Tomato Yellow Leaf Curl Virus Infection in a Monocotyledonous Weed (Eleusine indica)
Article information
Abstract
Tomato yellow leaf curl virus (TYLCV) is one of the most important plant viruses belonging to the genus Begomovirus of the family Geminiviridae. To identify natural weed hosts that could act as reservoirs of TYLCV, 100 samples were collected at a TYLCV-affected tomato farm in Iksan from 2013 to 2014. The sample weeds were identified as belonging to 40 species from 18 families. TYLCV was detected in 57 samples belonging to 28 species through polymerase chain reaction using root samples including five species (Eleusine indica, Digitaria ciliaris, Echinochloa crus-galli, Panicum dichotomiflorum, and Setaria faberi) from the family Poaceae. Whitefly Bemisia tabaci-mediated TYLCV transmission from TYLCV-infected E. indica plants to healthy tomatoes was confirmed, and inoculated tomatoes showed typical symptoms, such as leaf curling and yellowing. In addition, TYLCV was detected in leaf and root samples of E. indica plants inoculated by both whitefly-mediated transmission using TYLCV-viruliferous whitefly and agro-inoculation using a TYLCV infectious clone. The majority of mastreviruses infect monocotyledonous plants, but there have also been reports of mastreviruses that can infect dicotyledonous plants, such as the chickpea chlorotic dwarf virus. No exception was reported among begomoviruses known as infecting dicots only. This is the first report of TYLCV as a member of the genus Begomovirus infecting monocotyledonous plants.
Tomato yellow leaf curl virus (TYLCV, genus Begomovirus, family Geminiviridae) is a tomato (Solanum lycopersicum)-infecting virus with a single-stranded circular DNA genome of about 2.8 kb encapsulated in a twinned icosahedral virion (Czosnek and Laterrot, 1997; Moriones and Navas-Castillo, 2000). TYLCV can be transmitted by sweet potato whitefly (Bemisia tabaci) and infected seeds, which has been refuted through other studies (Ghanim et al., 1998; Kil et al., 2016; Pérez-Padilla et al., 2020). TYLCV infection induces severe symptoms such as interveinal yellowing and curling of leaves and severe stunting in tomato plants, which can lead to critical production losses (Papayiannis et al., 2011). Since TYLCV was reported from the Middle East in 1931, it has spread continuously and damaged tomato crops in tropical and subtropical areas (Czosnek and Laterrot, 1997). In Korea, after the first report of TYLCV-infected tomatoes in 2008, it spread rapidly throughout the country and the damages it causes have been reported continuously (Kil et al., 2014a; Lee et al., 2010).
Due to the importance of TYLCV in agriculture, multiple studies have been conducted on the host range of TYLCV. Not only tomatoes, but pepper (Capsicum spp.), common bean (Phaseolus vulgaris), soybean (Glycine max), and eustoma (Eustoma grandiflora) are known TYLCV hosts (Anfoka et al., 2009; Cohen et al., 1995; Kil et al., 2017; Moriones and Navas-Castillo, 2000; Navas-Castillo et al., 1999; Reina et al., 1999). Additionally, many weed species from families including Amaranthaceae, Asclepiadaceae, Caparidaceae, Compositae, Cucurbitaceae, Euphorbiaceae, Leguminosae, Malvaceae, Nyctaginaceae, and Solanaceae have been confirmed as hosts for TYLCV (Bedford et al., 1998; Jordá et al., 2001; Kil et al., 2014b; Papayiannis et al., 2011; Salati et al., 2002). Identification of weed hosts for TYLCV is essential to understanding its epidemiology and designing management strategies (Smith et al., 2015).
Geminiviruses have been shown to differ in the number and compositions of open reading frames depending on their genera, and they exhibit clear differences in host ranges and vector insects (Hanley-Bowdoin et al., 2013). Before the early 1990s, when geminivirus group was established, it was widely believed that begomoviruses (formerly subgroup II) can infect only dicotyledonous plants through mediation of the whitefly B. tabaci, and mastreviruses (formerly subgroup I) can infect only monocotyledonous plants of the family Poaceae through leafhoppers (Fauquet and Stanley, 2003; Horn et al., 1993; Rojas et al., 2018). These assurances in academia have begun to crack after a report of infection of a dicotyledonous plant by tobacco yellow dwarf virus in Australia (Morris et al., 1992). According to the updated report from the International Committee on Taxonomy of Viruses, eight dicot-infecting mastreviruses, including chickpea chlorotic dwarf virus (CpCDV) from the Indian subcontinent, the Arabic peninsula, and Africa have been reported (Akhtar et al., 2014; Horn et al., 1993; Liu et al., 1999; Nahid et al., 2008; Zerbini et al., 2017). However, in the case of begomoviruses, no exceptions have been identified.
