 |
 |
| Plant Pathol J > Volume 38(5); 2022 > Article |
|
Abstract
Planthopper infestation in rice causes direct and indirect damage through feeding and viral transmission. Host microbes and small RNAs (sRNAs) play essential roles in regulating biological processes, such as metabolism, development, immunity, and stress responses in eukaryotic organisms, including plants and insects. Recently, advanced metagenomic approaches have facilitated investigations on microbial diversity and its function in insects and plants, highlighting the significance of microbiota in sustaining host life and regulating their interactions with the environment. Recent research has also suggested significant roles for sRNA-regulated genes during rice-planthopper interactions. The response and behavior of the rice plant to planthopper feeding are determined by changes in the host transcriptome, which might be regulated by sRNAs. In addition, the roles of microbial symbionts and sRNAs in the host response to viral infection are complex and involve defense-related changes in the host transcriptomic profile. This review reviews the structure and potential functions of microbes and sRNAs in rice and the associated planthopper species. In addition, the involvement of the microbiota and sRNAs in the rice-planthopper-virus interactions during planthopper infestation and viral infection are discussed.
Rice (Oryza sativa L.) is one of the most important crops and is the primary food for nearly 50% of the world’s population (Nevita et al., 2018), providing approximately 20% of the caloric requirement (Zeigler and Barclay, 2008). By 2050, the world population will reach over nine billion; therefore, improving the sustainability and yield of major crops, including rice, is crucial (Carvalho, 2006; United Nations, 2005). New cultivation and farming techniques, including pest control strategies, are being developed and improved to enhance crop productivity. Several insect pests, including bugs, leaf folder worms, aphids, thrips, and planthoppers, infest rice fields. Planthoppers are a large group of phloem-feeder insects that infest rice; significant damage is caused mainly by three species: brown planthopper (BPH), Nilaparvata lugens; small brown planthopper (SBPH), Laodelphax striatellus; and white-backed planthopper (WBPH), Sogatella furcifera. In addition to causing direct feeding damage, planthoppers are the vectors of some of the most destructive rice viruses, including rice stripe virus (RSV), rice black-streaked dwarf virus (RBSDV), and southern rice black-streaked dwarf virus (SRBSDV) (Pathak and Khan, 1994). Therefore, understanding the interaction between rice, associated planthoppers, and viruses and the underlying mechanisms of interaction is essential for identifying potential targets of intervention for control and management of rice viral diseases.
During their life cycles, plants are strongly influenced by biotic and abiotic factors in their ecosystems. Biotic factors within the plant ecosystem are mainly the microbial communities associated with plants, known as the plant microbiome or phytobiome (Müller and Ruppel, 2014). The microbiome can be defined as the sum of all microbial communities and their genomes present in a specific ecosystem (Ursell et al., 2012). The plant microbiome is indispensable for sustaining the growth and development of host plants, but it also includes pathogenic microbes (Mendes et al., 2013). Microbial communities associated with plants reside mainly in one of three ecosystems: the rhizosphere (area affected by plant root exudates) (Walker et al., 2016), endosphere (inside plant tissues) (Hardoim et al., 2008), and phyllosphere (surfaces of leaves and stems) (Berlec, 2012). Advances in metagenomic approaches and their integration with other meta-omics tools have facilitated understanding of the phytobiome and drawn attention to its roles in the interactions with the surrounding environment, including pests (Mannaa and Seo, 2021). Moreover, insects are also greatly influenced by the associated microbiome, as the associated microbes may influence the host biology (Akami et al., 2019; Miller et al., 2019). Bacteria establish many types of symbiotic associations with the host (as either intra- or extra-cellular symbionts), ranging from mutualistic to parasitic to pathogenic (Sazama et al., 2019).
