 |
 |
| Plant Pathol J > Volume 38(4); 2022 > Article |
|
Abstract
In Japan, the P1 protein (S-type) encoded by leek yellow stripe virus (LYSV) isolates detected in Honshu and southward is shorter than the P1 (N-type) of LYSV isolates from garlic grown in Hokkaido due to a large deletion in the N-terminal half. In garlic fields in Hokkaido, two types of LYSV isolate with N- and S-type P1s are sometimes found in mixed infections. In this study, we confirmed that N- and S-type P1 sequences were present in the same plant and that they belong to different evolutionary phylogenetic groups. To investigate how LYSV with S-type P1 (LYSV-S) could have invaded LYSV with N-type P1 (LYSV-N)-infected garlic, we examined wild Allium spp. plants in Hokkaido and found that LYSV was almost undetectable. On the other hand, in Honshu, LYSV-S was detected at a high frequency in Allium spp. other than garlic, suggesting that the LYSV-S can infect a wider host range of Allium spp. compared to LYSV-N. Because P1 proteins of potyviruses have been reported to promote RNA silencing suppressor (RSS) activity of HC-Pro proteins, we analyzed whether the same was true for P1 of LYSV. In onion, contrary to expectation, the P1 protein itself had RSS activity. Moreover, the RSS activity of S-type P1 was considerably stronger than that of N-type P1, suggesting that LYSV P1 may be able to enhance its RSS activity when the deletion is in the N-terminal half and that acquiring S-type P1 may have enabled LYSV to expand its host range.
Virus infection of garlic results in yield loss and quality deterioration. In Japan, among the vigorous efforts to produce virus-free garlic, the Fukuchi-White (FW) garlic variety from Aomori Prefecture has met with some success, propelling the prefecture to the top of Japan in terms of production. FW garlic is also actively promoted for production in Hokkaido, the largest northern island of Japan.
Garlic virus diseases are intensified by mixed infections. In particular, mixed infections with two potyviruses, leek yellow stripe virus (LYSV), onion yellow dwarf virus (OYDV), and allexiviruses (garlic viruses A, B, C, D, and X), cause severe garlic mosaics. When garlic has obvious symptoms of viral infection, these three species are usually detected. In Hokkaido, LYSV is more commonly detected than OYDV, while OYDV is more common in other parts of Japan. In previous report, we found that OYDV isolates detected in garlics in Hokkaido mostly have a large deletion at the 5' end of the HC-Pro gene and have lost the ability to support aphid transmission of the virus (Kim et al., 2020). Therefore, mixed infection with LYSV has been suggested to be important for aphid transmission of OYDV (Jayasinghe et al., 2021).
LYSV infects garlic and leek and is detected in garlic-growing areas worldwide. Although onions generally are not considered to be natural hosts for LYSV (Katis et al., 2012; Vučurović et al., 2017), onion damage due to LYSV infection has also been reported in some regions (Santosa and Ertunç, 2020; Sharhraeen et al., 2008). LYSV detected in garlic-growing areas in Japan can be classified into two types, northern type (LYSV-N) and southern type (LYSV-S), based on a characteristic deletion of 200-300 bases in the P1 gene (Takaki et al., 2005; Yoshida et al., 2012). Mixed infections of LYSV-N and LYSV-S have also been reported in Honshu, main island of Japan (Takaki et al., 2005). LYSVs detected in Hokkaido are most, if not all, N-type, but in rare cases, mixed infections of N-type and S-type have been identified.
The P1 protein of potyvirus has been reported to bind to the P3 protein and to be involved in viral replication (Merits et al., 1999). In the interaction between the P1 protein and host factors, P1 of tobacco etch virus has been reported to bind to the 60S ribosomal subunit and to promote protein translation (Martínez and Daròs, 2014). Hu et al. (2020) used immunoprecipitation experiments with P1 of three potyviruses and comprehensively detected P1-binding proteins from Arabidopsis with liquid chromatograph-tandem mass spectrometer (LC-MS/MS). They thus identified many proteins involved especially in RNA silencing and defense response. Also, P1 of sweet potato mild mottle virus (family Potyviridae, genus Ipomovirus) has been reported to bind to the AGO1 protein, the key player in the RNA silencing pathway (Giner et al., 2010); thus, P1 appears to have the function of interfering with RNA silencing.
On the other hand, there have been some reports that P1 itself has no RNA silencing suppressor (RSS) activity but promotes RSS activity of the HC-Pro protein (Kasschau and Carrington, 1998; Valli et al., 2006). However, because there are also reports that P1 of watermelon mosaic virus inhibits the RSS activity of P25 of a non-potyvirus, cucurbit yellow stunting disorder virus (crinivirus) (Domingo-Calap et al., 2021), the viral proteins that P1 can promote in RSS activity may be limited to HC-Pro. In addition, P1 can assist the RSS activity of a heterologous HC-Pro, even if the P1 gene is exchanged between the two potyvirus species. However, when hybrid viruses were created in the P1 sequence between plum pox virus (PPV) and tobacco vein mottling virus, all hybrids could not infect Prunus percicae, which is a natural host of PPV, suggesting that the combination of P1 and HC-Pro may determine the host specificity of potyviruses (Salvador et al., 2008).
In the absence of cleavage between P1 and HC-Pro from a polyprotein by the P1 proteinase activity, the RSS activity of HC-Pro was not well exerted (Pasin et al., 2014). Similarly, the RSS activity of HC-Pro was stronger in transgenic plants that express a P1-HC-Pro fusion protein than in transgenic plants that co-express P1 and HC-Pro as separate proteins (Hu et al., 2020). Because RSS activity of the virus seems to be important for determining infectivity of a potential host (Shimura et al., 2022), the effect of P1 on HC-Pro might be important for the evolution of potyviruses. There are many studies on the P1 proteins of other potyviruses, but there are no reports on the function of the P1 gene of garlic-infecting potyviruses, so we do not know whether the P1 proteins of LYSV and OYDV promote RSS activity of HC-Pro as the P1 proteins of other potyviruses do.