According to our previous study, 25 species of weeds belonging to 11 families have been confirmed as TYLCV-infected in tomato-cultivating areas in Korea (Kil et al., 2014b). In that time, only dicotyledonous weeds were collected for analysis because begomoviruses have been known to be infectious only among dicotyledonous plants.
In this study, 100 samples from naturally occurring weeds in Iksan, including monocotyledonous weeds, were collected and analyzed to check for TYLCV infection. Among monocotyledonous weeds that showed TYLCV-specific amplicons in PCR analyses, E. indica, which is one of the representative summer annual weeds that occur nationwide in Korea, was used for further experiments, including experimental inoculation by agro-inoculation and viruliferous whitefly-mediated transmission and organ-specific PCR.
Materials and Methods
Weed sample collection
From 2013 to 2014, TYLCV-infected tomato plants and viruliferous whiteflies were found in tomato-cultivating greenhouses in Iksan (located in the southern part of Korean peninsula) (Fig. 1A and B). From there, 100 samples were collected and identified as belonging to 40 species from 18 families (Table 1). Sampling was randomized with or without virus symptoms, and both root and leaf samples were taken. Root samples were used to prevent the possibility of contamination by larvae and eggs of TYLCV-viruliferous whitefly and check systemic infection of the virus. The use of plant parts in this study complies with international, national and/or institutional guidelines.

Tomato yellow leaf curl virus (TYLCV)-infected tomato plants and weeds. (A) Symptomatic tomato plants infected by TYLCV. (B) Whitefly Bemisia tabaci adults on the abaxial side of tomato leaf. (C) Eleusine indica in the same tomato-cultivating greenhouse.
Viral DNA extraction and PCR analysis
Viral DNA extraction was conducted using the Viral Gene-spin Viral DNA/RNA Extraction Kit (iNtRON Biotechnology, Seongnam, Korea) from tissue samples following manufacturer’s protocol. PCR analysis was done with 1× AccuPower PCR Master Mix (Bioneer, Daejeon, Korea) and TYLCV-specific primer set, whose target is TYLCV partial sequence for intergenic region (IR) and V2 gene (5′-GATGGCCGCGCCTTTTCCTTTTATGTGG-3′, 5′-GCTGCTGTATGGGCTGTCGAAGTTCAG-3′) (Kil et al., 2016). Amplification was carried out under the following conditions: preheating at 94°C (3 min), then 35 cycles of 30 s at 94°C, 30 s at 58°C and 1 min at 72°C, followed by a final extension at 72°C (5 min). PCR products were visualized in 1% agarose gel electrophoresis.
IR sequencing
To confirm nucleotide sequences from amplicons from samples which showed positive results in previous PCR analyses, TYLCV IR was amplified by PCR with the IR-specific primer set (425 bp of target size) (Kil et al., 2015). Amplicons were ligated into pGEM-T easy vectors (Promega, Madison, WI, USA) and sequenced (Macrogen, Seoul, Korea). Sequencing data were analyzed using the MultAlin multiple alignment program (http://multalin.toulouse.inra.fr/multalin/) (Corpet, 1988).
Experimental inoculations for TYLCV with infectious clone and viruliferous whitefly
To confirm that E. indica is a host for TYLCV by artificial inoculation under controlled conditions, agro-inoculation was carried out using a TYLCV-IL infectious clone prepared previously (Kil et al., 2014a). Seedlings of E. indica were collected from an open field in Suwon where whitefly B. tabaci was free and TYLCV-free statuses of collected plants were checked by TYLCV IR and V2-specific PCR. After transplanting the seedling to pots, they were cultivated in a walk-in growth chamber in Sungkyunkwan University, Korea. Before agro-inoculation, some new shoots were removed from the crown, and cultivated agrobacterium containing TYLCV infectious clone (0.8 of OD600 value) were injected into the base of tillers twice in a different day in the same week to increase infection yield (500 μl of cultured soup of agro-bacteria used for each inoculation). Plants inoculated with agrobacterium containing pCAMBIA1303 empty vector were also prepared as a control. To perform insect vector–mediated inoculation, three TYLCV-free E. indica plants were put in BugDorm-4E4590DH cages (MegaView Science Co., Taichung, Taiwan) separately with TYLCV-viruliferous whiteflies (the Mediterranean [MED] cryptic species formerly called a Q-biotype) from existing insect cages for maintenance of viruliferous whiteflies. Root and shoot samples were collected from each plant and analyzed by PCR four weeks later as previously described.