Small RNAs (sRNAs), generally noncoding RNA molecules (<200 nucleotides in length), play significant roles in regulating diverse fundamental cellular processes and activities, such as regulation of gene expression, genome stability, and defense, in multicellular organisms, including plants and insects (Bonnet et al., 2006; Navarro et al., 2006). Plant endogenous sRNAs are noncoding RNAs, 20-24 nucleotides long, that regulate a wide range of biological processes. Plant sRNAs are classified mainly into micro RNAs (miRNAs) and endogenous small interfering RNAs (siRNAs) based on their origin (Xie et al., 2004). MiRNAs originate from single-stranded RNA precursors, whereas siRNAs originate from double-stranded RNA (Axtell 2013; Carthew and Sontheimer 2009; Feng et al., 2021). Based on their origin, plant siRNAs are further classified into four major types: transacting siRNAs, heterochromatic siRNAs, natural antisense transcript-derived siRNAs, and long siRNAs. These siRNAs are processed by dicer-like proteins (DCLs) and bind to argonaute proteins (AGOs) to form RNA-induced gene silencing complexes (Baulcombe, 2004).
In the insects, sRNAs have also been found to perform important roles in regulating and expressing genes involved in biological and physiological processes. For example, the mobility of sRNAs and sRNA silencing complexes may be critical in cellular functions and reproductive development processes, such as meiosis and germ cell fate regulation (Van Ex et al., 2011; Zhang et al., 2011). Insect sRNAs are classified based on their biogenesis, characteristics, and association with AGOs into various types, including miRNAs, endogenous siRNAs, and piwi-interacting RNAs (Golden et al., 2008). This review discusses the composition and roles of the microbiome and sRNA associated with rice and planthoppers, focusing on their involvement in rice-planthopper interaction and related molecular mechanisms. This review aims to improve the understanding of rice-planthopper interactions, and provide insights into the versatile roles of associated microbiota and sRNA facilitating effective control methods against this destructive pest.
Plants are holobionts surrounded by and continuously interact with diverse microbial communities in or around them; plants provide various nutrients for endophytic and epiphytic microorganisms (Arnold et al., 2003; Gordon et al., 2013; Lindow and Andersen, 1996). The microbiome composition depends on different environmental and plant-related factors (i.e., plant type and growth stage) that shape the structure of plant-associated microbial communities, which can vary even between individual plants in the same ecosystem (Baltrus, 2017). Advances in metagenomic approaches in integration with functional multi-omics have paved the way for the taxonomic identification and functional characterization and of microbes in the plant microbiome (Mannaa and Seo, 2021).
In rice, several culture-dependent and culture-independent studies utilizing molecular methods have been developed and utilized to investigate the composition and functions of rice-associated microbes (Kim and Lee, 2020). The rice microbiota of significance to the plant localize in three different parts, bulk soil, rhizosphere, and phyllosphere area. Some of these microbes play beneficial roles in supporting healthy plant growth, i.e., the symbiome, while others are disease-causing organisms, i.e., the pathobiome. The structure of the rice microbiome, including the prominent roles of the symbiome and pathobiome, as well as responses of rice plants, are summarized in Fig. 1.
Bulk soil in rice paddy fields represents the primary source of microbes associated with rice plants and has been extensively studied because of its involvement in trapping water and air, cycling different nutrients, and accumulating organic matter and minerals (Chaudhari et al., 2013). The bacterial community in the soil is one of the most diverse ecosystems encompassing a plethora of microbes, including Proteobacteria, Chloroflexi, Actinobacteria, and Acidobacteria (Hussain et al., 2011; Jiang et al., 2016). The fungal community in rice paddy fields includes Ascomycota, Basidiomycota, and Glomeromycota (Jiang et al., 2016; Yuan et al., 2018).
