 |
 |
| Plant Pathol J > Volume 38(4); 2022 > Article |
|
Abstract
Seed-borne pathogens in crops reduce the seed germination rate and hamper seedling growth, leading to significant yield loss. Due to the growing concerns about environmental damage and the development of resistance to agrochemicals among pathogen populations, there is a strong demand for eco-friendly alternatives to synthetic chemicals in agriculture. It has been well established during the last few decades that plant seeds harbor diverse microbes, some of which are vertically transmitted and important for plant health and productivity. In this study, we isolated culturable endophytic bacteria and fungi from soybean seeds and evaluated their antagonistic activities against common bacterial and fungal seed-borne pathogens of soybean. A total of 87 bacterial isolates and 66 fungal isolates were obtained. Sequencing of 16S rDNA and internal transcribed spacer amplicon showed that these isolates correspond to 30 and 15 different species of bacteria and fungi, respectively. Our antibacterial and antifungal activity assay showed that four fungal species and nine bacterial species have the potential to suppress the growth of at least one seed-borne pathogen tested in the study. Among them, Pseudomonas koreensis appears to have strong antagonistic activities across all the pathogens. Our collection of soybean seed endophytes would be a valuable resource not only for studying biology and ecology of seed endophytes but also for practical deployment of seed endophytes toward crop protection.
Seeds are the genetic vehicle of plants for dispersal, persistence, and initiation of developmental transition into seedlings (Berg and Raaijmakers, 2018). Diverse microbes including plant pathogens are associated with seeds, and some of the microbes appear to have co-evolved alongside them (Nelson, 2018). It is important to distinguish epiphytes and endophytes among seed-associated microbes, as the two often differ from one another in their mode of transmission (horizontal vs. vertical transmission) and sources of origin (environmental sources and seed tissues) (Nelson, 2018). In contrast to the epiphytes that are more likely to show fluctuation in their composition and abundance from one seed to the next in a stochastic and environment-dependent manner, the seed endophytes tend to show relatively stable existence owing to their ability to interact with seeds as well as other microbes.
It has been demonstrated in many studies that seed endophytes can play roles in seed germination, seedling growth, and tolerance to biotic and abiotic stresses (Berg et al., 2017; Compant et al., 2019). For example, recent work on a bacterial endophyte of rice seed nicely illustrates the potential impact and importance of roles that seed endophytes might play in keeping the crop plants healthy (Matsumoto et al., 2021). This work showed that a seed-endophytic bacterium Sphingomonas melonis can confer resistance to the seed-borne pathogen Burkholderia plantarii via the production of anthranilic acid, which interferes with the sigma factor RpoS of the pathogen. Over the last few years, an increasing number of studies aimed at understanding the origin, diversity, and functions of seed endophytes have been undertaken in an attempt to realize the full potential of endophytic microbes of seeds (Berg and Raaijmakers, 2018; Bziuk et al., 2021; Compant et al., 2019; Dai et al., 2020; Johnston-Monje et al., 2021; Kim and Lee, 2021; Nelson, 2018).
Seed endophytes are probably the first microbes that can interact with plants during the germination of seeds and development into seedlings before other microbes in the soil do. In ecological theory, it was suggested that the priority effect, in which the order and timing of species arrival to the given niche often shapes the way community develops, influencing the resulting community structure (Sprockett et al., 2018). In line with the priority effect, it was recently shown that seed-transmitted bacteria and fungi make up the majority of the juvenile plant microbiome (Johnston-Monje et al., 2021). In this context, it is reasonable to think that changing the seed-endophytic microbiome structure of a crop plant can have a long-lasting effect on plant microbiome structure, which in turn, plays important roles in crop health and productivity.