Whitefly-mediated transmission from Eleusine indica to tomato
Whitefly-mediated TYLCV transmission from TYLCV-infected E. indica plants to healthy tomato plants was performed in restricted conditions. Non-viruliferous Bemisia tabaci (the MED cryptic species) were bred with virus-free tomato plants and virus infection was checked by PCR as described in a previous study (Kil et al., 2015). E. indica plants which were inoculated with TYLCV infectious clone and confirmed as TYLCV-infected were placed in three individual BugDorm-4E4590DH cages (MegaView Science Co.) together with three-week-old healthy “Seogwang” tomato (TYLCV-susceptible cultivar) plants and non-viruliferous whiteflies at 25°C with a day length of 12 h. A negative control group was also set with TYLCV-free E. indica, non-viruliferous whiteflies and healthy tomatoes in an insect cage. All plants were observed to check for symptom development or abnormal status. After four weeks, plants and whiteflies were harvested and analyzed for TYLCV infection by PCR analysis.
Results
Identification of TYLCV infection from collected weed samples
From tomato greenhouses in Iksan where symptomatic (yellowing and curling of leaves and stunting) plants infected with TYLCV have been reported continuously, 100 samples of 40 different species and 18 families were collected, identified, and tested by PCR (Table 1, Fig. 1). Among them, according to PCR results with TYLCV-specific primers, TYLCV was detected in 57 samples belonging to 28 species, including Aster subulatus var. sandwicensis, Erigeron annuus, Youngia japonica, Myosoton aquaticum, Stellaria media, and Solanum americanum, which were listed as TYLCV-infected in our previous survey (Table 1) (Kil et al., 2014b). Solanum nigrum, considered