The rhizosphere is the area adjacent to plant roots affected by root exudates and is considered a complex environment that includes different microbial communities, such as fungi, algae, bacteria, nematodes, oomycetes arthropods, viruses, protozoa, and archaea (Buée et al., 2009; Raaijmakers et al., 2009). Root exudates play essential roles in shaping the microbial composition of the rhizosphere by facilitating plant colonization and selective recruitment of specific microbial species that help in plant adaptation, growth, and survival. The rhizosphere of rice plants is one of the most biodiverse environments in which rice plants and microbial communities continuously interact. This microbial diversity benefits plants by promoting growth or protecting against phytopathogens (Breidenbach et al., 2016; Hussain et al., 2012; Lee et al., 2015). In their review of previous studies on rice rhizosphere microbial communities, Ding et al. (2019) suggested that the rice rhizosphere is mainly composed of Proteobacteria (specifically, Alpha-, Beta-, and Deltaproteobacteria classes), Acidobacteria, Actinobacteria, and Chloroflexi bacterial phyla, while Crenarchaeota, Thaumarchaeota, and Euryarchaeota represent the main archaeal phyla. The fungal community in the rice rhizosphere differed significantly from other plants and was mainly enriched with the phylum Chytridiomycota, known as aquatic fungi, and hence paddy soils support their growth (Ding et al., 2019).
Beneficial microbes, such as plant growth-promoting rhizobacteria (PGPR) and arbuscular mycorrhiza fungi (AMF), support plant growth and nutrition by mobilizing nutrients. The processes involved in mineralization and decomposition of different organic matter are essential for enhancing plant growth through different mechanisms, such as chelation, solubilization, and oxidation or reduction reactions (Rincon-Florez et al., 2013). Bacteria, especially nitrogen-fixing bacteria, can facilitate nitrogen uptake by converting nitrogen found in the air into ionic nitrogen (Franche et al., 2009). Moreover, PGPR can stimulate plant growth by producing growth regulators, such as auxins, cytokinins, and gibberellins (Lugtenberg and Kamilova, 2009). Many studies have focused on isolating microbial antagonists and analyzing their antagonistic effects against rice pathogens, such as Magnaporthe oryzae, Rhizoctonia solani, and Xanthomonas oryzae (De Costa et al., 2006; Harsonowati et al., 2017; Thapa et al., 2018). The rhizosphere microbiota have also been found to enhance plant resistance to aboveground herbivore infestation. In the study of Jiang et al. (2020), a rich and diverse rhizosphere microbiota following treatment with organic amendments was found to enhance rice plant tolerance and resistance to planthoppers N. lugens. An earlier study indicated that treating rice seedlings by root drenching with Bacillus velezensis YC7010, an endophytic strain, induced systemic resistance against BPH (Harun-Or-Rashid et al., 2018). Another study reported the efficiency of decoyinine, a metabolite derived from the endophytic Streptomyces hygroscopicus in triggering induced systemic resistance in rice against the SBPH, L. striatellus and indirectly affect the fecundity and population life table parameters (Shah et al., 2022).
The phyllosphere refers to the aerial parts of plants, and the associated microbiota in the phyllosphere have not been studied extensively compared to the rhizosphere microbiota (Knief et al., 2012). It is considered a harsh environment for the survival of microorganisms, such as bacteria, archaea, and fungi, because of direct contact with ultraviolet radiation, rapid shifts in temperature from high to low, and depletion of nutrients (Müller and Ruppel, 2014). Previous studies have suggested a wide variation in the microbial composition of the rice phyllosphere microbial community. Ren et al. (2014) reported that Enterobacteriaceae (class Gammaproteobacteria) is the predominant microbial family in the rice phyllosphere. A metagenomic analysis of the microbiome in the rice phyllosphere showed that Actinobacteria and both Alphaproteobacteria and Betaproteobacteria are the main groups of bacteria found in the region (Knief et al., 2012). Harsonowati et al. (2017) reported that a total of 18 actinomycetes species isolated from leaves belonged to the genera Streptomyces, Saccharothrinx, Gordonia, and Lentzea and showed antagonistic activity against rice blast fungus. As discussed previously, a large group of microbes can harbor and colonize plants. These microbial communities play a vital role in plant growth and health, as some of these microbes can cause diseases, while others can promote growth, nutrient uptake, and adaptation to both biotic and abiotic stresses (Brader et al., 2017).
