas arboricola pv. juglandis strains used in this study
Supplementary Table 2.
List of primers used for PCR amplification of Xanthomonas arboricola pv. juglandis
Supplementary Table 3.
List of primers used for PCR amplification of housekeeping genes
Supplementary Table 4.
Biochemical characteristics of Xaj using Vitek 2
Fig. 1
Disease symptoms of bacterial blight caused by Xanthomonas arboricola pv. juglandis on leaves, fruits, and stem of walnut in field conditions. The symptoms on leaves appear as small brown spots in an irregular form that coalesced to form necrotic areas and brown to black lesions surrounded by green-yellow chlorotic haloes (A, B). Symptoms on fruits appeared as sunken and water-soaked lesions (C), later becomes expanded and black colored sap oozes out (D). The cankers appear on the stem (E), and the kernel from immature fruit becomes damaged which is shown in cross-section (F).

Fig. 2
Morphological characteristics of Xanthomonas arboricola pv. juglandis isolate ANU-Y25 cultured on tryptic soy agar plate and observed via transmission electron microscopy showing light yellow colonies with round shaped (A), dotted lines show enlarged images of individual colonies (B), rod shaped bacterial cells (C), enlarged image of an individual bacterium (D), a single bacterium with one flagellum (E), and dividing bacterium (F). The experiment was carried out at least twice with three replicates per treatment. Scale bars = 2 μm (C), 0.5 μm (D), 1 μm (E, F).
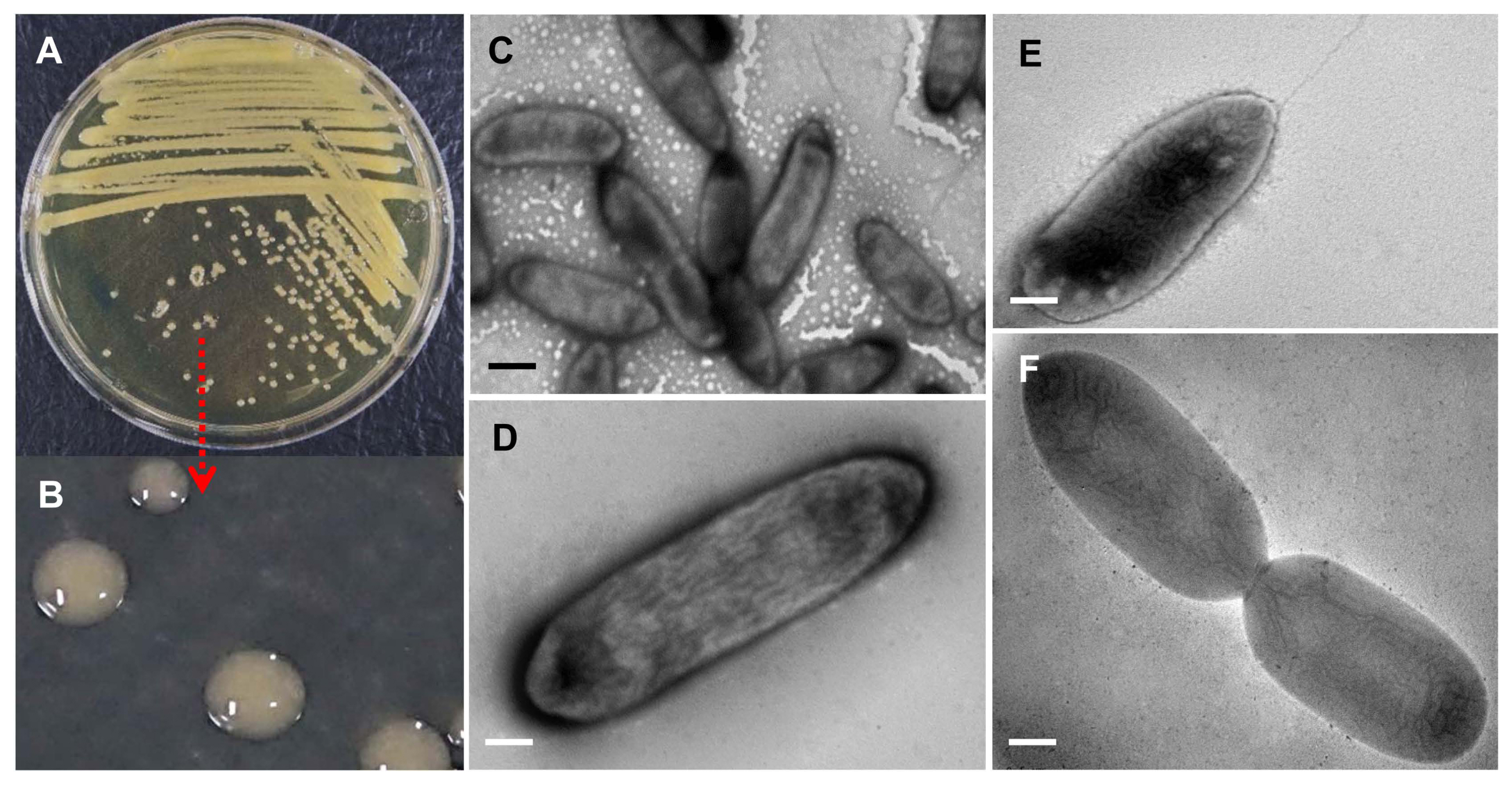
Fig. 3
Phylogenetic analysis of Xanthomonas arboricola pv. juglandis and other bacterial species based on 16S rDNA sequences. The tree was constructed using the neighbor-joining method (MEGA 4.0), based on the 16S rDNA gene amplification. The numbers above the branches represent the bootstrap values obtained for 100 replicates.
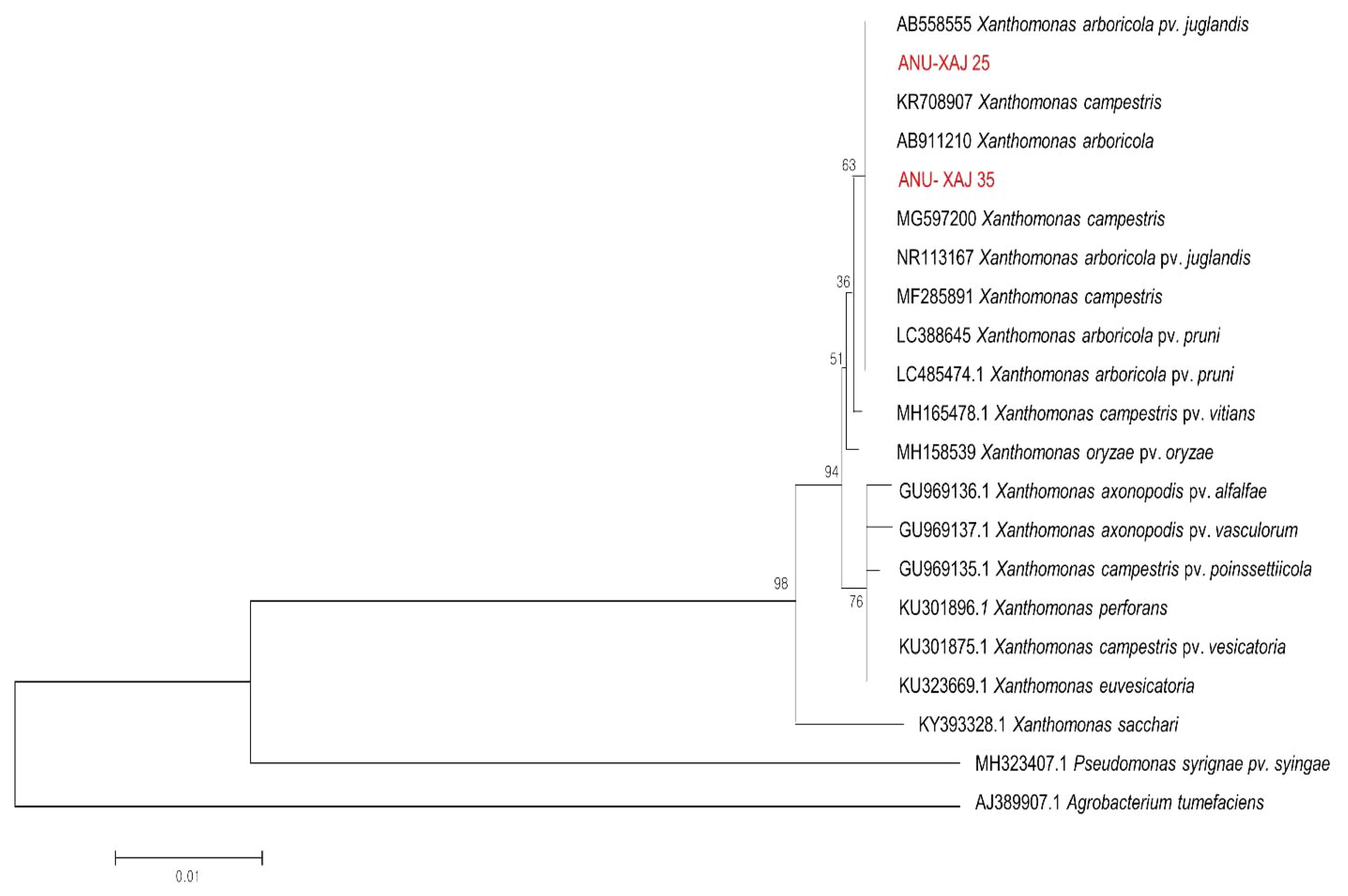
Fig. 4
Gel electrophoresis analysis of PCR products. (A) PCR amplicons of Xanthomonas arboricola amplified with universal primer pair XajF/XajR for all isolated pathovars. (B) PCR amplicons of Xanthomonas arboricola pv. juglandis (Xaj) amplified with Xaj-neF/ Xaj-neR-specific primers only for Xaj. Lanes 1 and 19, DNA molecular size marker (100-bp ladder); lane 2, ANU-XAJ002; lane 3, ANU-XAJ012; lane 4, ANU-XAJ013; lane 5, ANU-XAJ021; lane 6, ANU-XAJ022; lane 7, ANU-XAJ025; lane 8, ANU-XAJ026; lane 9, ANU-XAJ027; lane 10, ANU-XAJ028; lane 11, ANU-XAJ030; lane 12, ANU-XAJ031; lane 13, ANU-XAJ035; lane 14, ANU-XAJ036; lane 15, ANU-XAJ037; lanes 16-18, ANU-XAJ0 negative controls.