However, crop seeds are often infected with a seed-borne pathogen (Gitaitis and Walcott, 2007). Infested seeds result in the re-emergence of diseases, leading to significant economic losses and unnecessary use of synthetic chemicals worldwide. Although the early diagnosis of seed-borne pathogens is important, infected seeds often appear symptomless, rendering visual inspection of seeds ineffective (Gitaitis and Walcott, 2007). Even the polymerase chain reaction (PCR)-based detection methods with high sensitivity and specificity have limitations due to their dependence on seed sample size (Hariharan and Prasannath, 2021).
Soybean (Glycine max) is an important legume crop, providing low-fat and protein-rich diet for both human and animal consumption (Messina, 1999). Common seed-borne fungal pathogens in soybean include, but are not limited to, Cercospora kikuchii, Cercospora sojina, Diaporthe eres, and Septoria glycines, while Pseudomonas syringae pv. tabaci and Xanthomonas axonopodis pv. glycines are among commonly isolated seed-borne bacterial pathogens. To date, endophytes of soybean seed and their application potential for producing healthy seeds have not been fully explored. In this study, we isolated diverse bacterial and fungal endophytes of soybean seeds and evaluated their antagonistic activities against common seed-borne pathogens of soybean plants.
C. sojina (KACC No. 49847), S. glycines (KACC No. 43091), X. axonopodis pv. glycines (KACC No. 10491), and P. syringae pv. tabaci (KACC No. 17820) were acquired from the Korean Agriculture Culture Collection (KACC) in this study. D. eres and C. kikuchii were isolated from diseased soybean seeds and sequenced for identification. For identification of D. eres, the primers EF1-728F (5′-CATCGAGAAGTTCGAGAAGG-3′) and EF1-986R (5′-TACTTGAAGGAACCCTTACC-3′) were used to amplify part of the translation elongation factor 1-α gene (TEF1) (Gomes et al., 2013) and for identification of C. kikuchii, the primers cfp_F1 (5′-GCCGATCGATTGCAGGAGTTGGC-3′) and cfp_R1 (5′-TTGCTGATCCAAGTAGTCGGACG-3′) were used to amplify the cercosporin facilitator protein (CFP) gene region (Soares et al., 2015).
Daewon soybean seeds (Kyoungshin Seeds Co., Ltd., Uiseong, Korea), which are cultivated commonly on Korean farms, were used in this study. In order to obtain the next generation of seeds, the soybean seeds were planted in the experimental plot of Dong-A University (location: Gimhae-si, Gyeongsangnam-do, Republic of Korea). Soybean seeds were washed with 70% ethanol and dipped into 1% sodium hypochlorite solution for 5 min to disinfect the surface and rinsed with sterilized water. Surface-sterilized seeds were ground by using 1× phosphate buffered saline (PBS) buffer with mortar and pestle. The ground seeds were serially diluted with 1× PBS buffer, and the final solutions were serially diluted up to 10−5. One hundred microliters of each solution was spread on the medium for isolation of endophytic microbe. Potato dextrose agar (PDA; Difco, Detroit, MI, USA), Rose-bengal agar (RBA; MBcell, Seoul, Korea), Czapek Dox agar (CZA; MBcell), and V8 agar (V8A; 8% V8 juice, 1.5% agar) media were used to isolate endophytic fungi, while tryptic soy agar (TSA; Difco), Reasoner’s 2A (R2A; Difco) agar, Luria-Bertani (LB; Difco) agar, and nutrient agar (NA; Difco) media were used to isolate endophytic bacteria. The plates were then incubated at 25ºC for 3-7 days for fungi and 30ºC for 1-3 days for bacteria, depending on the number of colonies formed and how fast those colonies grow. Following incubation, colonies of the same color and morphology were grouped and considered as a single species.
For the identification of fungal DNA, the DNA was extracted using the quick method (Chi et al., 2009). PCR was performed to amplify internal transcribed spacer (ITS) region using forward primer ITS3 (5′-GCATCGATGAAGAACGCAGC-3′) and reverse primer ITS4 (5-TCCTCCGCTTATTGATATGC-3′) (White et al., 1990). The 20 μl reaction mixture consisted of 2 μl of template DNA, 1 μl of each primer, 10 μl of master mix (Takara Bio Inc., Shiga, Japan) and 6 μl of nuclease free water. The PCR was performed in a thermal cycler (Life Technologies Holdings Pte Ltd., Woodlands, Singapore) using cycling conditions that consisted of an initial denaturation at 96ºC for 5 min and then 35 cycles with denaturation at 96ºC for 1 min, annealing at 55ºC for 1 min, and extension at 72ºC for 1 min. A final extension was performed at 72ºC for 10 min.