In recent years, there has been considerable interest in the identification of sRNAs, such as miRNAs and siRNAs, and their functions in diverse biological processes. Most of the functional processes of sRNAs have been summarized in terms of their role in regulation of plant growth (Juarez et al., 2004), development (Zhu and Helliwell, 2011), root architecture, yield (Miura et al., 2010; Zhang et al., 2013), and response to biotic and abiotic stresses (Lu et al., 2008; Sun et al., 2020). In response to the constant infection pressure from insect and pathogen attacks, plants have evolved multiple levels of defense in the immune system conferring various protective effects, such as those mediated on sRNAs, as shown in Fig. 2 (Cook et al., 2015; Jones et al., 2016).
Plant AGO proteins are endonucleases that cut target RNAs. Many sRNAs are produced and used by the plant DCL/AGO system and are collectively termed siRNAs (Huang et al., 2019; Hudzik et al., 2020; Navarro et al., 2006; Niu et al., 2016). There is a variation in the abundance of sRNA species between different species and between individuals in the same species. The core components of sRNAs in rice immunity are DCLs, AGOs, and RNA-dependent RNA polymerase proteins. The rice genome encodes 8 DCLs, 19 AGOs, and 8 RNA-dependent RNA polymerase proteins. The DCLs include OsDCL1a, OsDCL1b, OsDCL1c, OsDCL2a, OsDCL2b, OsDCL3a, OsDCL3b/OsDCL5, and OsDCL4, which are responsible for the production of different sRNAs. For instance, OsDCL3b/OsDCL5 and OsDCL4 produce 21 or 24 nucleotide long siRNAs (Song et al., 2012; Wu et al., 2010), and OsDCL1 and OsDCL3a are involved in the biosynthesis of 21- or 24 long nucleotide miRNAs (Yan et al., 2011). Besides, OsDCL2 contributes to the accumulation of virus-derived small interfering RNAs (vsiRNAs) derived from O. sativa endornavirus (Urayama et al., 2010).
Interestingly, Wang et al. (2015) described a unique concept called “expression quantitative locus” analysis to examine how sRNAs control additional encoded genes in rice plants. Different plants of the same species often express different amounts of sRNAs, but the underlying reason behind such variation remains unclear. The analysis identified more than 53 million sRNA molecules from different rice populations, which varied in abundance between different plants within the same population. The study also showed that thousands of individual rice genes could increase or decrease the abundance of associated sRNA molecules (Table 1). Some sRNA molecules are translated to specific proteins, whereas others have different functional roles. For example, sRNA molecules affect plant growth, shape, and structure and affect survival under stressful conditions, such as drought, salt stress, insect infestation and disease (Wang et al., 2015). Several factors affect variations in sRNA abundance, such as elements located in the promoter region of sRNAs themselves that influence sRNA transcription levels, the mother gene or long noncoding RNAs, and sequence polymorphisms.
MiRNAs are a type of sRNAs that play essential roles in regulating plant defense and morphological characterization, such as leaf structure and root and flower development. Over 4,600 miRNAs have been identified from 50 plant species since the invention of high-throughput sequencing techniques, such as next-generation sequencing and the MiSeq Illumina system. Hundreds of miRNAs were characterized from different plants, including rice, barley, wheat, and Arabidopsis. O. sativa and Glycine max have the most identified miRNAs (661 and 395 miRNAs, respectively) (Guo et al., 2012; Sunkar et al., 2008). Under abiotic stress, such as drought and salt stress, 23 novel miRNAs from sRNAs of control rice seedlings and treated plants were identified (Sunkar et al., 2008). Another 24 novel miRNA libraries were discovered from rice calli by Chen et al. (2011), 43 novel sRNA families from rice spikelets by Peng et al. (2011), and 75 novel miRNAs from developing pollen of rice seedlings by Wei et al. (2011). Furthermore, a novel rice miRNA, OsmiR530, was reported by Sun et al. (2020), who demonstrated that OsmiR530 negatively regulates grain yield as its expression significantly decreases both grain size and panicle branching. These sRNAs have potential targets for improving rice development and differentiation under stressful conditions, and breeding high-yielding rice varieties.
