RNA Modification and Its Implication in Plant Pathogenic Fungi
Article information
Abstract
Interaction of a pathogen with its host plant requires both flexibility and rapid shift in gene expression programs in response to environmental cues associated with host cells. Recently, a growing volume of data on the diversity and ubiquity of internal RNA modifications has led to the realization that such modifications are highly dynamic and yet evolutionarily conserved system. This hints at these RNA modifications being an additional regulatory layer for genetic information, culminating in epitranscriptome concept. In plant pathogenic fungi, however, the presence and the biological roles of RNA modifications are largely unknown. Here we delineate types of RNA modifications, and provide examples demonstrating roles of such modifications in biology of filamentous fungi including fungal pathogens. We also discuss the possibility that RNA modification systems in fungal pathogens could be a prospective target for new agrochemicals.
Gene expression is regulated by a large number of cellular factors comprising multiple layers that are entwined with one another. At chromatin level, epigenetic factors such as DNA methylation and histone modifications control accessibility of transcription factors and transcriptional machineries to the portion of DNA sequences encoding genetic information (Razin and Kantor, 2005). Following transcription, the fate of RNA molecules is determined by several regulatory mechanisms including modification of coding RNAs as well as non-coding RNAs (ncRNAs) (Boo and Kim, 2020; Frye et al., 2018; Roundtree et al., 2017). The most well-known examples of RNA modifications are 5′ capping, 3′ polyadenylation, and splicing of pre-mRNAs, which control export via nuclear pore, stability, degradation and translation of mRNAs (Gagliardi and Dziembowski, 2018; Proudfoot et al., 2002; Wilkinson et al., 2020). One of the key players that enable and mediate such control is RNA binding proteins (RBPs) (Corley et al., 2020). These proteins usually bind to RNA molecules via their recognition of cis-acting elements in the target RNA. The RBPs also interact with other RBPs, leading to the formation of ribonucleoprotein complexes (RNPs) (Tartaglia, 2016). It is the RNPs that regulate the distribution and amount of RNAs across cellular compartments and across time.
Since the report on RNA modifications as early as 1958 (Adler et al., 1958), 172 RNA modifications other than the cap and tail modification of mRNA have been documented to date, according to the database dedicated to archival of RNA modifications (Boccaletto et al., 2018). RNA modifications have been observed in all domains of life and across many RNA types. Such expansion of RNA modification catalogue is indebted to the rapid advances in approaches including sequencing technology for detection of modification sites in a diverse array of RNA molecules. Technical aspects of RNA modification detection are extensively reviewed elsewhere (Helm and Motorin, 2017; Motorin and Marchand, 2021; Ovcharenko and Rentmeister, 2018), and thus are not covered in our review. RNA modifications that have been identified up to date include internal modifications such as pseudouridine and N6-methyladenosine (m6A) on mRNA and ncRNAs. It is now well established that almost all types of RNAs including tRNAs and rRNAs are heavily modified (Decatur and Fournier, 2002; Krutyhołowa et al., 2019; Lyons et al., 2018; Sloan et al., 2017). Discoveries of these RNA modifications have opened up new research avenues for understanding the roles of RNA modifications during fungal pathogenesis. This review briefly describes the recent advances in our understanding of RNA modifications, and discuss potential roles of such RNA modifications in fungal development and pathogenesis that might hint at new targets for development of agrochemicals. However, we do not discuss processing of pre-mRNAs (5′ capping, 3′ polyadenylation, and splicing), siRNA and miRNAs, which are well known and have been extensively reviewed elsewhere (Gagliardi and Dziembowski, 2018; Ghildiyal and Zamore, 2009; O’Brien et al., 2018; Shelton et al., 2016; Yu and Chen, 2010).