For the identification of bacterial DNA, the DNA was extracted using bacterial 16S rDNA PCR Kit Fast (Takara Bio Inc.). PCR was performed to amplify 16S rRNA using forward primer V3 (5′-CCTACGGGNGGCWGCAG-3′) and reverse primer V4 (5′-GACTACHVGGGTATCTAATCC-3′) (Klindworth et al., 2013). The composition of the PCR reaction mixture was the same as described above. The PCR was performed using cycling conditions that consisted of an initial denaturation at 95ºC for 15 min and then 30 cycles with denaturation at 95ºC for 1 min, annealing at 50ºC for 30 s, and extension at 72ºC for 30 s. A final extension was performed at 72ºC for 5 min. PCR amplicons were then purified with ExoSAP-IT purification reaction system (Thermo Fisher Scientific, Waltham, MA, USA) (Bell, 2008). The purified PCR products were analyzed by 1% agarose gel electrophoresis, and the samples were sent for sequencing by the company (Bionics, Seoul, Korea). The resulting sequences were subjected to nucleotide BLAST against NCBI for identification of bacterial and fungal species (https://blast.ncbi.nlm.nih.gov).
A cylindrical agar plug (5 mm in diameter) of the seed-borne pathogenic fungi was placed in direct opposition to the test fungi isolated from soybean seeds, which are inoculated ~2 cm away from the edge of the agar plate (9 cm in diameter). After incubating the plates at 25ºC, fungal mycelial growth was measured. D. eres was incubated for 5 days, while C. kikuchii, C. sojina, and S. glycines were incubated for 7 days. The inhibition rate was calculated as ((Rc - Rexp)/Rc) × 100%, where Rc represents the radius of control fungal mycelium and Rexp is the radius of the seed-borne pathogenic fungus. Each test was replicated five times. For testing the antifungal activity of bacterial isolates, a cylindrical agar plug (5 mm in diameter) of the seed-borne pathogenic fungi was placed at the center of a Petri dish containing mixture of PDA and TSA in a 1:1 ratio. Bacterial test strains were streaked to make a circle around the fungal pathogen (~6 cm in diameter). After incubating the plates at 27.5ºC, fungal mycelial growth was measured. D. eres was incubated for 4 days, while C. kikuchii and S. glycines were incubated for 9 days, and C. sojina was incubated for 7 days. The inhibition rate was calculated following the formula described above.
A cylindrical agar plug (5 mm in diameter) containing the test strain of fungi was placed at ~1 cm away from each side of the seed-borne pathogenic bacterial streak (~1 cm in width) located in the middle of the Petri dish. These tests were performed using agar plates containing 1:1 mixture of PDA and TSA. After incubating for 3 days at 27.5ºC, the inhibition rate was calculated as ((Wc - Wexp)/Wc) × 100%, where Wc represents the width of control bacteria and Wexp represents the width of the seed-borne pathogenic bacteria. Each test was replicated five times. For testing the antibacterial activity of bacterial isolates, the individual seed-borne bacterial pathogens were streaked onto the TSA agar plates to form a strip (~1 cm in width) on the center. A single streak of test bacteria was made perpendicular to the pathogenic bacterial strip in a way that test strains do not initially directly touch the pathogens. Plates were incubated for 3 days at 30ºC, and inhibition rate was calculated following the formula described above.
In order to screen seed endophytes for potential biocontrol activity against different seed-borne pathogens, we procured some of the important seed-borne pathogens of soybean. Among the pathogens, X. axonopodis pv. glycines (bacterial pustule), P. syringae pv. tabaci (wildfire), C. sojina (leaf spot), and S. glycines (brown spot) were obtained from the Korean Agriculture Culture Collection (KACC) (Fig. 1A and B). In addition, we tried to isolate fungal pathogens from the seeds displaying obvious disease symptoms (purple spot and seed decay). Following isolation of a few fungi from the seeds, fungal colonies were classified into two groups based on their colony morphology (panels i and iv in Fig. 1B). Sequencing of the ITS region in these fungi showed that they