a TYLCV weed host in previous works, has persistently showed positive results in TYLCV-specific detection efforts (Bedford et al., 1998; Papayiannis et al., 2011; Sánchez-Campos et al., 1999). Twenty-eight species were verified as infected, while TYLCV infection from Achyranthes fauriei, Conyza canadensis, Erechtites hieracifolia, Cardamine flexuosa, Rorippa palustris, Cerastium glomeratum, Trifolium repens, Vicia angustifolia var. segetilis, V. tetrasperma, Leonurus japonicus, Cocculus trilobus, Phytolacca americana, D. ciliaris, E. crus-galli, E. indica, P. dichotomiflorum, S. faberi, Galium spurium, and Mazus pumilus was newly identified in this study (Table 1).
Five monocotyledonous weeds verified as TYLCV-infected
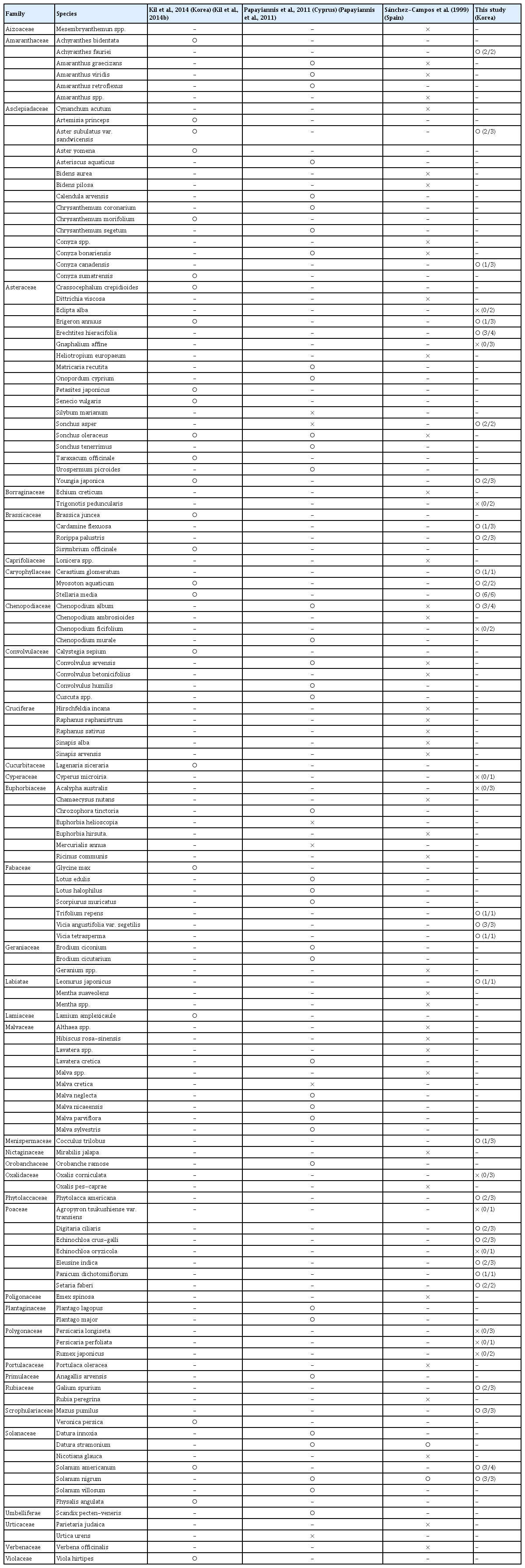
Viral infection in five species in the family Poaceae (Digitaria ciliaris, Echinochloa crus-galli, Eleusine indica, Panicum dichotomiflorum, and Setaria faberi) was confirmed from leaf and root samples with a TYLCV IR-specific primer set by PCR and amplicon sequencing. All amplicons showed the same size of target bands and DNA sequences as amplicons from TYLCV-infected tomato. Systemic infection of TYLCV in infected E. indica plants was examined with leaf, spikelet, node, crown, and root tissues (Fig. 2A). According to PCR results, target-specific amplicons were detected in all organ samples (Fig. 2B).
Organ-specific collection and tomato yellow leaf curl virus (TYLCV) detection from TYLCV-infected Eleusine indica plants. (A) A TYLCV-infected E. indica plant. a, leaves; b, spikelet; c, node; d, crown; e, root. (B) Organ-specific PCR results from an E. indica plant. N, no template control; P, positive control with TYLCV dimer construct.