In this study, we analyzed the P1 gene of LYSV to determine how the LYSV-S was able to invade the garlic fields in Hokkaido. We also discuss how short P1 of LYSV-S (S-type P1) may have been generated and how LYSV with S-type P1 may have emerged.
The Hokkaido garlic varieties tested for virus were all derived from FW, which had been originally developed in Aomori Prefecture, Japan. In this paper, they are all abbreviated as FW. Garlic plants other than FW, such as American garlic (USA garlic) and Chinese garlic (China garlic), were purchased in the market, as were the seeds of the seven onion varieties (Sapporo-Yellow [SY], Fresh-Red [FR], Heian, Kitano-Daichi, Neo, Focus, Kaizuka). Seedlings of SY were purchased from a commercial producer in Hokkaido. Viruses in wild Allium plants (nobiru, asatsuki, gyoja-garlic) and Allium vegetables other than garlic (rakkyo, wakegi) were investigated mainly by J. Sasaki and C. Masuta for plants grown in Hokkaido, and by K. Yamashita and O. Kim for those from Honshu.
Total RNA was extracted from garlic leaves using TRIzol Reagent (Invitrogen, Carlsbad, CA, USA). If necessary, the RNA was further purified through a column using NucleoSpin RNA (Takara, Otsu, Japan) especially for RNA-sequencing (RNA-seq).
Reverse transcription polymerase chain reactions (RT-PCRs) were either two-step or one-step reactions. For two-step, cDNA was first synthesized with avian myeloblastosis virus (AMV) reverse transcriptase (Nippon Gene, Tokyo, Japan). The cDNA sequences were then amplified with Takara Ex Taq (Takara). For one-step RT-PCRs, Takara One Step RNA PCR kit (AMV) (Takara) was used. For nested-PCRs, 1 μl of the first-round PCR reaction mixture was added to the second PCR preparation. The manufacturer’s specifications were used for all the PCRs. All primer sequences are given in Supplementary Table 1.
Nucleotide sequences of the P1 regions of LYSV determined in this study and from GenBank were used for the analysis with two isolates of OYDV (GenBank accession nos. KJ451436 and NC_005029) as outgroups. The multiple sequence alignment was generated using the software mafft v7.429 (Katoh and Standley, 2013) via the corresponding amino acid sequence. Before the phylogenetic reconstruction, recombination analysis was performed using the software RDP5 (Martin et al., 2021), and any isolates that might potentially contain recombination sites in the sequences were discarded. A phylogenetic tree was inferred using the Bayesian method implemented in MrBayes v3.2.7 (Ronquist and Huelsenbeck, 2003) under the general time-reversible model with gamma-distributed among-site rate variation and a proportion of invariable sites. Markov chain Monte Carlo sampling was performed for 3,000,000 iterations sampling every 3,000 steps. The first 25% of samples were discarded as burn-in state. The resulting consensus tree was visualized using the R package ggtree (Yu et al., 2017). Genetic differentiation was analyzed using an analysis of molecular variance (AMOVA), and nucleotide diversity were calculated using the software Arlequin v3.5.2 (Excoffier and Lischer, 2010). Nucleotide similarity across the P1 region was analyzed using the software SimPlot v3.5.1 (Lole et al., 1999). LYSV Tunisian isolate 2.1 (MH890561) was used as the query sequence. Similarity was estimated under the Kimura (two-parameter) model and plotted against the query with a window size of 100 nt and a step size of 10 nt. Five isolates from the Brazil + Tunisia clade (KP236097, KP236101, KP236102, KP236103, MH890559) and the China + Japan clade (AB636327, MN059500, MN059520, MN059534, MN059543) were included. The isolates for which the P1 nucleotide sequence were determined in this study were also included as representative isolates of LYSV-N and LYSV-S.
Aphid transmission was tested following the method described by Jayasinghe et al. (2021) with slight modifications. Individuals of peach aphid, Myzus persicae, were raised on Nicotiana tabacum plants with 16 h/8 h light/dark at 25°C. Apterous aphids were collected in a petri dish and starved for 3 h, then fed on LYSV-S-infected onion plants (14 days after inoculation) for 5 min. Twelve aphids were then transferred to LYSV-N-infected garlic plants (USA garlic), and then killed with a pesticide spray after 24 h. The transmission of LYSV was checked at 20 dpi by nested-RT-PCR using primer pair LY-5P/LY-2M for the first-round RT-PCR and LYSV-5-278/LYSV-3-278 for the nested-PCR.
RSS activity was assayed as described before (Kim et al., 2020; Shimura et al., 2022). The DNA fragments coding for the HC-Pro and FLAG-tagged P1 proteins were cloned into the pBE2113 plasmid between XbaI and SacI sites, and then Agrobacterium was transformed with each of those plasmids. The concentration of each bacterial culture was adjusted to OD600 = 1.0 for the assay using Nicotiana benthamiana and to 0.2 for the assay using onion. N. benthamiana leaves or onion epidermis were then infiltrated with a mixed bacterial suspension of the green fluorescence protein (GFP) construct, P1 and HC-Pro constructs (1:1:1 ratio). GFP fluorescence was assessed under UV light at 5 days post agroinfiltration (dpa) in N. benthamiana and at 2 dpa in onion. The GFP fluorescence intensity in onion epidermis was quantified using LAS AF software (Leica, Wetzlar, Germany). For confirming P1 expression, total protein extracts from N. benthamiana leaf tissues infiltrated with Agrobacterium carrying the P1 construct were separated by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis, and the P1-FLAG was detected by western blot analysis using anti-FLAG monoclonal antibody (Kim et al., 2020; Shimura et al., 2022).