Types and Biological Implication of RNA Modifications
RNA modifications are as diverse as chemical alteration of existing bases or ribose, and cleavage of RNA molecules at specific sites. The most common modification in cellular RNA is isomerization of the uridine base that leads to the formation of pseudo-uridine (ψ) (Fig. 1), which in turn affect the secondary structure of RNA (Cohn, 1960). The ψ is particularly abundant in rRNA and tRNA, and dynamically regulated by either RNA-guided mechanism or the Pus (Pseudouridine synthase) family enzymes (Rintala-Dempsey and Kothe, 2017). Another modification that is abundant in tRNA and rRNA is 2′-O-methylation of the ribose (2′-O-Me). Considering that 2-hydroxyl of ribose is frequently involved in formation of higher order RNA structures, 2′-O-Me might have significant impact on RNA structure itself and subsequent interaction with proteins (Schibler et al., 1977). Methylation of adenosine and cytosine is also commonly found in many different types of RNAs. Such methylation of bases occurs as methyl-6-adenosine (m6A), methyl-1-adenosine (m1A) and methyl-5-cytosine (m5C).
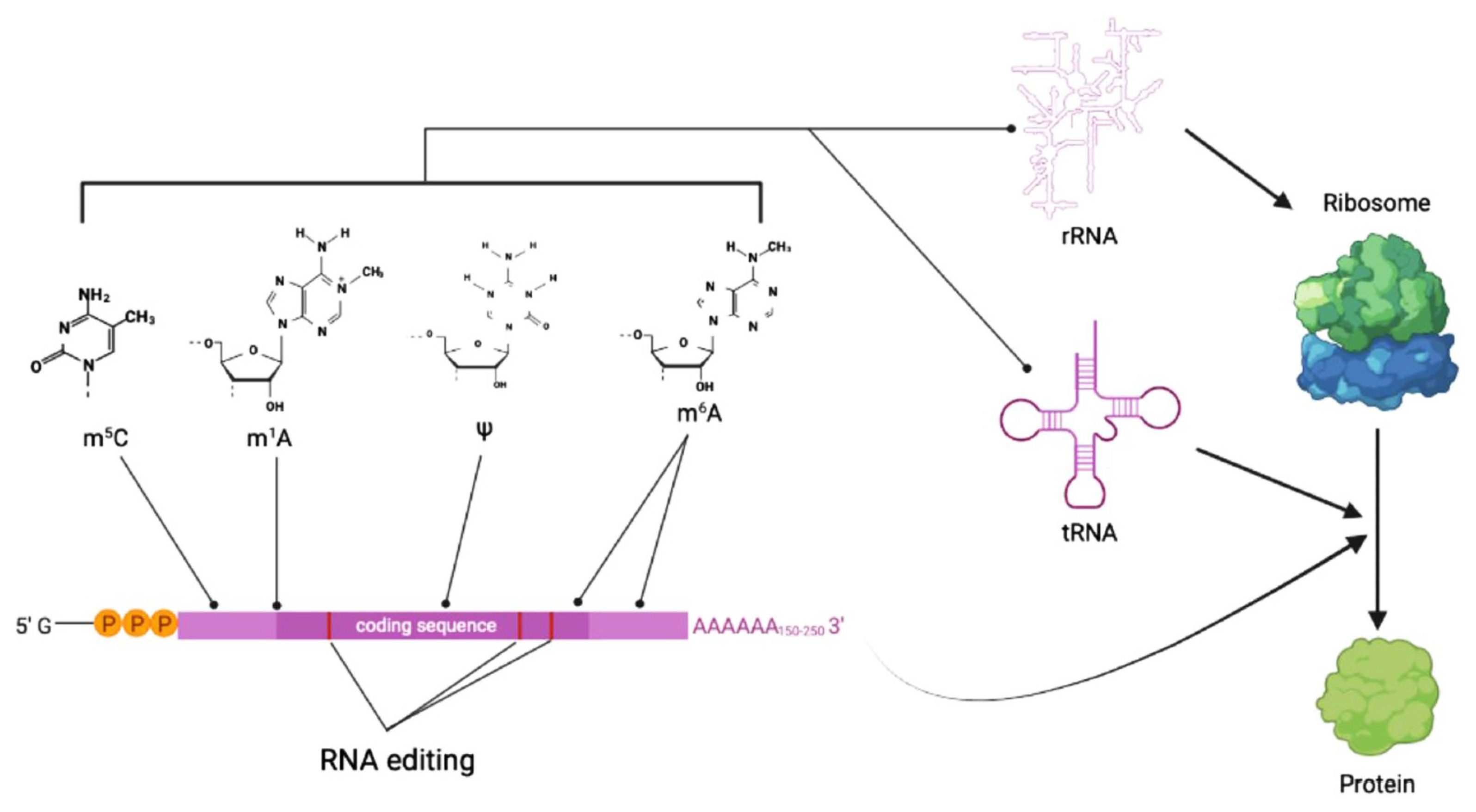
Summary diagram for types of RNA modifications and their impact on gene expression. RNA modifications such as methylation of adenosine and cytosine (denoted as a line with a dot at the end) affect stability and secondary structure of RNA molecules. RNA editing of mRNA (denoted as red bars inside coding sequence block in purple) can have impact on RNA processing and translation. Overall, all these RNA modifications contribute to spatiotemporal regulation of gene expression in a coordinated manner. m5C, 5-methylcytosine; m1A, N1-methyladenosine; ψ, pseudo-uridine; m6A, N6-methyladenosine.