Experimentally inoculated Eleusine indica plants showing systemic infection
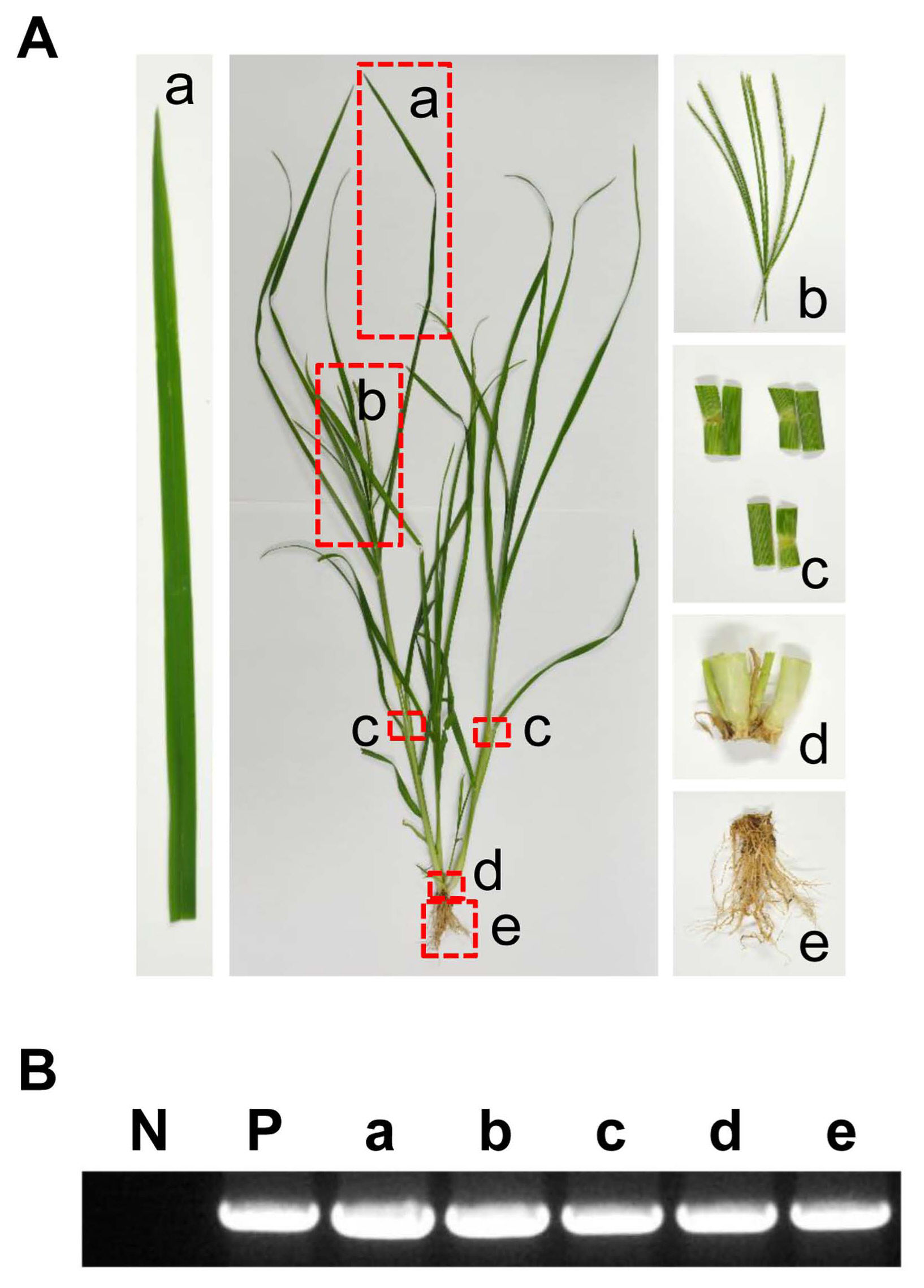
Using the TYLCV infectious clone prepared in the previous study, artificial inoculation of six healthy E. indica plants was conducted (Fig. 3A). No specific symptoms such as leaf curling or yellowing were observed in the inoculated plants. Four weeks after inoculation, virus infection was confirmed in all inoculated plants (6/6). To check systemic infection in inoculated E. indica plants, four different organs (flowers, leaves, stems, and roots) were collected from inoculated plants and their results showed clear target bands from TYLCV-specific PCR (Fig. 3B). When three different healthy E. indica plants were placed in separate cages and TYLCV-viruliferous whitefly B. tabaci was added, it was confirmed that all the leaves and roots of the three plants were infected when they were diagnosed after four weeks. Taken together, and using two different inoculation methods (agro-inoculation and whitefly-mediated inoculation), it was demonstrated that E. indica plants could be infected with TYLCV.
Agro-inoculation of tomato yellow leaf curl virus (TYLCV) in Eleusine indica plants. (A) E. indica plants transplanted from open field to pots after confirming TYLCV-free status (left) and E. indica plants of which new shoots were removed from the crown before agro-inoculation (right). Red arrow shows piercing and injecting point for agrobacterium culture media with TYLCV infectious clone. (B) Four different organ samples from agro-inoculated E. indica plants (top) and TYLCV-specific PCR results from organ samples (bottom). Lane N, no template control; lane P, positive control with TYLCV dimer construct; lane F, flowers; lane L, leaves; lane S, stems; lane R, roots.