belong to two genera: Cercospora and Diaporthe. However, the ITS did not provide enough resolution for species identification of the two. We, therefore, did a literature search to identify additional markers and found that CFP and translation elongation factor 1-alpha (TEF1-α) could serve as a secondary identification barcode for Cercospora and Diaporthe, respectively (Gomes et al., 2013; Soares et al., 2015). Sequencing of the second marker genes and BLAST search indicated that the two fungal isolates belong to C. kikuchii (purple spot) and D. eres (seed decay), respectively. Our phylogenetic analyses with the known species strongly support our species identification results (Fig. 1C and D).
To isolate the seed endophytes, soybean seeds were surface-sterilized and grounded to be plated on a variety of media following serial dilution (Fig. 2A). For isolation of bacteria, TSA, R2A agar, LB agar, and NA media were used, while PDA, RBA, CZA, and V8A were employed for isolation of fungi. This whole process was repeated several times in order to isolate as many and phylogenetically diverse seed endophytes as possible. Over a thousand colonies were formed on plates. As the molecular identification of more than a thousand isolates is not feasible, they were grouped and selected based on their pigmentation and morphology. As a result, a total of 66 fungal colonies and 87 bacterial colonies were selected for sequencing. Fungal isolates were subject to ITS sequencing, while bacterial isolates were to 16S rDNA sequencing. The results showed that 66 fungal isolates belong to 15 species from 9 genera, while 87 bacterial isolates are assigned to 30 different species from 22 genera (Fig. 2B and C, Supplementary Tables 1 and 2). Interestingly, only a few species including Staphylococcus and Glutamicibacter species took up a large portion of bacterial isolates, while Cladosporium species were predominant among the fungal isolates.
To evaluate potential antagonistic activities of the seed endophytes we have isolated, the dual culture assay was performed to test and measure the inhibition of pathogen growth by the endophytes. We have optimized dual culture assay conditions such as temperature and incubation time, as the different pathogens have different growth rates and optimal temperatures. Following the optimization of assay conditions, antagonistic activity of individual isolates was assayed through the dual culture method with modifications that are made depending on the combinations of pathogens and endophytes (Fig. 3A and ‘Materials and Methods’ for the detailed description).
With six pathogen species and 50 species of endophytes isolated from the seeds, there were three hundred combinations of assays possible. Five replicates were used for each combination of the pathogens and endophytes, leading to one thousand and five hundred assays in total. During our assay, we arbitrarily chose 20% inhibition rate, compared to the control, as a threshold to consider an isolate as having an inhibitory potential. Our results showed that some of the endophytes clearly possess the ability to suppress the growth and/or alter the physiology of the seed-borne pathogens, although they appeared to differ in the mode of action (Fig. 3B). For example, Phomopsis sp. tends to suppress the growth of pathogens by outgrowing them, while Penicillium steckii, Pseudomonas koreensis, and two Kosakonia species appear to exert inhibitory effects without direct contact with the pathogens, suggesting the production of antimicrobial metabolites or volatiles.
We also noticed that the endophytes display differences in the range of pathogens that they can inhibit. Overall, 13 species out of 50 endophytes showed antagonistic activity against at least one pathogen (26%). Among those 13, four were fungi, and the remaining nine were bacterial endophytes. Many endophytes including Alternaria hungarica and Stenotrophomonas maltophilia were antagonistic against a broad range of pathogens, in contrast to some endophytes such as Clasdosporium pseudocladosporioides and Staphylococcus sciuri having inhibitory effects only against one or two species of pathogens (Fig. 4).