Ribosomal RNAs (rRNAs) are the main cellular RNA species comprising more than ~80% of total cellular RNAs (Dammann et al., 1993, Roundtree et al., 2017). In the budding yeast, Saccharomyces cerevisiae, more than 100 sites are known to be modified in rRNA molecules, although most of them are found inside the ribosome, suggesting that modifications precede folding of the rRNA and maturation of ribosome. However, two dominant modification types found in rRNAs, as briefly mentioned above, are 2′-O-Me and ψ. Rather than being evenly distributed across rRNA molecule, modification sites cluster around functional sites of rRNA. This distribution pattern strongly suggests that modifications have functional implication for ribosome (Decatur and Fournier, 2002). Indeed, deletion of such modifications was shown to have considerable effects on phenotypes in yeast (Sloan et al., 2017).
Transfer RNAs (tRNAs) are estimated to take up about 10% of total cellular RNAs and the most heavily modified RNA species. They are reported to have on average 3 modifications per molecule and to undergo cleavage in the anticodon loop under various cellular stresses (Lyons et al., 2018; Roundtree et al., 2017; Thompson and Parker, 2009). Types of modification found in tRNAs include methylation of base or sugar, thiolation, and addition of sugars and amino acids (Roundtree et al., 2017). In tRNAs, modifications are centered around sequences comprising anticodon loop of tRNA, and such modifications are known to facilitate the correct interaction of anticodon with codon, thereby increasing efficiency and accuracy of translation (Lyons et al., 2018). Modifications that occur outside the anticodon loop are reported to contribute to the formation of overall cloverleaf-like structure of tRNAs (Lyons et al., 2018, Roundtree et al., 2017). At least in yeast, it was shown that other than translation and structure, tRNA modifications play a variety of roles in tRNA biogenesis and function (Alexandrov et al., 2006; Engelke and Hopper, 2006). It was also shown that overall level of tRNA modifications can be modulated by cellular stresses, indicating that they are not static but dynamic entities (Chan et al., 2010).
Discovery of internal modifications of mRNA followed shortly after the discovery of the cap and tail modifications. However, low cellular abundance of mRNA itself and low stoichiometry of mRNA internal modifications made it difficult to do quantitative analyses, and thus casted doubt on their biological implication for the last few decades. Only recently it has been possible to map internal modifications of mRNAs at single-nucleotide resolution and to investigate their biological functions, owing to improved detection methods. These modifications include m6A, m1A, m5C, 2′-O-Me, and ψ, into which investigation was fueled by identification of enzymes installing (methyltransferase) and removing methylation (demethylase) in combination with advances in high-throughput sequencing technology (Roundtree et al., 2017).
Among these modifications, m6A is the most prominent and abundant internal modification of mRNA. In mammals, m6A on mRNAs is deposited by enzymatic action of METTL3/METTL14 complex (Śledź and Jinek, 2016). m6A can be removed either passively by mRNA degradation or by m6A demethylases FTO or ALKBH5, both of which belong to AlkB family of dioxygenases (Jia et al., 2013; Zheng et al., 2013). It was shown that m6A is mainly distributed in the upstream region of coding sequences and 3′-untranslated region of mRNA molecules (Fig. 1). Interestingly, m6A seems to play contrasting roles between plants and mammals in terms of transcript stability, although it is an evolutionarily conserved mechanism of gene expression. In Arabidopsis, a study showed that m6A generally acts as a stabilizing mark, while in mammals, m6A tend to accelerate degradation of mRNAs (Anderson et al., 2018; Du et al., 2016). However, its role in fungal species has not been investigated in detail.