Fig. 5
Multi-locus sequence analysis-based phylogenetic relationship between the isolates from this study and selected Xanthomonas arboricola pv. juglandis (Xaj) isolates from other countries. (A) The tree was constructed using the neighbor-joining method, based on the concatenated housekeeping sequence data of atpD, dnaK, efp, and rpoD. For comparison, the sequences of the genes of other Xaj strains are included. The numbers above the branches represent the bootstrap values obtained for 50,000 replicates. Bar = estimated nucleotide substitutions per site = 0.0005. The source of the Korean isolates are indicated by the following colors: isolates from Andong (red), isolates from Yeongyang (blue). (B) Amplified PCR products of four housekeeping genes from Xaj. M, 1.0 kb ladder; 1, atpD (ATP synthase β subunit); 2, dnaK (70-kDa heat shock protein); 3, efp (elongation factor P); 4, rpoD (RNA polymerase sigma 70 factor).

Fig. 6
Pathogenicity test with Xanthomonas arboricola pv. juglandis on detached walnut leaves and stem by artificial inoculation. (A) Development of disease symptoms on wounded leaves 2 weeks after inoculation with yellow halos around the black spots. (B) No disease symptoms were developed on non-inoculated leaves. (C) Development of disease symptoms on inoculated stem 1 week after inoculation (the sites of inoculation noted by arrows). A cross-section of the stem shown at the corner represents the disease spread inside the stem tissue. (D) No symptoms were developed on the non-inoculated stem (control).