There is a pressing need for reducing the over-use of agricultural chemicals that can pose a serious threat to the environment and human health. One promising and much-explored remedy to the issue is probably the utilization of microbial biological agents. They are applied to crops for biological control of a wide range of plant pathogens, promotion of plant growth, and resilience to abiotic stresses. Many microbial biological agents have been isolated from diverse environments not limited to soils (Pirttilä et al., 2021; Whipps, 2001). However, only a handful of seed endophytes has been explored for the potential as microbial biological agents for crops (Kumar et al., 2021; Matsumoto et al., 2021). In this work, we isolated seed endophytes of the soybean plant and tested their antagonistic activities against a set of seed-borne pathogens.
The bacterial pustule and wildfire disease are representative seed-borne bacterial diseases (Kang et al., 2021; Myung et al., 2009), while purple spot, leaf spot, and seed decay are representative seed-borne fungal diseases of soybean (Li et al., 2019; Sun et al., 2013). Two of the fungal pathogens we have as target species require an explanation for including them in our study. First, the brown spot, which is caused by Septoria glycines and thus also known as Septoria leaf spot, is not particularly a seed-borne disease, as it primarily overwinters on leaf residues of the plants (Cruz et al., 2010). However, S. glycines is also known to infect and overwinter on the seeds without visible symptoms. Such asymptomatic nature of seed infection by S. glycines makes it difficult to remove contaminated seeds with visual inspection alone. In addition, the most common causal agents of seed decay are known as Phomopsis longicolla and Diaporthe sojae (note that Phomopsis and Diaporthe species form Phomopsis/Diaporthe species complex). Despite our repeated efforts, we were not able to isolate either P. longicolla or D. phaseolum from the seeds showing decay symptoms. Instead, we isolated D. eres, which is also known to cause seed decay in soybean (Petrović et al., 2015). The occurrence and distribution of Diaporthe/Phomopsis species complex in naturally infected soybean has not been surveyed recently. Therefore, it is not clear that our failure to isolate P. longicolla or D. phaseolum resulted from the change in major seed decay pathogen population of Korea.
The bacteria and fungi isolated from the soybean seeds in our study showed taxonomically limited distribution. Such a pattern was expected, since it is well documented that the seed endophyte community is reduced in terms of abundance and diversity compared to the soil or rhizosphere communities (Rochefort et al., 2021). However, it should be noted that such low abundance and diversity is not an accurate reflection of seed endophyte community structure, as it does not account for microbes unamenable to the in-vitro culture. More accurate information on the community structure should be obtained from a culture-independent approach. The low diversity of culturable seed endophytes in our work is attributed to the predominance of particular taxa: Staphylococcus and Cladosporium species for bacteria and fungi, respectively. Interestingly, these species, despite their high abundances, were not very effective in their antagonistic activity against the set of pathogens tested in our study. This strongly suggests that they are merely some endophytes that have evolved the ability to enter the seeds and/or have adapted well to the seed environment. However, it should not be ruled out that they could express antagonism in interactions with other seed endophytes, as our dual culture assay setting does not allow such interactions.
One of the fungal endophytes showing a strong antagonism against a broad range of the seed-borne pathogens in our study is Phomopsis sp. Many Phomopsis spp. are known as seed pathogens, although some seem to be endophytes capable of producing biologically active metabolites and volatiles (Petrović et al., 2021; Singh et al., 2011). Therefore, there is a possibility that the Phomopsis sp. isolated here might also be a seed-borne pathogen. More works need to be done to identify the fungus beyond the genus level down to a species. However, two observations should be noted regarding the issue: (1) Phomopsis sp. were all isolated from apparently healthy seeds. (2) Colony morphology of Phomopsis sp. is distinct from that of the common seed-borne pathogen, P. longicolla. Furthermore, there have been conceptual changes about mutualism and pathogenicity, leading to the awareness that they might not be inherent properties of microbes and are only expressed within certain contexts (Nelson, 2018; Pirofski and Casadevall, 2012). This idea, which states that there is no clear borderline between the lifestyles, suggests the possibility of Phomopsis sp. being either commensal or pathogen depending on biotic and/or abiotic environments in the soybean seeds.