Planthoppers are sucking insects among the destructive insect pests found in rice fields worldwide. During severe infestations, planthoppers are usually causing deadly effects, such as wilting and complete drying of rice crops (Bottrell and Schoenly, 2012). In addition, planthoppers can damage rice by transmitting viruses during phloem-sucking (Yang et al., 2018). Planthoppers also excrete honeydew on rice leaves that triggers microbial growth and causes plant leaf deformation during severe infestations. It is noted that BPH honeydew-associated microbes reduce rice defense responses and cause specific plant volatiles release, such as momilactones (Wari et al., 2019a). Insect and plant microbiota structure has been examined under different conditions, including soil type and host plants (Griffiths et al., 2011). The interaction between plants, microbes, insects, and the surrounding environment in the ecosystem is summarized in Fig. 3, showing the beneficial and harmful effects on each other. Many studies have reported that microbes living in insect bodies play a vital role in facilitating insect adaptation to host plants (Engel and Moran, 2013; Xia et al., 2017).
The characterization of microbial diversity and structure is necessary for understanding the complicated relationships between microbes and insect hosts. The bacterial community in BPH has been investigated at various growth stages using the next generation sequencing amplicon sequencing technique. It was reported that nymphal insects have the highest richness in bacterial species, while Proteobacteria was the highest abundant phylum at all of the developmental stages (Wang et al., 2020). At the genus level, the abundance of Acinetobacter in eggs was significantly lower than in other developmental stages. Additionally, microbial structure in male and female adults was different, indicating the gender-dependent microbial communities (Wang et al., 2020).
Moreover, the microbial diversity in several tissues during BPH different developmental stages was observed (Wang et al., 2020; Zhang et al., 2019). Although Firmicutes and Proteobacteria were dominant in all samples, significant variation in the relative abundance of Firmicutes and Proteobacteria was found between BPH ovaries and fat bodies (Zhang et al., 2019). Insect virulence is also found to be correlated with microbial composition. An in vivo study showed that different bacterial and fungal symbionts differed in BPH populations with different virulence levels (Xu et al., 2014a). Comparative transcriptomic analyses by Yu et al. (2014) between BPH populations with different virulence levels showed that many genes belonging to BPH symbionts, such as yeast-like symbionts and Wolbachia spp., were expressed. There is a big gap in understanding the roles of symbionts in the adaptation process of BPH to rice plants, and the complete structure of symbionts and their roles in the adaptation process remains unclear (Wang et al., 2021).
The bacterial communities in L. striatellus and S. furcifera were dominated by the genera Wolbachia, Cardinium, Rickettsia, and Pantoea when studied using 16S rRNA amplicon sequencing (Bing et al., 2020). The Wolbachia and Cardinium abundance was negatively correlated within the community of both planthoppers. Moreover, the dynamic of the symbiont Asaia spp. in WBPH-laboratory and field-collected populations was investigated using 16S RNA phylogenetic analysis and transmission electron microscopy (Li et al., 2019). Asaia spp. were found in all of laboratory WBPH adults and tissues, while field-collected WBPH showed different infection rates (around 45%). Similarly, BPH microbial profile was studied in two geographically different populations, the indoor population (NlIP) and field population (NlFP), using 16S rRNA amplicon sequencing (Zhang et al., 2018b). It was found that NlIP and NlFP had different microbial community compositions. The phyla, Proteobacteria, Actinobacteria, and Firmicutes, were the most dominant within the whole community (>75%). NlIP group is also shown to have a higher relative abundance of Pantoea and Stenotrophomonas spp., NlFP harbored more of Wolbachia, Actinobacteria, and Herbaspirillum spp. Moreover, the BPH gut microbiota was also evaluated after growing in two different conditions; laboratory-insecticide-susceptible (IS) and field-insecticide-resistant (IR) (Vijayakumar et al., 2018). In IS population, Proteobacteria was the dominant phylum (99.86%), whereas Firmicutes was the dominant phyla in the IR population (46.06%). The genera, Weissella, Morganella, and Enterococcus, were shared within the bacterial community of the two groups, indicating that they may represent the core microbiota of BPH insects. The microbial composition of the SBPH alimentary canal was collected from different regions in China and characterized by 16S rRNA sequencing (Liu et al., 2020). The microbial community with rich diversity was mainly composed of the phyla; Proteobacteria, Actinobacteria, Firmicutes, Bacteroidetes, and Tenericutes. The bacterial abundance and structure were highly similar as the geographic distance between the populations classified into three categories: temperate, subtropical, and tropical, based on the geographic location (Liu et al., 2020).