Fig. 7
Pathogenicity test on walnut fruits. The immature walnut fruits were artificially inoculated with bacterial suspensions (108 cfu/ml) of Xanthomonas arboricola pv. juglandis (A) or C. gloeosporioides spores (105 spores/ml) (B) or combined inoculation of bacterial and fungal pathogens in 1:1 ratio (C). Disease symptoms were recorded at 5, 7, 11, and 13 days after inoculation. (D) No symptoms were developed on non-inoculated (control) fruits. The experiment was performed two times.
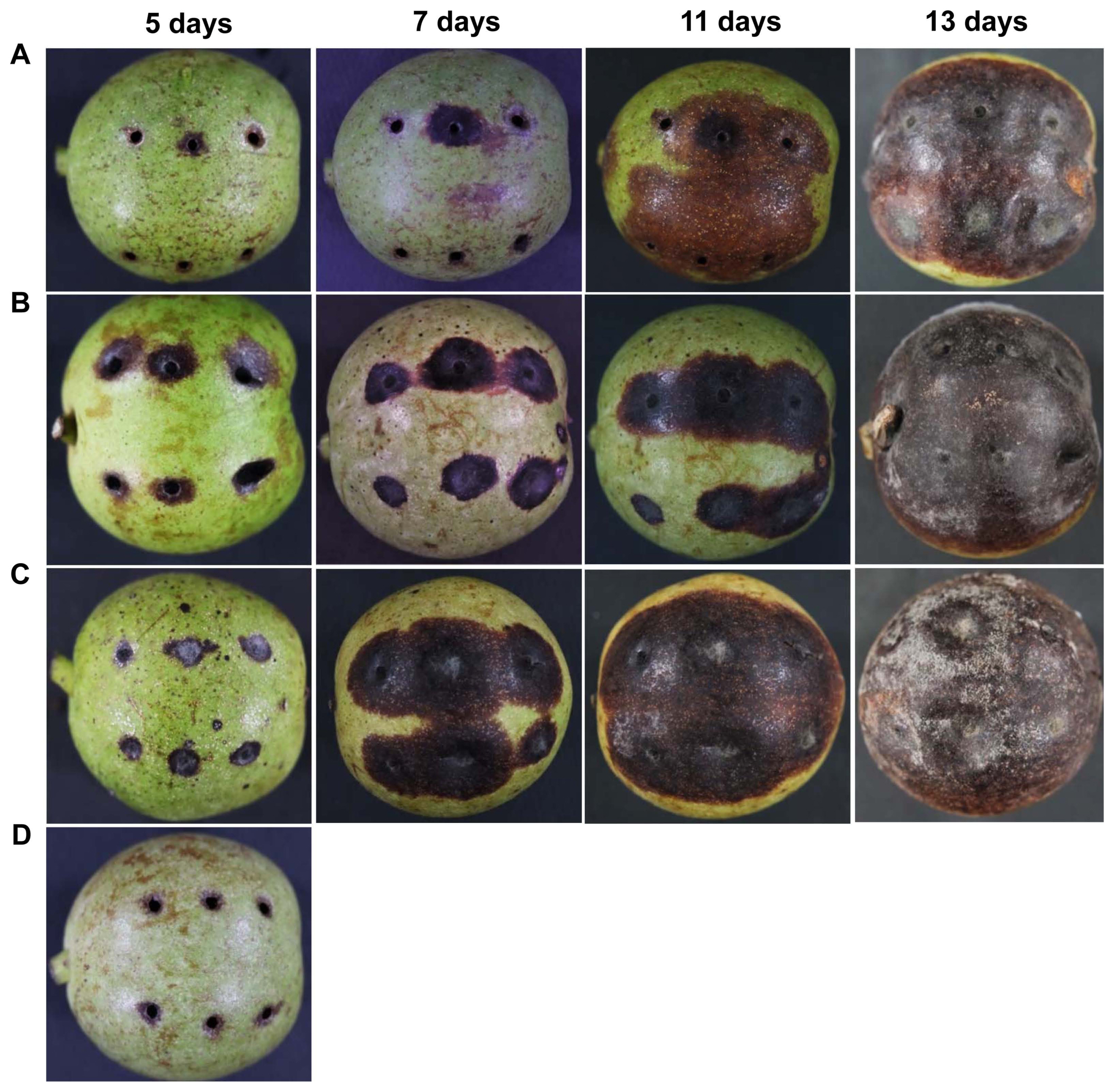
Fig. 8
Population size of Xanthomonas arboricola pv. juglandis (Xaj) recovered from dormant walnut flower bud (FB) and leaf bud (LB) in orchard during March and April months. Line graph represents the percentage of Xaj from all recovered cells. Twenty samples (replicates) were collected from each orchard.
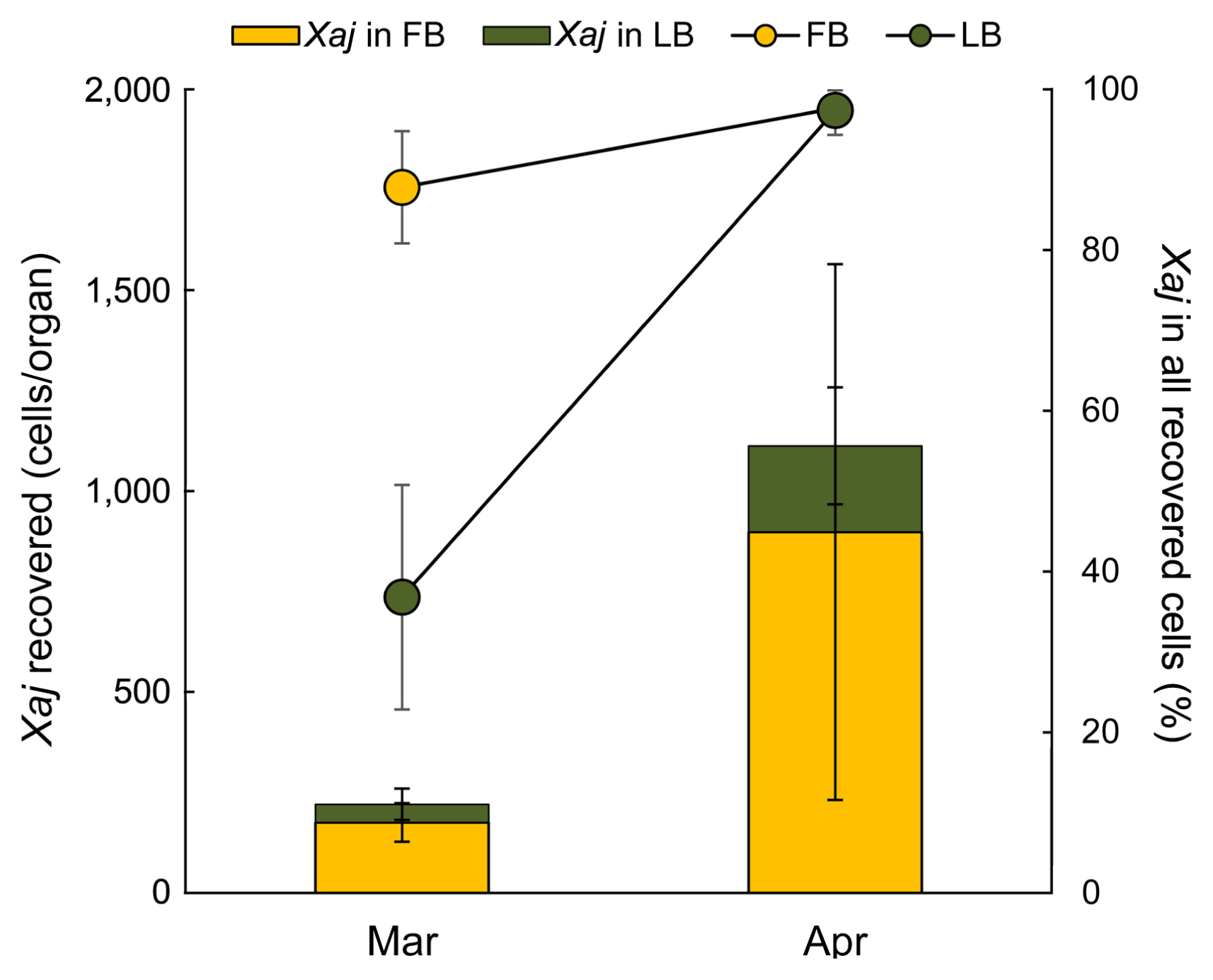
Table 1
Biochemical characteristics of Xanthomonas arboricola pv. juglandis (Xaj) and X. oryzae pv. oryzae
| Test | Reaction | Xaj (Eskandiari et al., 2018) | X. oryzae pv. oryzaea (Padmaja et al., 2017) |
|---|---|---|---|
| Gram reaction | − | − | − |
| Mucoid growth on YDC | Yellow pigment | Yellow pigment (in NSA medium) | Yellow pigment |
| Oxidative | Aerobe | Aerobe | Oxidative |
| Oxidase | ND | ND | ND |
| Growth at 35°C | + | ND | + |
| Starch hydrolysis | + | + | + |
| Esculin hydrolysis | + | + | + |
| Levan production | + | + | + |
| Arginine hydrolysis | ND | ND | + |
| Indole production | ND | ND | + |
Table 2
Growth characteristics of two Xanthomonas arboricola pv. juglandis (Xaj) isolates on diverse carbon sources assessed using Biolog
Table 3
Antibiotic sensitivity test for Xanthomonas arboricola pv. juglandis isolate ANU-Y25 using various concentrations
References
Balaž, J., Ivanović, Ž., Davidović, A., Iličić, R., Janse, J. and Popović, T. 2016. Characterization of Xanthomonas hortorum pv. pelargonii isolated from geranium in Serbia. Plant Dis. 100:164-170.