In this work, we have isolated the seed endophytes to use them for protection of soybean seeds and eventually plants. We are aware that in-vitro antagonism of endophytes toward pathogens does not predict protection in live plant tissue, because the activity of biocontrol agents can depend on environmental conditions such as temperature, humidity, and other microbes (Cray et al., 2016). However, it is certain that our endophytes and screening results provide candidate microbial agents and guiding information with which rational synthetic communities can be designed, tested, and formulated for the seed microbiome-based protection of soybean plants.
Acknowledgments
This work was supported by a grant from the Rural Development and Administration, Republic of Korea (PJ0157682021).
Electronic Supplementary Material
Electronic Supplementary Material
Supplementary materials are available at The Plant Pathology Journal website (http://www.ppjonline.org/).
Fig. 1
Isolation and identification of seed-borne pathogens in soybean plant. (A) Seed-borne pathogens causing seed-borne diseases in soybean plant. (B) Morphology of seed-borne pathogens on agar media: (i) Cercospora kikuchii; (ii) Cercospora sojina; (iii) Septoria glycines; (iv) Diaporthe eres; (v) Xanthomonas axonopodis pv. glycines; (vi) Pseudomonas syringae pv. tabaci. (C, D) Maximum-likelihood phylogenetic analysis of Diaporthe and Cercospora species based on the marker gene sequences (translation elongation factor 1-alpha and cercosporin facilitator protein, respectively). The numbers above the nodes are the bootstrap values obtained from 1,000 replicates. The trees are drawn to scale, with branch lengths measured in the number of substitutions per site. The accession number from NCBI is shown next to species name. The isolates obtained in this study are shown in boldface.
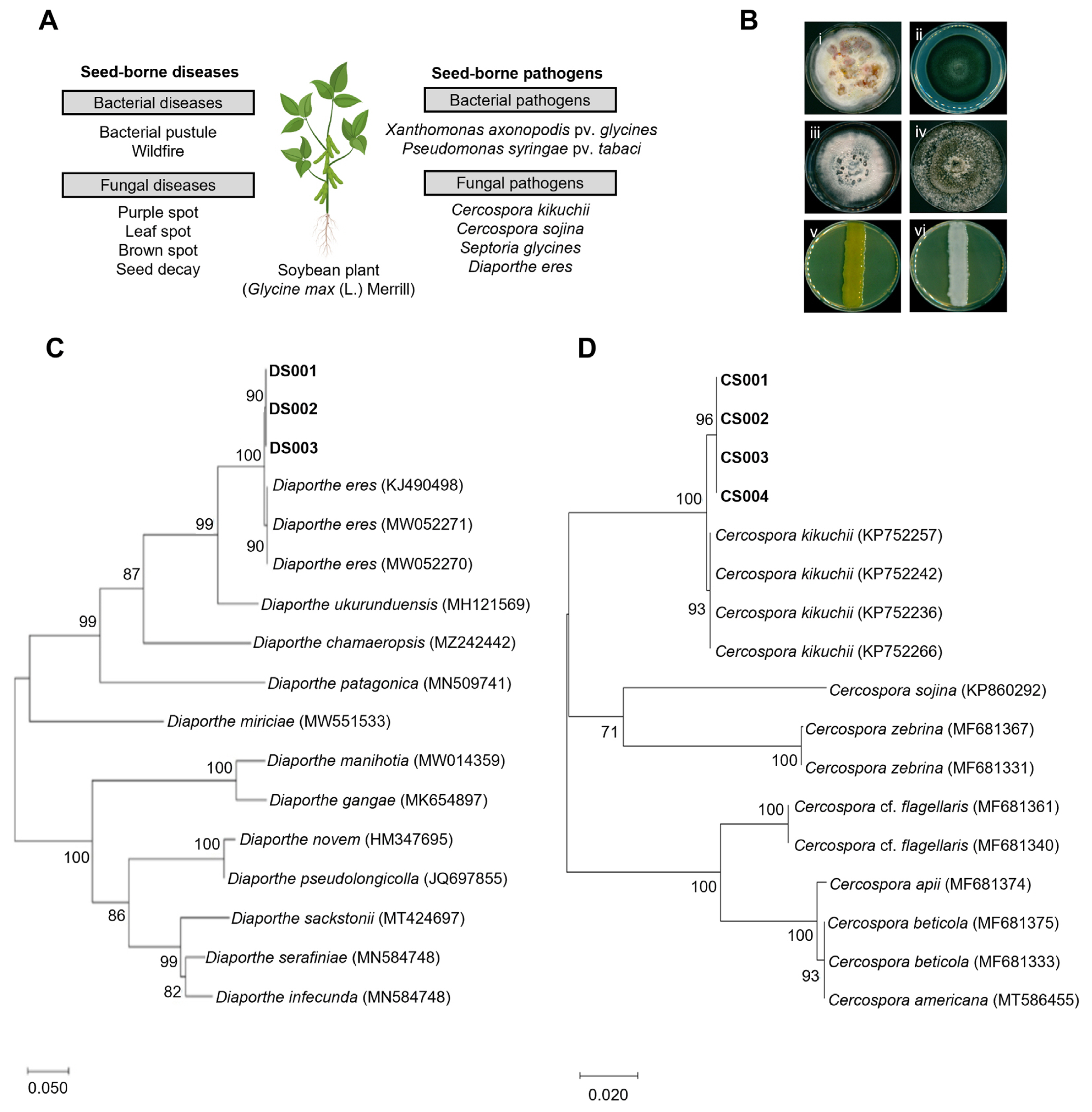
Fig. 2
Isolation of endophytes from soybean seeds. (A) Step-by-step procedure of isolating microorganisms from soybean seeds. The surface-sterilized seeds were ground using 1× phosphate buffered saline (PBS) and serially diluted up to 10−5. One hundred microliters of each dilution was spread on agar media and incubated for growth of endophytic bacteria and fungi. Eighty-seven bacterial isolates and 66 fungal isolates were selected for sequencing. (B) Genus-level identification of bacterial isolates. (C) Genus-level identification of fungal isolates.
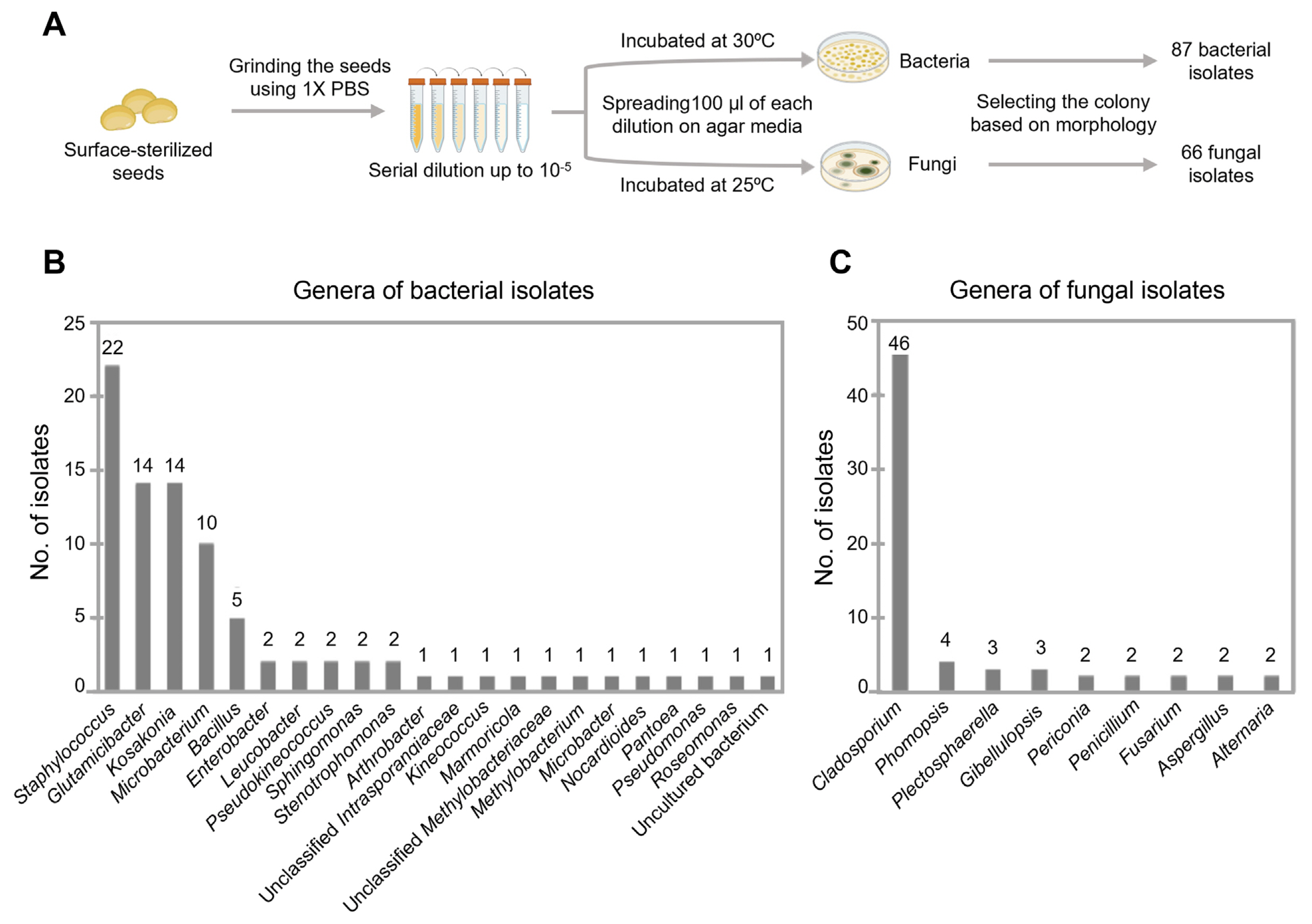
Fig. 3
Dual culture assay for evaluation of antagonistic activities of endophytes. (A) Schematic diagram showing dual culture assay strategies for antifungal and antibacterial activities. PF, pathogenic fungi; TF, test fungi; PB, pathogenic bacteria; TB, test bacteria. (B) Representative images of antifungal and antibacterial activity results. The inoculated strain is mentioned at the bottom of each plate, and the first plate on the left of each group is the control. C. kikuchii, Cercospora kikuchii; C. sojina, Cercospora sojina; D. eres, Diaporthe eres; S. glycines, Septoria glycines; P. syringae, Pseudomonas syringae pv. tabaci; X. axonopodis, Xanthomonas axonopodis pv. glycines, A. hungarica, Alternaria hungarica; P. steckii, Penicillium steckii; P. koreensis, Pseudomonas koreensis; K. cowanii, Kosakonia cowanii; G. nicotianae, Glutamicibater nicotianae.
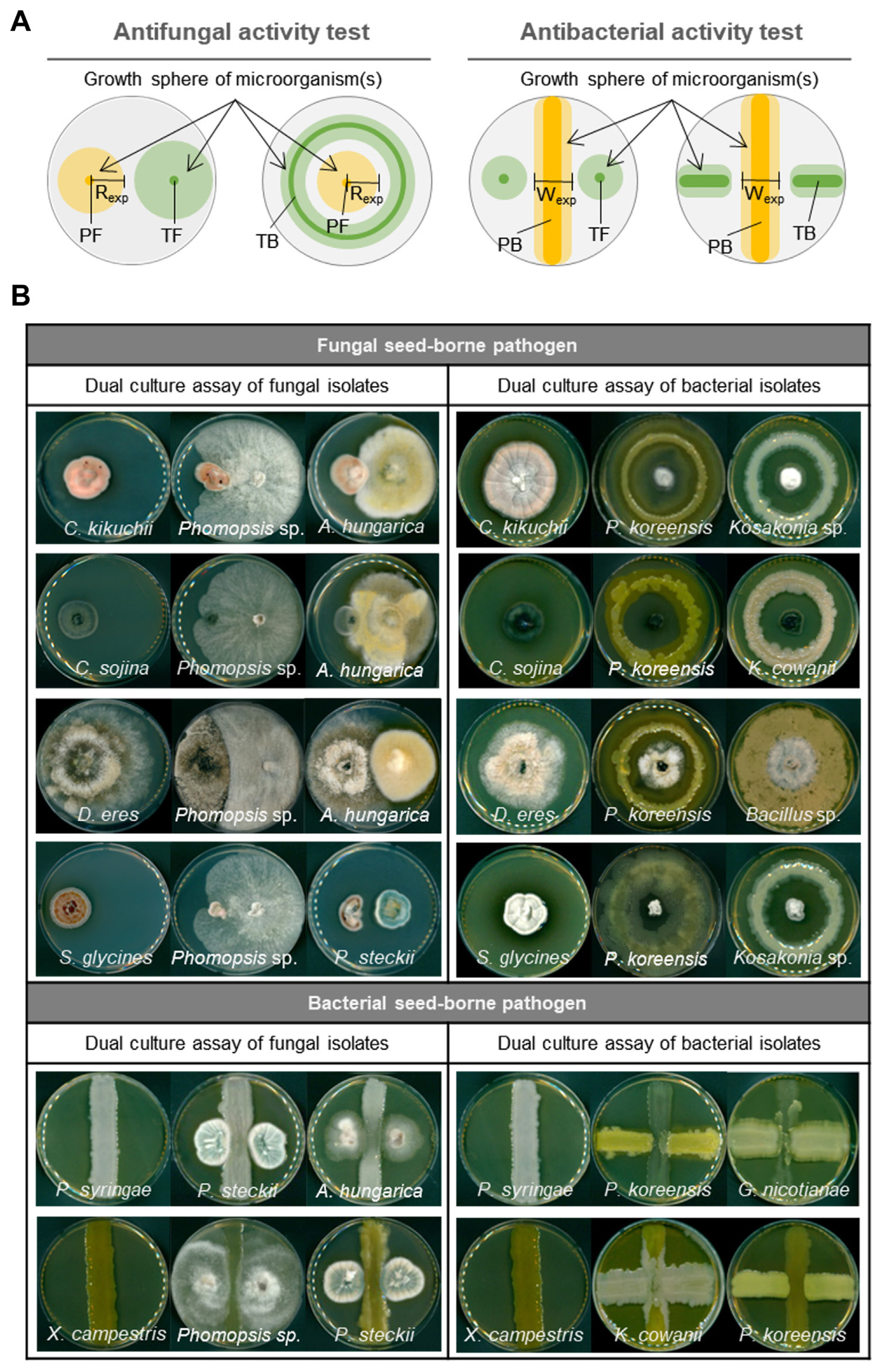
Fig. 4
Heatmap summary of antifungal and antibacterial activity assay results. Isolated endophytes were used as test strains against seed-borne pathogens. Ck, Cercospora kikuchii; Cs, Cercospora sojina; De, Diaporthe eres; Sg, Septoria glycines; Pst, Pseudomonas syringae pv. tabaci; Xag, Xanthomonas axonopodis pv. glycines. The inhibition rate was represented by color gradients in five discrete scales.