We first identified the isolates of LYSV in FW garlic in the Tokachi and Okhotsk regions of Hokkaido and detected several mixed infections of LYSV-N and LYSV-S, albeit at low frequency (Fig. 1). In the Tokachi region, FW virus-free garlic seedlings are distributed to growers every year. To ensure the number and size of seedlings to be distributed, the seedlings are normally grown for at least 2 years in the fields but often re-infected with the virus by aphids during the 2-year period; they thus cannot remain completely virus-free. After surveying LYSV infection in the same fields in 2019 and 2020, three mixed infections out of 40 individuals surveyed were detected in 2019. In 2020, however, no mixed infections were detected among 60 individuals surveyed in the same fields (Table 1), perhaps due to the distribution of virus-free seedlings each year, suggesting that there were no more LYSV-S-infected source plants in the surrounding area in 2020.
Total RNA including viral RNAs of both LYSV N-type (Okhotsk-N) and S-type (Okhotsk-S) were isolated from the mixed-infected garlic from the Okhotsk region, and the P1 genes were amplified by RT-PCR and then sequenced. The deletion of amino acids in the P1 proteins was shown in the P1 protein sequence alignment (Supplementary Fig. 1). We also sequenced the P1 genes of LYSV-N and LYSV-S (N-type P1 and S-type P1, respectively) isolated from garlics grown in other parts of Japan (Okinawa) and imported garlics (from USA and China). We then used these sequences and those of numerous P1 genes from the GenBank database to construct a phylogenetic tree (Supplementary Fig. 2); Fig. 2 shows the simplified phylogenetic tree by collapsing some clades to focus on our primary interests. In the tree, N-type P1 (Okhotsk-N) and S-type P1 (Okhotsk-S) from the mixed infection in garlic from Okhotsk were clearly placed in different groups; thus, S-type P1 did not originate from the N-type P1 in the mixed infection of LYSV-N and LYSV-S. We then examined the P1 sequence population variation between N-type P1 and S-type P1 using AMOVA and found that these two types significantly differed by 41.6% (P < 0.001). Moreover, the nucleotide diversity of N-type P1 (mean 0.413, SD 0.198) was higher than that of S-type P1 (mean 0.313, SD 0.149). These results indicate that S-type P1 was newly introduced from outside, probably by aphid transmission, into the LYSV-N-infected garlic. Our phylogenetic tree also indicated that two clades of the N-type P1 (Brazil + Tunisia clade and China + Japan clade) were evolutionarily independent and formed a sister lineage to the S-type P1 even though these isolates did not have common deletions shown in S-type P1 but had sequences of short stretches of various lengths (Fig. 2, Supplementary Fig. 2). These results therefore suggest that the distinct N-type P1 clades might be the ancestral population of S-type P1 before the occurrence of P1 deletion.
As mentioned, LYSV has been reported to infect plants of the genus Allium, but it has rarely been analyzed in detail except for garlic and leek. LYSV is known not to infect most onion varieties. In this study, we focused on onions, which are cultivated extensively in Hokkaido. Each onion variety was mechanically inoculated with sap from garlic leaves infected with LYSV containing either LYSV having N-type P1 (LYSV-N USA) or LYSV having S-type P1 (LYSV-S China). In two onion cultivar, SY and FR, OYDV was also detected by RT-PCR in the upper non-inoculated leaves at 10 days postinoculation (dpi) (Table 2). OYDV must have been eliminated from the sap so that co-infection of LYSV and OYDV does not affect the result of the aphid transmission experiment. LYSV had been detected continuously up to 30 dpi only in FR (Table 2). On the other hand, OYDV in FR plants inoculated with the LYSV-S China garlic sap was no longer detected at 20 dpi (Table 2, Supplementary Fig. 3). We then tested LYSV-S transmission by aphids to LYSV-N USA-infected garlic using sap from the LYSV-S-infected FR plant prepared as explained above (Fig. 3A). As a result, the garlic plants were infected with both LYSV-N and LYSV-S, albeit at a low rate (2/12), providing experimental evidence that it is possible to infect LYSV-N-infected garlic with LYSV-S by aphid transmission (Fig. 3). To determine whether we can detect LYSV in onion grown in Hokkaido, we purchased commercial SY onion seedlings to test them for LYSV infection using RT-PCR. The LYSV-S was present in 1 of 40 of the tested seedlings (Supplementary Fig. 4). We also confirmed the absence of LYSV in those onion plants by RNA-seq. As a result, no reads were mapped to LYSV out of a total of 19,492,475 paired-end reads (Supplementary Table 2). However, because we used nested-PCR, it is unlikely that LYSV-S was introduced from onion to garlic by aphid transmission at this trace level of virus. In Hokkaido, onions are planted from May to June due to the cold climate, and onions are sometimes planted near garlic fields. Because LYSV can also infect some onion varieties as shown in Table 2, potential LYSV infection of onions must be considered to be able to eradicate LYSV from garlic.
We then investigated whether any wild Allium species in Hokkaido could serve as reservoirs of LYSV. In Hokkaido, wild asatsuki (Allium schoenoprasum var. foliosum) and gyoja-garlic (Allium victorialis subsp. platyphyllum) are widely gathered from the wild and sold or even cultivated. In Honshu, nobiru (Allium macrostemon), wakegi (Allium fistulosum var. caespitosum), and rakkyo (Allium chinense) grow in wild or farm field. We tested for LYSV infection in these plants collected from various parts of Japan and detected LYSV in wild Allium plants including nobiru, asatsuki, rakkyo, wakegi, and gyoja-garlic in Honshu by single RT-PCR (without nested-PCR). However, in Hokkaido, LYSV was not detected by single RT-PCR in wild asatsuki (four samples from each of two locations) and gyoja-garlic (four samples from each of five locations); LYSV-S was only detected by nested-PCR in one wild asatsuki plant (from Takikawa region in Hokkaido). Here, we wondered if the reason LYSV was not detected in wild asatsuki and gyoja-garlic in Hokkaido, compared to Allium species such as nobiru, wakegi, rakkyo, and asatsuki from Honshu, might be due to the PCR primers we used. Therefore, RNA was extracted from four individuals of wild asatsuki (from the Takikawa region) and subjected in bulk to RNA-seq to analyze all virus sequences. As a result, a total of 23,636,713 paired-end reads were obtained with an average Q30 of 89.6%. No reads were mapped to LYSV, and also few reads were mapped to the genomes of other garlic viruses (allexiviruses, OYDV, garlic latent virus, garlic common latent virus, and shallot yellow stripe virus) (Supplementary Table 2). In other words, wild plants of the genus Allium in Hokkaido are not considered to be sources of these viruses. On the other hand, in Honshu, LYSV was easily detected in these wild Allium plants by single RT-PCR, so they could potentially serve as reservoirs for aphid transmission to garlic.