Barionovi, D. and Scortichini, M. 2008. Integron variability in Xanthomonas arboricola pv. juglandis and Xanthomonas arboricola pv. pruni strains. FEMS Microbiol Lett. 288:19-24.


Belisario, A., Maccaroni, M., Corazza, L., Balmas, V. and Valier, A. 2002. Occurrence and etiology of brown apical necrosis on Persian (English) walnut fruit. Plant Dis. 86:599-602.


Boudon, S., Manceau, C. and Nottéghem, J.-L. 2005. Structure and origin of Xanthomonas arboricola pv. pruni populations causing bacterial spot of stone fruit trees in Western Europe. Phytopathology. 95:1081-1088.


Chisholm, S. T., Coaker, G., Day, B. and Staskawicz, B. J. 2006. Host-microbe interactions: shaping the evolution of the plant immune response. Cell. 124:803-814.


Djoussé, L., Levy, D., Cupples, L. A., Evans, J. C., D'Agostino, R. B. and Ellison, R. C. 2001. Total serum bilirubin and risk of cardiovascular disease in the Framingham offspring study. Am J Cardiol. 87:1196-1200.


El-Banoby, F. E. and Rudolph, K. 1979. Induction of water-soaking in plant leaves by extracellular polysaccharides from phytopathogenic pseudomonads and xanthomonads. Physiol Plant Pathol. 15:341-349.

Eskandari, S., Khakvar, R. and Torchi, M. 2018. Detection of bacterial blight of walnut (Xanthomonas arboricola pv. juglandis) in northwest of Iran and investigation of its pathogenicity on different native walnut cultivars. Biochem Cell Arch. 18:1785-1788.
Fargier, E., Fischer-Le Saux, M. and Manceau, C. 2011. A multi-locus sequence analysis of Xanthomonas campestris reveals a complex structure within crucifer-attacking pathovars of this species. Syst. Appl. Microbiol. 34:156-165.


Fischer-Le Saux, M., Bonneau, S., Essakhi, S., Manceau, C. and Jacques, M.-A. 2015. Aggressive emerging pathovars of Xanthomonas arboricola represent widespread epidemic clones distinct from poorly pathogenic strains, as revealed by multilocus sequence typing. Appl. Environ. Microbiol. 81:4651-4668.