References
Bell, J. 2008. A simple way to treat PCR products prior to sequencing using ExoSAP-IT. Biotechniques 44:834.


Berg, G., Köberl, M., Rybakova, D., Müller, H., Grosch, R. and Smalla, K. 2017. Plant microbial diversity is suggested as the key to future biocontrol and health trends. FEMS Microbiol. Ecol 93:fix050.

Bziuk, N., Maccario, L., Straube, B., Wehner, G., Sørensen, S.J., Schikora, A. and Smalla, K. 2021. The treasure inside barley seeds: microbial diversity and plant beneficial bacteria. Environ. Microbiome 16:20.




Chi, M.-H., Park, S.-Y. and Lee, Y.-H. 2009. A quick and safe method for fungal DNA extraction. Plant Pathol. J 25:108-111.

Compant, S., Samad, A., Faist, H. and Sessitsch, A. 2019. A review on the plant microbiome: ecology, functions, and emerging trends in microbial application. J. Adv. Res 19:29-37.



Cray, J.A., Connor, M.C., Stevenson, A., Houghton, J.D.R., Rangel, D.E.N., Cooke, L.R. and Hallsworth, J.E. 2016. Biocontrol agents promote growth of potato pathogens, depending on environmental conditions. Microb. Biotechnol 9:330-354.



Cruz, C.D., Mills, D., Paul, P.A. and Dorrance, A.E. 2010. Impact of brown spot caused by Septoria glycines on soybean in Ohio. Plant Dis 94:820-826.


Dai, Y., Li, X.-Y., Wang, Y., Li, C.-X., He, Y., Lin, H.-H., Wang, T. and Ma, X.-R. 2020. The differences and overlaps in the seed-resident microbiome of four Leguminous and three Gramineous forages. Microb. Biotechnol 13:1461-1476.




Gitaitis, R. and Walcott, R. 2007. The epidemiology and management of seedborne bacterial diseases. Annu. Rev. Phytopathol 45:371-397.


Gomes, R.R., Glienke, C., Videira, S.I.R., Lombard, L., Groenewald, J.Z. and Crous, P.W. 2013.
Diaporthe: a genus of endophytic, saprobic and plant pathogenic fungi. Persoonia 31:1-41.