P1 of potyviruses has been reported to promote RSS activity of HC-Pro, but there have been no reports of P1 itself having RSS activity. Because LYSV has the N-type and S-type P1s that differ in length, we first analyzed whether P1 itself really has no RSS activity, and whether the N- and the S-type P1 can each promote RSS activity of HC-Pro. Although the RSS ability of LYSV is supposed to be primarily determined by HC-Pro, we considered that if the HC-Pro-assisting activity of P1 differs between N-type and S-type, then deletions in P1 would be quite important.
First, we used the conventional method for RSS activity assay using agroinfiltration of N. benthamiana. As expected, HC-Pro had strong RSS activity, and the activity of the LYSV-S-derived HC-Pro (O-HC) was weaker than that of the LYSV-N-derived HC-Pro (K-HC) in N. benthamiana (Fig. 4A). On the other hand, we found that N-type P1 and S-type P1 had little or no RSS activity, or if one of them did, it was extremely weak, as was the case with P1 of other potyviruses (Fig. 5A). When P1 and K-HC were co-expressed, the agroinfiltrated patch with S-type P1 was found to have a little stronger GFP intensity than that with N-type P1 (Fig. 5B), in spite of the fact that the two patches without P1 must be observed to have the same level of GFP intensity maintained by the K-HC RSS activity. Therefore, S-type P1 seemed to promote the RSS activity of HC-Pro more strongly than N-type P1, considering that both N-type P1 and S-type P1 have little RSS activity in N. benthamiana. Similarly, co-expression of O-HC with S-type P1 also slightly enhanced RSS activity (Supplementary Fig. 5). Expression of the P1 protein in N. benthamiana was confirmed by western blot analysis (Supplementary Fig. 6).
We then used a similar agroinfiltration assay with onion for two reasons. The first is because we believe, based on our previous studies, that host or closely related plants should be used, if possible, to detect the RSS activity of viral proteins (Kim et al., 2020; Shimura et al., 2022). Second, based on the report by Shan et al. (2018), we thought that S-type P1 might have evolved from N-type P1 in the favor of the virus. They reported that trimming the N-terminal portion of P1 of PPV increased the replication ability of the virus, allowing it to infect a non-permissive host plant (cucumber). Therefore, we thought that we might detect different RSS activity using a different assay plant. The results of the agroinfiltration assay using onion showed that HC-Pro showed strong RSS activity as shown in N. benthamiana (Fig. 4B and C). Contrary to our expectations, in onion epidermal cells, P1 itself had strong RSS activity, and the S-type P1 provided considerably stronger activity than the N-type P1 did (Fig. 5C and D). When P1 and HC-Pro were co-expressed, little additive or synergistic RSS activity was observed, comparable to the activity with P1 alone in onion.
It is noteworthy that LYSV detected in onion (cv. Sapporo-Yellow) was LYSV-S with S-type P1 (Supplementary Fig. 4) because the LYSV-S was not detected in garlic in Hokkaido. However, the possibility that aphids transmitted LYSV from onion to garlic must be very low because it could only be detected in onion by nested-PCR. Based on the results of the inoculation test, LYSV infected only two onion cultivars at very low frequency, so there is no need to be particularly concerned about onions as a reservoir of LYSV at least in Hokkaido. Nevertheless, our experiments have demonstrated that aphid transmission does, in fact, occur from LYSV-S-infected onion to LYSV-N-infected garlic if LYSV-S is at sufficiently high titer in onion. If we happen to select an onion variety that is very susceptible to LYSV, we may still need to be careful that LYSV is not transmitted from onion to garlic.
A few plants collected in the Okhotsk region of Hokkaido were found to be infected with a mixture of LYSV-N (Okhotsk-N) and LYSV-S (Okhotsk-S). In the phylogenetic analysis using the Bayesian method to clarify the evolutionary relationship between the two isolates, the topology of the tree indicated that LYSV-N (Okhotsk-N) and LYSV-S (Okhotsk-S) belong to different groups, so it is unlikely that S-type P1 of Okhotsk-S emerged from N-type P1 of Okhotsk-N as a defective interfering RNA in the fields. In addition, this phylogenetic tree and the population variation test suggest that S-type P1s evolved from N-type P1s as we previously demonstrated using the maximum likelihood method (Yoshida et al., 2012). In addition, while LYSV-N are globally distributed, including North and South America and Europe, LYSV-S seem to be in more limited areas in Asian countries. This distribution suggests that LYSV-S may have emerged through the adaptation of the ancestral LYSV-N to hosts in Asian countries. The lower nucleotide diversity of S-type P1 compared with that of N-type P1 also supports this hypothesis. Moreover, the distinct N-type P1 clades (Brazil + Tunisia clade and China + Japan clade) that were a sister lineage to the S-type P1 clade suggests the existence of an ancestral LYSV-N population before the deletion in the P1 region had occurred. When we additionally examined the nucleotide similarity of these evolutionarily distinct N-type P1 isolates to the S-type P1 isolates using the software, SimPlot, the isolates in the distinct N-type P1 clades (Brazil + Tunisia clade and China + Japan clade) had even higher similarity to the S-type P1 isolates rather than to the other N-type P1 isolates especially around the nucleotide position 250 numbered from the first nucleotide of the P1 region (Supplementary Fig. 7). Bampi et al. (2015) reported that Brazilian LYSV isolates are possible ancestors of LYSV-S, consistent with our results. Our analysis also revealed that the Tunisian LYSV isolates might be another possible ancestral source of LYSV-S because the P1 sequences from Tunisian LYSV isolates were placed as a sister lineage to those from the Brazilian LYSV isolates (Supplementary Fig. 2).