TYLCV transmission by Bemisia tabaci from Eleusine indica to tomato plants
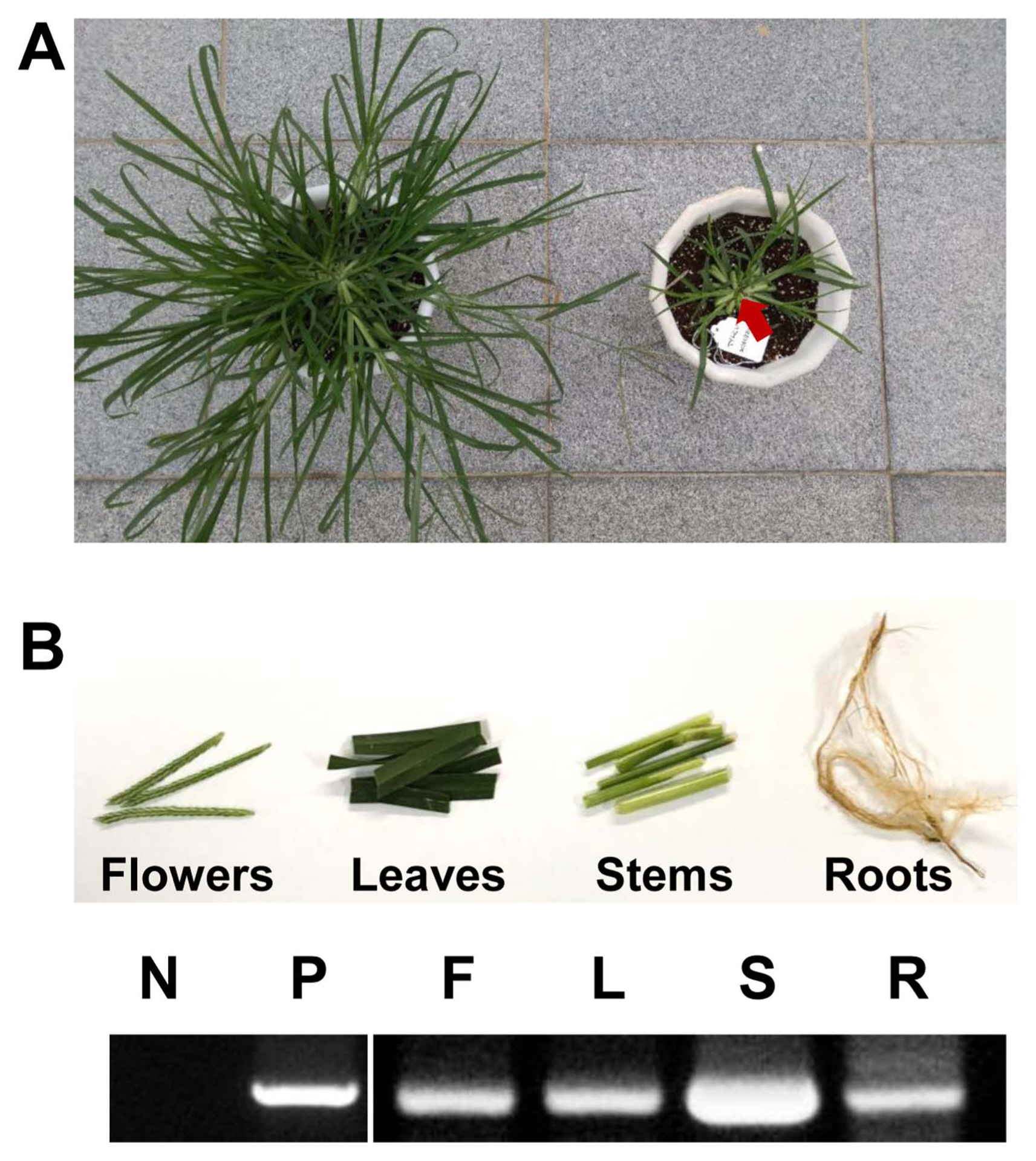
To determine whether E. indica can act as an inoculum of TYLCV for whitefly-mediated transmission, TYLCV-free whiteflies and tomato plants were placed with TYLCV-infected E. indica in the same insect cage and TYLCV detection from insects and plants was conducted using PCR analysis (Fig. 4A). After four weeks, non-viruliferous whiteflies and TYLCV-free tomato plants in three individual tents with TYLCV-infected E. indica became TYLCV-viruliferous and infected (3/3) (Fig. 4B). TYLCV-typical symptoms were also observed in TYLCV-infected tomato plants. Additionally, according to IR-specific PCR results, amplicons from insects and two plants showed identical sequences. In the control group with TYLCV-free E. indica and tomato plants, and non-viruliferous whiteflies, E. indica and tomato plants did not show any virus symptoms and TYLCV was not detected in all insects and plants in those cages of control group.

Whitefly-mediated tomato yellow leaf curl virus (TYLCV) transmission from Eleusine indica plants. (A) Experimental scheme of whitefly-mediated transmission analysis (left) and TYLCV-infected E. indica plants (donor) and TYLCV-free tomato plants (recipient) with non-viruliferous whiteflies in insect cage (right). (B) PCR results with TYLCV primers from insect vector (B. tabaci, V), weed (E. indica plant, W), and tomato plant (T) before and after transmission tests. Lane N, no template control; lane P, positive control with TYLCV dimer construct.