Methylation of N1 position of adenosine (m1A) is an another important but less abundant adenosine methylation in mRNA. Unlike m6A, m1A, almost without exception, maps near the translation start site and first splicing site in mRNA (Fig. 1), and positively correlates with increased translation efficiency (Li et al., 2016). This modification, which can be removed by a demethylase ALKBH3, is known to respond to diverse cellular stresses in mammals, suggesting its role in fine-tuning gene expression under stress conditions (Dominissini et al., 2016). Adenosine methylations such as m6A and m1A are in general read by the YT521-B homology (YTH) domain family proteins, which can mediate methylation-dependent regulation of RNA processing, translation, and decay (Dominissini et al., 2012).
Cytosine methylations such as 5-methylcytosine (m5C) and 5-hydroxymethylcytosine (hm5C) are the less abundant type of RNA methylation than adenosine methylations. m5C is usually found in untranslated regions of mRNA (Fig. 1), and might be involved in nuclear export of mRNA (Yang et al., 2017). The m5C methyltransferase targeting tRNA, NSUN2 was shown to be also responsible for m5C methylation in some mRNAs (Hussain et al., 2013). Unlike m5C, hm5C is observed in exon and intron regions of mRNA, and not much has been known about the roles of hm5C yet (Roundtree et al., 2017).
Other than adenosine and cytosine methylation, the 2′-O-Me and ψ are also found in regions near 5′-cap structure or coding sequences of mRNA, respectively (Fig. 1) (Roundtree et al., 2017). They appear to affect the secondary structure of RNA, but their biological roles are not clear, although data suggest that ψ functions in altering stop codon readthrough (Fernández et al., 2013).
Last but not least important, and unique type of RNA modification is RNA editing, which is the post-transcriptional change of a nucleotide sequence at one or more positions within an RNA transcript, leading to the different version of transcript from that originally encoded in the genome (Fig. 1) (Samuel, 2003). The first example of RNA editing was provided by the study on the four extra uridine that restore frame-shifted transcript to the functional gene transcript in mitochondrial cytochrome C oxidase gene of a unicellular protozoa, trypanosome (Benne et al., 1986). RNA editing is primarily catalyzed by adenosine (adenosine deaminase acting on RNA [ADAR] and adenosine deaminase acting on tRNA [ADAT] protein family) and cytidine deaminases (AID/APOBEC family) that result in adenosine-to-inosine (A-to-I) and cytidine-to-uridine (C-to-U) changes, respectively. It has been shown that RNA editing can influence such diverse processes as pre-mRNA splicing, mRNA translation and RNA degradation (Christofi and Zaravinos, 2019).
RNA Modifications in Biology of Filamentous Fungi
Given the extensive efforts dedicated to understanding RNA modifications mainly of mammals, plants and yeast, relatively little is known about RNA modifications in filamentous fungi. However, recent works, which we describe below, shed light on occurrence, regulation, function and even evolution of mRNA modifications in filamentous fungi.
In the rice blast fungus, Magnaporthe oryzae (also known as Pyricularia oryzae), Shi et al. characterized the function of m6A methylation in the development and virulence (Shi et al., 2019). They identified genes encoding enzymes involved in regulation and recognition of m6A in the genome of M. oryzae: PoIME4, which is an orthologue of S. cerevisiae Ime4 encoding N6-methyladenosine methyltransferase; PoALKB1, an orthologue of m6A demethylase Alkbh1; PoYTH1 and PoYTH2, orthologues of a human gene encoding YTH-domain containing protein, Ythdc1. This study showed that deletion of PoIME4 results in decreased m6A RNA methylation, and that deletion of all individual m6A-related genes lead to decrease in virulence. Interestingly, deletion of PoYTH1 and PoYTH2 caused defect in asexual reproduction. Overall, these data indicate that m6A methylation of RNA is important for fungal development and pathogenesis.