Inoculation of reproductive symbionts affects the microbial community in planthoppers. For instance, when the microbiota of BPH was characterized with and without Cardinium infection, it was observed that microbial diversity was lower in BPH different stages, mainly in female midguts, and male testes than that in the controls (Li et al., 2020). From one side, a negative correlation is noted to be between Cardinium and most of its related genera, and between Bacteroidetes and Proteobacteria from another side. It was clear that the microbial composition changed after Cardinium inoculation, but Acinetobacter spp. was a core genus within the microbial community. The BPHs adult was treated with some antimicrobials and water to investigate the variation in the structure of gut microbes (Song et al., 2021). Antimicrobial treatment lead to increasing the gut microbial diversity by inhibiting the growth of Acinetobacter which considered as the dominant genus in adult gut. Also, antimicrobials decreased the abundance of microbial whole community, however, Serratia spp. absolute abundance was significantly increased under the antimicrobial exposure. Furthermore, the tetracycline effects on the diversity and structure of bacterial community in SBPH various tissues were studied using 16S rRNA amplicons sequencing (Zhang et al., 2020). The results showed that the phyla; Proteobacteria, Bacteroidetes, Firmicutes and Actinobacteria were the highest abundant in SBPH population. Regarding to genera, Asaia and Wolbachia had the dominance at all body parts. It was noted also that tetracycline cause inhibition for Firmicutes, Tenericutes, Bacteroidetes and Fusobacteria, and nearly prohibited the genera; Bacteroides, Wolbachia, and Abiotrophia spp. from SBPH bacterial community.
Regarding the influence of bacterial symbionts on the physiological features of the planthoppers, in WBPH, it was noted that Asaia-infected planthoppers had a shorter nymphal duration and increased adult weight compared to Asaia-uninfected insects (Li et al., 2019), even though Asaia-uninfected WBPH fed more, suggesting that Asaia spp. plays a role in improving planthopper fitness through involvement in host nutrition. Higher diversity in ecological factors in various habitats may cause an increase in the microbial diversity of SBPH, which help them in the adaptation or tolerance to different extreme conditions (Liu et al., 2020). Cardinium infection was found to enhance the relative density of midgut-associated Acinetobacter spp., with both bacteria exhibiting tissue-specific tropism in BPH (Li et al., 2020). In addition, Cardinium infection caused changes in the core microbial composition. Interestingly, the susceptibility of BPH to some pesticides such as imidacloprid, chlorpyrifos, and clothianidin was significantly increased after being pretreated with antibiotics. Simultaneously, significant inhibition was found for the detoxifying enzyme activities of some enzymes, including cytochrome P450 and glutathione S-transferase (Tang et al., 2021). These results indicate the hidden role of gut microbes in planthopper resistance to pesticides through enzymatic detoxification.