Because most of the LYSV isolates in Hokkaido had the N-type P1, the mixed infection of the LYSV-N and LYSV-S is most likely due to the introduction of LYSV-S into the LYSV-N-infected population. We thus consider that garlic farmers may have brought LYSV-S-infected garlic from Honshu or even overseas and cultivated it in Hokkaido.
We had not previously thought that potyvirus P1 had RSS activity per se, although it could promote RSS activity of HC-Pro. However, when we tested for RSS activity using onion as the assay plant instead of N. benthamiana, P1 had clear RSS activity (Fig. 5C and D). In this case, P1 with a deletion in the N-terminal half (S-type P1) had RSS activity comparable to that of HC-Pro. In fact, Shan et al. (2018) previously reported that HC-Pro fused to the C-terminal half of P1 of PPV had RSS activity in Cucumis sativus; HC-Pro fused to the intact P1 had no RSS activity. In this experiment, it is conceivable that not only HC-Pro but also the short P1 portion had its own RSS activity. In addition, the authors showed that trimming the N-terminal half of P1 allowed PPV to infect Cucumis sativus, a non-permissive host of PPV, suggesting that P1 plays an important role in determining host range of potyviruses. This observation is supported also by the finding by Shimura et al. (2022) that the presence or absence of RSS activity of partitivirus may determine whether the virus can switch the host.
We previously observed that viral RSS activity is differentially detected in a host-dependent manner. For example, the 2b protein of cucumber mosaic virus (CMV)-Sj (soybean strain) suppresses naturally occurring RNA silencing against the soybean chalcone synthase gene, but the 2b of CMV-Y (ordinary strain isolated from tobacco) cannot (Senda et al., 2004). For potyviruses, the HC-Pro protein of OYDV showed more efficient RSS activity in onion as the RSS assay plant than in N. benthamiana (Kim et al., 2020). Considering these observations, we presume that LYSV and perhaps other potyviruses may have modified P1 to make it strong enough to adjust RSS activity for new host adaptation by trimming the N-terminal region of P1; as a result, LYSV may have succeeded in expanding its host range. Therefore, the generation of short P1 proteins may function as the driving force for host adaptation of potyviruses.
The finding by Shan et al. (2018) that trimming the N-terminus of P1 significantly affects viral host adaptation was just an experimental demonstration. On the other hand, our results were obtained by analyzing the P1 of field isolates of LYSV, proving the hypothesis by Shan et al. (2018). Curiously, only the LYSV-N has been observed in Hokkaido, while the LYSV-S with deletion in the P1 is widely distributed in Honshu and Okinawa. In Hokkaido, wild alliums die in winter; thus, LYSV cannot survive in wild to overwinter. In addition, the Allium crop grown in the same region as garlic is only onion in Hokkaido; LYSV rarely infects most onion varieties. On the other hand, because P1 is the S-type with RSS activity, LYSV-S in Honshu and Okinawa can quickly adapt to different plants and may be able to infect many Allium spp. According to the fact that some crops are grown even in winter and that wild alliums also thrive in the vicinity of the garlic field in Honshu and Okinawa, then LYSV-S-infected plants must be widespread in the area. We have one good observation that supports this idea. In Okinawa, garlic and rakkyo (A. chinense) are grown at the same time and in the same areas, so we investigated whether rakkyo was infected with LYSV. As shown in the phylogenetic tree in Fig. 2 and Supplementary Fig. 2, the LYSV isolate from rakkyo was found to be identical to the LYSV isolate from garlic in Okinawa, suggesting that the LYSV from garlic can also infect rakkyo and vice versa.
Acknowledgments
We thank Ms. Kuniko Konno for providing wild Allium plants. We also thank Ms. Chieko Hirata for viral detection by RT-PCR. This work was partially supported by JSPS KAKENHI Grant Number 21H02190.
Electronic Supplementary Material
Supplementary materials are available at The Plant Pathology Journal website (http://www.ppjonline.org/).
Fig. 1
Representative gels show N- and S-type of P1 from leek yellow stripe virus (LYSV) isolates indicating mixed infection in garlic plants (var. Fukuchi-White [FW]) from Tokachi (A) and Okhotsk (B) regions in Hokkaido. We detected only two mixed infections out of 40 samples tested in Okhotsk. For Tokachi, please see Table 1. P1 sequences were polymerase chain reaction-amplified using primer pairs, LY-5P/LY-2M. M, 100 bp DNA ladder (A, B).
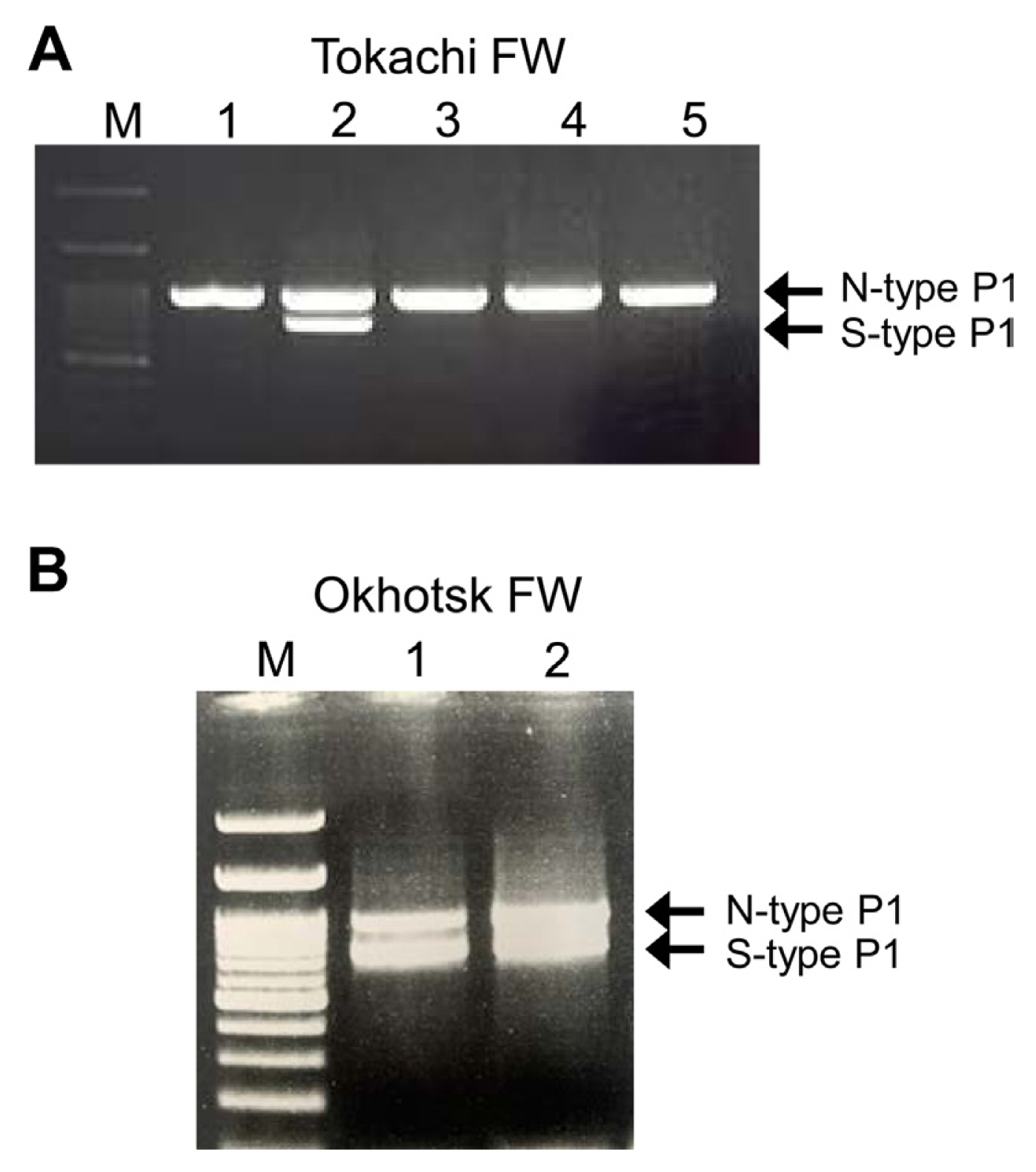
Fig. 2
Phylogenetic tree inferred from P1 region of leek yellow stripe virus (LYSV) using the Bayesian method in the MrBayes program. Node values indicate posterior probability (only those ≥ 50% are shown). Triangles denote collapsed clades, with countries of collection. The original tree is shown in Supplementary Fig. 2. Onion yellow dwarf virus (OYDV) isolates (KJ451436 and NC_005029) were used as outgroups. Labels in red indicate sequences obtained in this study. Note that the LYSV isolate from garlics in Okinawa (labeled as Okinawa) are almost identical to the LYSV isolate from rakkyo (Allium chinense) in Okinawa (labeled as Rakkyo).