Frutos, D. and López, G. 2012. Search for Juglans regia genotypes resistant/tolerant to Xanthomonas arboricola pv. juglandis in the framework of cost action 873. J. Plant Pathol. 94(1 Suppl):S1.37-S1.46.
Galán, J. E. 2009. Common themes in the design and function of bacterial effectors. Cell Host Microbe. 5:571-579.



Garita-Cambronero, J., Palacio-Bielsa, A. and Cubero, J. 2018. Xanthomonas arboricola pv. pruni, causal agent of bacterial spot of stone fruits and almond: its genomic and phenotypic characteristics in the X. arboricola species context. Mol Plant Pathol. 19:2053-2065.



Gevers, D., Cohan, F. M., Lawrence, J. G., Spratt, B. G., Coenye, T., Feil, E. J., Stackebrandt, E., Van de Peer, Y., Vandamme, P., Thompson, F. L. and Swings, J. 2005. Re-evaluating prokaryotic species. Nat. Rev. Microbiol. 3:733-739.


Giovanardi, D., Bonneau, S., Gironde, S., Fischer-Le Saux, M., Manceau, C. and Stefani, E. 2016. Morphological and genotypic features of Xanthomonas arboricola pv. juglandis populations from walnut groves in Romagna region, Italy. Eur. J. Plant Pathol. 145:1-16.

Gironde, S., Guillaumes, J. and Manceau, C. 2009. Specific detection of Xanthomonas arboricola pv. juglandis pathogen on walnut. EPPO Conference on Diagnostics. 1 Food and Environment Research Agency, York, UK.
Gray, J. 2013. Nuts and seeds. In: Encyclopedia of human nutrition, 3rd ed. eds. by B. Caballero, 329-335. Academic Press, Amsterdam, The Netherlands.

Hajri, A., Meyer, D., Delort, F., Guillaumès, J., Brin, C. and Manceau, C. 2010. Identification of a genetic lineage within Xanthomonas arboricola pv. juglandis as the causal agent of vertical oozing canker of Persian (English) walnut in France. Plant Pathol. 59:1014-1022.

Jami, F., Kazempour, M. N., Elahinia, S. A. and Khodakaramian, G. 2005. First report of Xanthomonas arboricola pv. pruni on stone fruit trees from Iran. J Phytopathol. 153:371-372.

Jeon, Y. H., Kim, S. G., Hwang, I. and Kim, Y. H. 2010. Effects of initial inoculation density of Paenibacillus polymyxa on colony formation and starch-hydrolytic activity in relation to root rot in ginseng. J. Appl. Microbiol. 109:461-470.



Kałużna, M., Pulawska, J., Waleron, M. and Sobiczewski, P. 2014. The genetic characterization of Xanthomonas arboricola pv. juglandis, the causal agent of walnut blight in Poland. Plant Pathol. 63:1404-1416.

Korea Forest Service 2017. 2017 Statistical yearbook of forestry. Korea Forest Service, Daejeon, Korea. 438.
Lamichhane, J. R. 2014. Xanthomonas arboricola diseases of stone fruit, almond, and walnut trees: progress toward understanding and management. Plant Dis. 98:1600-1610.


Lang, M. D. and Evans, K. J. 2010. Epidemiology and status of walnut blight in Australia. J. Plant Pathol. 92(1 Suppl):S1.49-S1.55.
Lindow, S. E. and Brandl, M. T. 2003. Microbiology of the phyllosphere. Appl. Environ. Microbiol. 69:1875-1883.



Loreti, S., Pucci, N., Perez, G., Catara, V., Scortichini, M., Bella, P., Ferrante, P., Giovanardi, D. and Stefani, E. 2015. Detection and identification of Xanthomonas arboricola pv. pruni from symptomless plant material: results of an Italian test performance study. EPPO Bull. 45:41-51.

Ma, L., Cao, Y. H., Cheng, M. H., Huang, Y., Mo, M. H., Wang, Y., Yang, J. Z. and Yang, F. X. 2013. Phylogenetic diversity of bacterial endophytes of Panax notoginseng with antagonistic characteristics towards pathogens of root-rot disease complex. Antonie Van Leeuwenhoek. 103:299-312.