In metazoans, it was shown that A-to-I RNA editing is common and correlates with abundance of dsRNAs formed from genomic repeats. Unlike metazoans, yeasts and filamentous fungi had been believed to lack A-to-I RNA editing of mRNA, as their genomes do not harbor genes encoding adenosine deaminase (ADAR: adenosine deaminase acting on RNA) enzymes (Liu et al., 2016). It should be noted that A-to-I editing of tRNA by ADAT enzymes are found across all domains of life (Torres et al., 2014). Due to such evolutionary pattern, A-to-I RNA editing of mRNA was considered as a metazoan innovation. During their study on PUK1 (pseudokinase 1) kinase gene in Fusarium graminearum, Li et al. found that 90% transcripts of a pseudokinase gene (PUK1) undergo A-to-I RNA editing (note that inosine base-pairs with cytosine) at tandem stop codons (UA1831GUA1834G to UG1831GUG1834G). Since PUK1 gene is highly expressed in perithecia, the authors carried out RNA-seq experiments to compare RNAs isolated from conidia, hyphae, and perithecia. This comparative transcriptome analysis revealed that more than 26,000 perithecium-specific A-to-I editing sites. Unlike metazoan A-to-I editing sites found mainly in introns and untranslated regions, 70.5% of editing sites occur in coding regions with two-thirds of editing events leading to the recoding (changes in amino acid or change to stop codon). The authors extended their search to PUK1 orthologues in Neurospora crassa and Fusarium verticillioides, and found that corresponding orthologues also show stage-specific expression and A-to-I editing.
Based on such data, Liu et al. investigated A-to-I editing in N. crassa at the transcriptomic level (Liu et al., 2017). This led to the finding that even in N. crassa, A-to-I editing occurs during sexual reproduction and is common in coding sequences. The authors also showed that many of editing sites are conserved among F. graminearum, N. crassa, and Neurospora tetrasperma. Taken together with a study on early diverging ascomycete fungus, Pyronema confluens (Teichert et al., 2017), these results suggest that A-to-I editing is a conserved evolutionary feature of filamentous ascomycetes. Later, Wu et al. (2019, 2021) demonstrated that in contrast to ascomycetes, basidiomycetes fungi show C-to-U, U-to-C, and G-to-A editing in addition to A-to-I editing, indicating deeper origin of RNA editing in fungal species. However, RNA editing mechanisms in filamentous fungi are not clear and still under debate, since enzymes responsible for editing have not been identified yet.
Potential of RNA Modifications as a Target for Agrochemicals
It has long been known that RNA editing correlates with pathogenesis in parasites of vertebrates, trypanosomes such as Tripanosoma brucei and Ttripanosoma cruzi (Barrett et al., 2003). The study on M. oryzae, which is described above, also points to implication of m6A mRNA modification during fungal development and pathogenesis, although it is not clear exactly which cellular processes are affected by RNA modifications. Data from F. graminearum and N. crassa indicate that RNA editing is involved primarily in sexual reproduction of these species. Considering that ascospores are primary inoculum for fungal pathogens such as F. graminearum, interference of RNA editing in these fungi may wreck havocs in pathogenesis. A particularly interesting observation is that RNA modifications including RNA editing appear to occur through different mechanisms (enzymes) than plants or metazoans. Thus, it is tempting to envisage that RNA modification systems could serve as novel targets for development of future agrochemicals.
Concluding Remarks
There is no doubt that scarcity of data for RNA modifications in filamentous fungi demands more research efforts to be put into this uncharted area. To date, only a handful of filamentous fungi were studied. Therefore, diverse fungal species including pathogenic fungi should be sampled for investigation on presence/absence of RNA modifications and comparison among them. Furthermore, enzymes responsible for RNA modifications should be identified and studied for underlying mechanisms to be elucidated and targeted for control of those fungal pathogens. Despite such lack of data, it is clear that RNA modifications are new research avenue that would help us better understand fungal biology and open numerous possibilities from both conceptual and practical perspectives.
Acknowledgments
This work was supported by 2019 Yeungnam University Research Grant.
Notes
Conflict of interest
No potential conflict of interest relevant to this article was reported.