Fig. 3
Protocol for aphid transmission of leek yellow stripe virus (LYSV) with S-type P1 (LYSV-S) to garlic infected with LYSV with N-type P1 (LYSV-N) and reverse transcription polymerase chain reaction (RT-PCR) detection of S- and N-type P1 transmitted by aphids. (A) Steps for aphid transmission and detection of LYSV-S to LYSV-N-infected garlic. Sap from garlic (China garlic) infected with LYSV-S was used to mechanically inoculate onion variety Fresh-Red (FR), then 14 days postinoculation (dpi), aphids were allowed to feed for 5 min on FR and transferred to another garlic (USA garlic), which was infected with LYSV-N, for 24 h. The aphids were then killed with a pesticide spray. At 20 dpi, garlic samples were processed for RT-PCR to detect the N- and S-types of P1. (B) Representative result of PCR detection. Primer pair LY-5P/LY-2M was used for the first-round RT-PCR, and primer pair LYSV-5-278/LYSV-3-278 was used for the nested-PCR. Among 12 plants, two had a mixed infection with both P1 types as confirmed by sequencing of the excised P1 bands. M, 100 bp DNA ladder.

Fig. 4
RNA silencing suppressor (RSS) activity of leek yellow stripe virus (LYSV) HC-Pro. (A) RSS activity of HC-Pro in Agrobacterium-infiltrated leaves of Nicotiana benthamiana. Two HC-Pro proteins (K-HC and O-HC) were tested: K-HC, derived from LYSV-N; O-HC from LYSV-S. HC-Pro proteins from LYSV, onion yellow dwarf virus (OYDV) or tobacco etch virus (TEV) were co-expressed with green fluorescence protein (GFP) in agroinfiltrated leaves. RSS activity of the HC-Pros was assessed by comparing the intensity of GFP at 5 days post agroinfiltration (dpa). N-terminal truncated HC-Pro from OYDV that lacks RSS activity (Kim et al., 2020) and GUS were used as negative controls. TEV HC-Pro was included as a positive control. (B, C) RSS activity of HC-Pro in onion epidermis at 3 dpa. HC-Pro proteins were co-expressed with GFP in onion epidermal tissues after agroinfiltration. GFP images of three different onion scales (scale 1 to 3) are shown. (C) Mean fold-change in GFP intensity in onion tissues when the value of GUS was set to 1.0. GFP intensity was calculated by LAS AF software (Leica). Fold-change values were log-transformed then compared using Tukey’s multiple comparison test (P < 0.05). Different letters above bars indicate a significant difference.