Discussion
One of the most important plant viruses in the world, TYLCV has a long history of damaging tomato crops (Scholthof et al., 2011). During TYLCV spread, some kinds of weeds can act as intermediate hosts to make the whitefly B. tabaci viruliferous. Since this is an insect vector for TYLCV, managing weeds is essential for the prevention and control of TYLCV. Many researchers around the world have consistently identified many weed species as hosts for TYLCV (Bedford et al., 1998; Jordá et al., 2001; Kil et al., 2014b; Papayiannis et al., 2011; Salati et al., 2002). We demonstrated in our previous study that Lamium amplexicaule can act as an intermediate host in the transmission by whitefly B. tabaci experimentally, and a virus-infected winter-hardy weed (Stellaria aquatica) can become an inoculum source for new tomato plants in the next season (Kil et al., 2014b, 2015). In this study, for the analysis of the weeds with TYLCV in Korea, major weeds were sampled and analyzed at a tomato farm in Iksan, where TYLCV occurrence had been reported. Among 28 species that were confirmed as TYLCV-positive from PCR results (Table 1), monocotyledonous weeds drew attention because begomoviruses including TYLCV have been known to infect only dicotyledonous plants–without exception until now–as opposed to eight species of mastreviruses infecting dicotyledonous hosts (Fauquet and Stanley, 2003; Rojas et al., 2018). We performed three independent trials to confirm that these results were not errors by contamination in the experimental procedure, and we were able to rule out the possibility of contamination and errors by confirming their infection in all the repeated results. We focused on one weed species (E. indica) that is one of the most common weeds in South Korea and can be found in different regions easily. We thought there is a high possibility that this weed can have an important role in TYLCV transmission. To check that the TYLCV infection caused viral systemic infection in E. indica as in other hosts, the root, leaf, stem and flower tissues of TYLCV-inoculated E. indica were individually sampled and analyzed, and as a result, infection was confirmed in all tissues (Fig. 2). To re-examine these outcomes in a controlled environment, an artificial inoculation with a TYLCV infectious clone was conducted for a healthy E. indica specimen. As a result, TYLCV infection was confirmed in individual organ samples, which meant that systemic infection was induced in artificially inoculated plants (Fig. 3). Whitefly-mediated inoculation also showed infection in virus-free E. indica with viruliferous whiteflies. Based on these results, it was verified that E. indica, a monocotyledonous plant, was the host of TYLCV. To see if E. indica, the TYLCV