Maes, M. 1993. Fast classification of plant-associated bacteria in the Xanthomonas genus. FEMS Microbiol Lett. 113:161-165.

Ménard, M., Delort, F., Baudry, A. and Le Saux, M. 2004. First report of bacterial canker of walnut caused by Brenneria nigrifluens in France. Plant Dis. 88:220.

Moragrega, C. and Özaktan, H. 2010. Apical necrosis of Persian (English) walnut (Juglans regia): an update. J. Plant Pathol. 92(1 Suppl):S1.67-S1.71.
Mulrean, E. N. and Schroth, M. N. 1982. Ecology of Xanthomonas campestris pv. juglandis on Persian (English) walnut. Phytopathology. 72:434-438.

Özaktan, H., Erdal, M., Aslan, E., Akköprü, A. and Bozkurt, A. 2010. Evaluation of bacterial antagonists and some chemicals to control of bacterial blight of walnut in Turkey. In: Annual Meeting of COST Action 873 on Bacterial Diseases of Sone Fruits and Nuts; pp 20Riga, Latvia.
Ozkan, G. and Koyuncu, M. A. 2005. Physical and chemical composition of some walnut (Juglans regia L) genotypes grown in Turkey. Grasas y Aceites. 56:141-146.
Padmaja, G., Devi, G. U., Yugander, A., Srinivas, C., Durga Rani, Ch. V., Sundaram, R. M. and Laha, G. S. 2017. Biochemical characterization of Xanthomonas oryzae pv. oryzae isolates collected from Telangana and Andhra Pradesh. Indian J Plant Prot. 45:193-199.
Palacio-Bielsa, A., Roselló, M., Cambra, M. A. and López, M. M. 2010. First report on almond in Europe of bacterial spot disease of stone fruits caused by Xanthomonas arboricola pv.pruni. Plant Dis. 94:786.

Pérez-Losada, M., Cabezas, P., Castro-Nallar, E. and Crandall, K. A. 2013. Pathogen typing in the genomics era: MLST and the future of molecular epidemiology. Infect Genet Evol. 16:38-53.


Roach, R., Mann, R., Gambley, C. G., Shivas, R. G. and Rodoni, B. 2018. Identification of Xanthomonas species associated with bacterial leaf spot of tomato, capsicum and chilli crops in eastern Australia. Eur. J. Plant Pathol. 150:595-608.

Roselló, M., Santiago, R., Palacio-Bielsa, A., García-Figueres, F., Montón, C., Cambra, M. A. and López, M. M. 2012. Current status of bacterial spot of stone fruits and almond caused by Xanthomonas arboricola pv. pruni in Spain. J. Plant Pathol. 94(1 Suppl):S1.15-S1.21.
Schaad, N. W., Jones, J. B. and Chun, W. 2001. Laboratory guide for identification of plant pathogenic bacteria. 3rd ed. American Phytopathological Society Press, St. Paul, MN, USA. 373.
Shen, Y. M., Huang, T. C., Chao, C. H. and Liu, H. L. 2013. First report of bacterial spot caused by Xanthomonas arboricola pv. pruni on Japanese plum in Taiwan. Plant Dis. 97:835.

Timilsina, S., Potnis, N., Newberry, E. A., Liyanapathiranage, P., Iruegas-Bocardo, F., White, F. F., Goss, E. M. and Jones, J. B. 2020. Xanthomonas diversity, virulence and plant-pathogen interactions. Nat. Rev. Microbiol. 18:415-427.



Webber, J. B., Putnam, M., Serdani, M., Pscheidt, J. W., Wiman, N. G. and Stockwell, V. O. 2020. Characterization of isolates of Xanthomonas arboricola pv. corylina, the causal agent of bacterial blight, from Oregon hazelnut orchards. J. Plant Pathol. 102:799-812.

Weisburg, W. G., Barns, S. M., Pelletier, D. A. and Lane, D. J. 1991. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 173:697-703.



Yirgou, D. 1964. Xanthomonas guizotiae sp. nov. on Guizotia abyssinica. Phytopathology. 54:1490-1491.



 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Supplement1
Supplement1 Print
Print