Fig. 5
RNA silencing suppressor (RSS) activity of leek yellow stripe virus (LYSV) P1 protein. (A) RSS activity of P1 proteins in Agrobacterium-infiltrated Nicotiana benthamiana leaves. Either N- or S-type P1 was co-expressed with green fluorescence protein (GFP) in leaves transfected by agroinfiltration. The RSS activity was assessed by comparing GFP intensity at 5 dpa. GUS was used as a negative control. (B) Effect of co-expression of P1 and HC-Pro on RSS activity at 5 dpa. HC-Pro from LYSV-N (K-HC) was co-expressed with P1 and GFP after agroinfiltration. GUS was used as a negative control. (C) RSS activity of P1 in agrobacterium-infiltrated onion epidermal tissues at 3 dpa. P1 and GFP were co-expressed with or without K-HC in onion tissues by agroinfiltration. GFP images of four different onion scales (Scale 1 to 4) are shown. (D) Mean fold-change in GFP intensity in onion tissues when the value of GUS was set to 1.0. GFP intensity was calculated by LAS AF software (Leica). Fold-change values were log-transformed then compared using Tukey’s multiple comparison test (P < 0.05). Different letters above bars indicate a significant difference.

Table 1
Survey of garlic plants in Tokachi garlic-propagation fields to detect LYSV-N and LYSV-S using PCR
| Year | No. of garlic plants infected with LYSV with N- and S-type P1 | |||
|---|---|---|---|---|
|
|
||||
| LYSV-positive (total tested) | N-type | S-type | N + S | |
| 2019 | 23 (40) | 20 | 0 | 3 |
| 2020 | 20 (60) | 20 | 0 | 0 |
Table 2
Detection of garlic viruses in onion cultivars Sapporo Yellow (SY) and Fresh Red (FR) at different days after mechanical inoculation (dpi) with sap from virus-infected garlic tissues
| Inoculum | Onion | Plant no. | Virus detectiona | |||||
|---|---|---|---|---|---|---|---|---|
|
|
||||||||
| 10 dpi | 20 dpi | 30 dpi | ||||||
|
|
|
|
||||||
| LYSV | OYDV | LYSV | OYDV | LYSV | OYDV | |||
| China garlic | SY | 1 | − | − | − | − | − | − |
| 2 | − | + | − | + | − | + | ||
| 3 | − | + | − | + | − | − | ||
| FR | 1 | − | (+) | + | − | + | − | |
| 2 | − | (+) | + | − | + | − | ||
| 3 | − | − | + | − | + | − | ||
| USA garlic | SY | 1 | − | − | − | − | − | − |
| 2 | − | − | − | − | − | − | ||
| 3 | − | − | − | − | − | − | ||
| FR | 1 | + | (+) | + | − | + | − | |
| 2 | + | (+) | + | − | + | − | ||
| 3 | + | (+) | + | − | + | − | ||
References
Bampi, D., Mituti, T., Pavan, M.A., Hammond, J. and Krause-Sakate, R. 2015. Leek yellow stripe virus isolates from Brazil form a distinct clade based on the P1 gene. J. Plant Pathol. 97:457-463.
Domingo-Calap, M.L., Chase, O., Estapé, M., Moreno, A.B. and López-Moya, J.J. 2021. The P1 protein of Watermelon mosaic virus compromises the activity as RNA silencing suppressor of the P25 protein of Cucurbit yellow stunting disorder virus
. Front. Microbiol. 12:645530.