host, could be a viral inoculum for whitefly B. tabaci as an intermediate host like any other weed hosts, we put TYLCV-infected E. indica plants with non-viruliferous whitefly B. tabaci and healthy tomatoes in insect cages and checked virus infection from whiteflies and tomato plants (Fig. 4). This result showed that E. indica plants acted as intermediate hosts for whitefly-mediated TYLCV transmission. Together with cases of viral dissemination from TYLCV-infected seeds and whitefly-mediated transmission from TYLCV-infected tomato plants, vector-mediated transmission from TYLCV-infected E. indica plants can be established as one possible route for TYLCV transmission (Fig. 5).
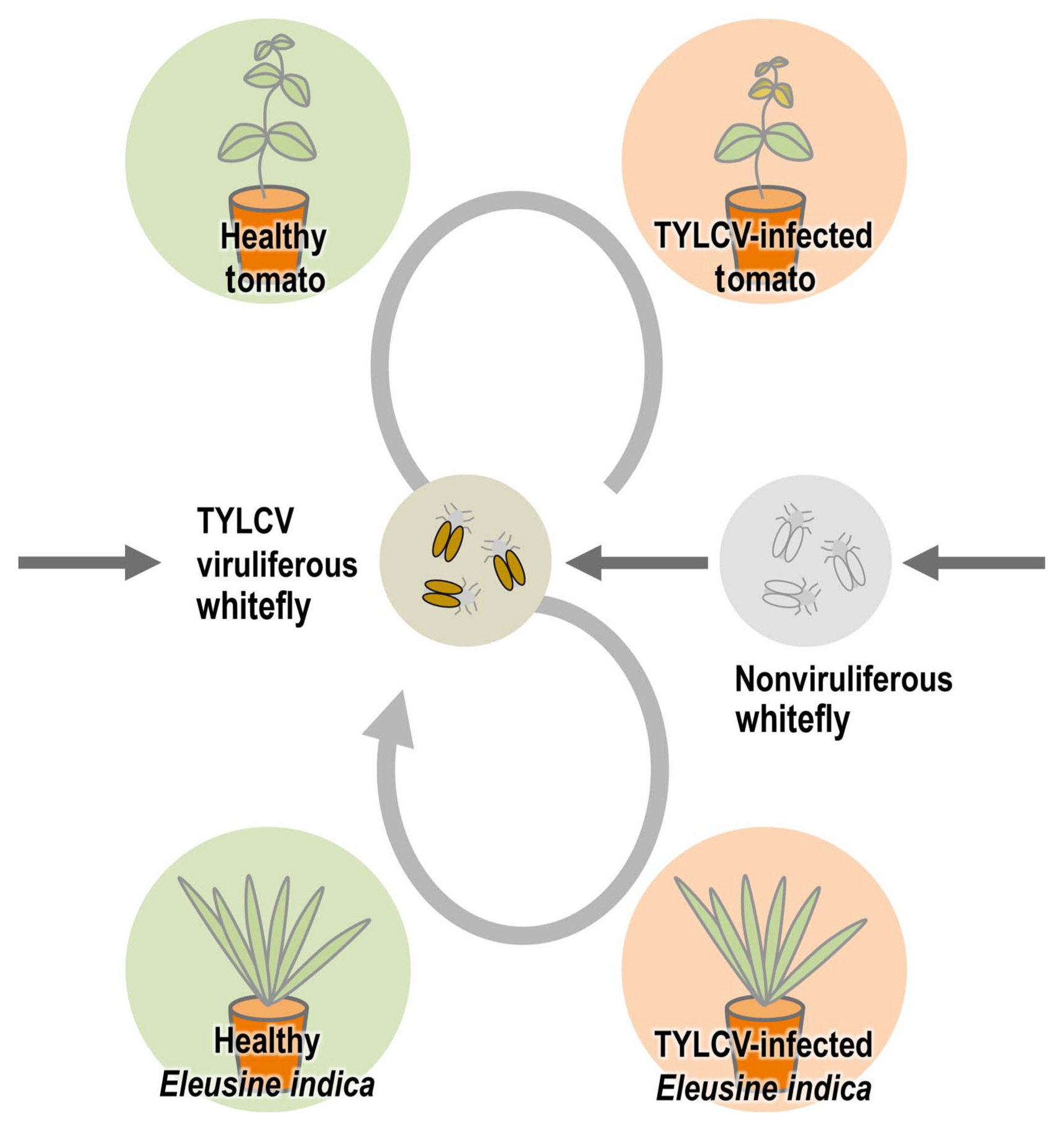
Schematic diagram of whitefly Bemisia tabaci-mediated tomato yellow leaf curl virus (TYLCV) transmission with infected tomato seeds, tomato plants, and Eleusine indica.
This is the first report of monocotyledonous hosts for a begomovirus. This result breaks long-accepted conventions regarding begomoviruses and monocotyledonous plants. Nucleotide sequences from dicotyledon-infecting mastreviruses showed phylogenetic differences from previously reported mastreviruses (Pande et al., 2012). Dicotyledon-infecting mastreviruses are believed to be the result of host adaptation of monocotyledon-infecting mastreviruses (Liu et al., 1998). Few cases of transmission of mastreviruses in monocotyledons (especially crops such as maize and wheat) to dicotyledons have been confirmed, and hosts of some dicotyledon-infecting mastreviruses (such as chickpea and soybean) have been reported, and vice versa (Zerbini et al., 2017). However, this may be because studies with artificial inoculation tests using infectious clones or host range surveys from a large number of dicotyledon plants have been relatively rare in the case of mastreviruses. This may explain why the initially detected dicotyledon-infecting mastreviruses showed local isolation from existing mastreviruses (Liu et al., 1998). In contrast to these mastreviruses, TYLCV, which is identified in the monocotyledonous weed in this study, is an easily infectious virus in dicotyledonous plants and transmission of TYLCV from monocotyledonous weeds to tomato plant was also observed.
Because of high reproductive rates and environmental adaptability, monocotyledonous weeds, including E. indica, are easily observed in or around a tomato-cultivating farmhouse in Korea. However, established assumptions have discouraged screening for TYLCV and other begomoviruses. Our findings challenge these assumptions.
Acknowledgments
This work was supported by a research grant of Andong National University.
Notes
Conflict of interest
No potential conflict of interest relevant to this article was reported.