Hariharan, G. and Prasannath, K. 2021. Recent advances in molecular diagnostics of fungal plant pathogens: a mini review. Front. Cell. Infect. Microbiol. 10:600234.



Johnston-Monje, D., Gutiérrez, J.P. and Lopez-Lavalle, L.A.B. 2021. Seed-transmitted bacteria and fungi dominate juvenile plant microbiomes. Front. Microbiol 12:737616.



Kang, I.-J., Kim, K.S., Beattie, G.A., Chung, H., Heu, S. and Hwang, I. 2021. Characterization of Xanthomonas citri pv. glycines population genetics and virulence in a national survey of bacterial pustule disease in Korea. Plant Pathol. J. 37:652-661.




Kim, H. and Lee, Y.-H. 2021. Spatiotemporal assembly of bacterial and fungal communities of seed-seedling-adult in rice. Front. Microbiol. 12:708475.



Klindworth, A., Pruess, E., Schweer, T., Peplies, J., Quast, C., Horn, M. and Glöckner, F.O. 2013. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 41:e1.



Kumar, K., Verma, A., Pal, G., Anubha White, J.F. and Verma, S.K. 2021. Seed endophytic bacteria of pearl millet (Pennisetum glaucum L.) promote seedling development and defend against a fungal phytopathogen. Front. Microbiol. 12:774293.



Li, S., Sciumbato, G., Boykin, D., Shannon, G. and Chen, P. 2019. Evaluation of soybean genotypes for reaction to natural field infection by Cercospora species causing purple seed stain. PLoS ONE 14:e0222673.



Matsumoto, H., Fan, X., Wang, Y., Kusstatscher, P., Duan, J., Wu, S., Chen, S., Qiao, K., Wang, Y., Ma, B., Zhu, G., Hashidoko, Y., Berg, G., Cernava, T. and Wang, M. 2021. Bacterial seed endophyte shapes disease resistance in rice. Nat. Plants 7:60-72.



Messina, M.J. 1999. Legumes and soybeans: overview of their nutritional profiles and health effects. Am. J. Clin. Nutr. 70(3 Suppl):439S-450S.


Myung, I.-S., Kim, J.-W., An, S.H., Lee, J.H., Kim, S.K., Lee, Y.-K. and Kim, W.G. 2009. Wildfire of soybean caused by Pseudomonas syringae pv. tabaci, a new disease in Korea. Plant Dis. 93:1214.

Petrović, K., Skaltsas, D., Castlebury, L.A., Kontz, B., Allen, T.W., Chilvers, M.I., Gregory, N., Kelly, H.M., Koehler, A.M., Kleczewski, N.M., Mueller, D.S., Price, P.P., Smith, D.L. and Mathew, F.M. 2021. Diaporthe seed decay of soybean [Glycine max (L.) Merr.] is endemic in the United States, but new fungi are involved. Plant Dis 105:1621-1629.


Petrović, K., Vidić, M., Riccioni, L., Đorđević, V. and Rajković, D. 2015. First report of Diaporthe eres species complex causing seed decay of soybean in Serbia. Plant Dis. 99:-1186.
Pirofski, L.-A. and Casadevall, A. 2012. Q&A: What is a pathogen? A question that begs the point. BMC Biol 10:6.




Pirttilä, A.M., Mohammad Parast Tabas, H., Baruah, N. and Koskimäki, J.J. 2021. Biofertilizers and biocontrol agents for agriculture: how to identify and develop new potent microbial strains and traits. Microorganisms 9:817.



Rochefort, A., Simonin, M., Marais, C., Guillerm-Erckelboudt, A.-Y., Barret, M. and Sarniguet, A. 2021. Transmission of seed and soil microbiota to seedling. mSystems 6:e0044621.




Singh, S.K., Strobel, G.A., Knighton, B., Geary, B., Sears, J. and Ezra, D. 2011. An endophytic Phomopsis sp. possessing bioactivity and fuel potential with its volatile organic compounds. Microb. Ecol. 61:729-739.



Soares, A.P.G., Guillin, E.A., Borges, L.L., da Silva, A.C.T., de Almeida, A.M.R., Grijalba, P.E., Gottlieb, A.M., Bluhm, B.H. and de Oliveira, L.O. 2015. More Cercospora species infect soybeans across the Americas than meets the eye. PLoS ONE 10:e0133495.



Sprockett, D., Fukami, T. and Relman, D.A. 2018. Role of priority effects in the early-life assembly of the gut microbiota. Nat. Rev. Gastroenterol. Hepatol. 15:197-205.




Sun, S., Kim, M.Y., Chaisan, T., Lee, Y.-W., Van, K. and Lee, S.-H. 2013. Phomopsis (Diaporthe) species as the cause of soybean seed decay in Korea. J. Phytopathol 161:131-134.


Whipps, J.M. 2001. Microbial interactions and biocontrol in the rhizosphere. J. Exp. Bot. 52:487-511.


White, T.J., Bruns, T., Lee, S. and Taylor, J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: PCR protocols: a guide to methods and applications, eds. by M.A. Michael, D.H. Gelfand, J.J. Sninsky and T.J. White, pp. 315-322. Academic Press, San Diego, CA, USA.

- TOOLS



 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Supplement1
Supplement1 Print
Print