Excoffier, L. and Lischer, H.E.L. 2010. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10:564-567.



Giner, A., Lakatos, L., García-Chapa, M., López-Moya, J.J. and Burgyán, J. 2010. Viral protein inhibits RISC activity by argonaute binding through conserved WG/GW motifs. PLoS Pathog. 6:e1000996.



Hu, S.-F., Wei, W.-L., Hong, S.-F., Fang, R.-Y., Wu, H.-Y., Lin, P.-C., Sonobar, N., Wang, H.-P., Sulistio, M., Wu, C.-T., Lo, H.-F. and Lin, S.-S. 2020. Investigation of the effects of P1 on HC-pro-mediated gene silencing suppression through genetics and omics approaches. Bot. Stud. 61:22.




Jayasinghe, W.H., Kim, H., Sasaki, J. and Masuta, C. 2021. Aphid transmissibility of onion yellow dwarf virus isolates with an N-terminal truncated HC-Pro is aided by leek yellow stripe virus. J. Gen. Plant Pathol. 87:178-183.


Kasschau, K.D. and Carrington, J.C. 1998. A counterdefensive strategy of plant viruses: suppression of posttranscriptional gene silencing. Cell 95:461-470.


Katis, N.I., Maliogka, V.I. and Dovas, C.I. 2012. Viruses of the genus Allium in the Mediterranean region. Adv. Virus Res. 84:163-208.


Katoh, K. and Standley, D.M. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30:772-780.



Kim, H., Aoki, N., Takahashi, H., Yoshida, N., Shimura, H. and Masuta, C. 2020. Reduced RNA silencing suppressor activity of onion yellow dwarf virus HC-Pro with N-terminal deletion may be complemented in mixed infection with another potyvirus in garlic. J. Gen. Plant Pathol. 86:300-309.


Lole, K.S., Bollinger, R.C., Paranjape, R.S., Gadkari, D., Kulkarni, S.S., Novak, N.G., Ingersoll, R., Sheppard, H.W. and Ray, S.C. 1999. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 73:152-160.




Martin, D.P., Varsani, A., Roumagnac, P., Botha, G., Maslamoney, S., Schwab, T., Kelz, Z., Kumar, V. and Murrell, B. 2021. RDP5: a computer program for analyzing recombination in, and removing signals of recombination from, nucleotide sequence datasets. Virus Evol 7:veaa087.




Martínez, F. and Daròs, J.-A. 2014.
Tobacco etch virus protein P1 traffics to the nucleolus and associates with the host 60S ribosomal subunits during infection. J. Virol. 88:10725-10737.




Merits, A., Guo, D., Järvekülg, L. and Saarma, M. 1999. Biochemical and genetic evidence for interactions between potato A potyvirus-encoded proteins P1 and P3 and proteins of the putative replication complex. Virology 263:15-22.


Pasin, F., Simón-Mateo, C. and García, J.A. 2014. The hypervariable amino-terminus of P1 protease modulates potyviral replication and host defense responses. PLoS Pathog. 10:e1003985.



Ronquist, F. and Huelsenbeck, J.P. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572-1574.


Salvador, B., Saénz, P., Yangüez, E., Quiot, J.B., Quiot, L., Delgadillo, M.O., García, J.A. and Simón-Mateo, C. 2008. Host-specific effect of P1 exchange between two potyviruses. Mol. Plant Pathog. 9:147-155.



Santosa, A.I. and Ertunç, F. 2020. Serological and molecular detection of Leek yellow stripe virus infecting onion (Allium cepa L.) and leek (Allium ampeloprasum L.) in Ankara, Turkey. Plant Prot. Bull. 60:49-56.

Senda, M., Masuta, C., Ohnishi, S., Goto, K., Kasai, A., Sano, T., Hong, J.-S. and MacFarlane, S. 2004. Patterning of virus-infected Glycine max seed coat is associated with suppression of endogenous silencing of chalcone synthase genes. Plant Cell 16:807-818.



Shahraeen, N., Lesemann, D.E. and Ghotbi, T. 2008. Survey for viruses infecting onion, garlic and leek crops in Iran. EPPO Bull. 38:131-135.

Shan, H., Pasin, F., Tzanetakis, I.E., Simón-Mateo, C., García, J.A. and Rodamilans, B. 2018. Truncation of a P1 leader proteinase facilitates potyvirus replication in a non-permissive host. Mol. Plant Pathol. 19:1504-1510.




Shimura, H., Kim, H., Matsuzawa, A., Akino, S. and Masuta, C. 2022. Coat protein of partitiviruses isolated from mycorrhizal fungi functions as an RNA silencing suppressor in plants and fungi. Sci. Rep 12:7855.




Takaki, F., Sano, T., Yamashita, K., Fujita, T., Ueda, K. and Kato, T. 2005. Complete nucleotide sequences of attenuated and severe isolates of Leek yellow stripe virus from garlic in northern Japan: identification of three distinct virus types in garlic and leek world-wide. Arch. Virol. 150:1135-1149.



Valli, A., Martín-Hernández, A.M., López-Moya, J.J. and García, J.A. 2006. RNA silencing suppression by a second copy of the P1 serine protease of Cucumber vein yellowing ipomovirus, a member of the family Potyviridae that lacks the cysteine protease HCPro. J. Virol. 80:10055-10063.




Vučurović, I., Nikolić, D., Radović, N., Vučurović, A., Ristić, D., Krstić, B. and Stanković, I. 2017. Incidence and distribution of Leek yellow stripe virus in Allium crops in Serbia. Pestic. Phytomed. 32:145-155.

- TOOLS
-
METRICS

- ORCID iDs
-
Chikara Masuta

https://orcid.org/0000-0003-3129-9167 - Related articles



 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Supplement1
Supplement1 Print
Print