A body of research has focused on identifying and characterizing sRNAs in planthoppers. For instance, sRNA libraries of BPH have been constructed and sequenced (Chen et al., 2012). Through this, a list of conserved miRNAs in three developmental stages, including male adult (452), female adult (430), and last instar female nymph (381), were identified, of which 71 were novel miRNAs. The phylogenic evolution of miRNA families in BPH is consistent with that of other species, and the sRNA library of the SBPH population has been constructed and sequenced (Zhou et al., 2014). The identified miRNAs were 501 in total, including conserved and novel miRNAs belonging to various families. The target genes of conserved and novel miRNAs were analyzed from the perspective of its functions and have shown to be similar. MiRNAs, conserved and novel, have also been reported in the rice pest WBPH (Chang et al., 2018). Deep sequencing identified 171 novel miRNAs biased towards female insects as they modulated the functionality and targets of miRNAs involved in sex differentiation.
Several physiological characters were found to be regulated by sRNAs in planthoppers such as wing polyphenism, molting and virus transmission. In adaptation to different conditions, several insects show two wing phenotypes. The sRNAs in BPH nymph instars were isolated and characterized to identify the miRNAs related to wing polyphenism (Xu et al., 2020). The sRNA libraries from long and short wing strains were then sequenced and analyzed to assign the conserved and novel miRNAs (158 and 96, respectively). It was also found that 122 miRNAs have different expressions between BPH strains. To understand the molecular mechanism underlying wing polyphenism in BPH, these miRNAs target genes were predicted to observe the differentially expressed miRNA-targeted genes which were found in the insulin signaling and insect hormone biosynthesis pathways (30 and 45, respectively, involved in wing dimorphism were identified) (Xu et al., 2020). Among these miRNAs, Nlu-miR-9a-5p, Nlu-miR-14-3p, and Nlu-miR-315-5p, were shown to interact with insulin receptors (NlInRs). The conserved miRNA, miR-34, was also found to play a role in controlling wing polyphenism through modulating a positive regulatory feedback loop of juvenile hormone and signaling pathway of insulin/IGF (Ye et al., 2019). Interestingly, Nlu-miR-34 is abundant in BPH short-wing strain and targets two binding sites in the 3′ untranslated region of NlInR1 to suppress NlInR1. The injection of agomir-34 to overexpress miR-34 in long-wing strain triggers the short-wing development while injecting miR-34 agomir to knockdown miR-34 in short-wing strain leads to more long-wing strains.
Molting is a vital physiological process in arthropod development. Recently, molting and chitin biosynthesis have been suggested to be regulated by miRNAs, which has drawn the attention of many researchers in determining the specific miRNAs involved in molting. In a study by Chen et al. (2013), conserved miR-8-5p, miR-2a-3p and their target genes in BPH were essential for ecdysone-induced chitin biosynthesis. The bioinformatic and experimental analyses were employed to investigate that phosphoacetyl glucosamine mutase (PAGM) and membrane-bound trehalase (Tre-2) in the pathway of chitin biosynthesis were targets of miR-2a-3p and miR-8-5p, respectively (Chen et al., 2013). As a response to the treatment of the insect hormone, 20-hydroxyecdyson (20E), which is reported to be contributing in molting, miR-2a-3p and miR-8-5p levels were decreased, while the PAGM and Tre-2 levels were upregulated. Additionally, the transcription of miR-2a-3p and miR-8-5p was repressed by the Broad-Complex (BR-C) gene, in the signaling pathway of 20E hormone. The overexpression of miR-2a-3p and miR-8-5p also significantly reduced the BPH survival rate due to a reduction in a molting obstacle phenotype caused by miR-2a-3p, which simulates feeding, and simultaneously reduced the BPH chitin content (Chen et al., 2013). This study indicated that miR-2a-3p and miR-8-5p have a role as molecular regulators that control the chitin biosynthesis pathway through the signaling of 20E hormone. In another study, four samples obtained from the BPH nymph instars, 2nd-3rd and 4th-5th, were sequenced and analyzed, yielding 61 conserved miRNAs and 326 novel miRNAs (Chen et al., 2018). The expression profile analyses reported that 36 miRNAs targets showed significantly different expression levels and most of these miRNAs targets were involved in energy and hormone pathways. Among the putative novel miRNAs, nlu-miR-173, interacted with 20E signaling through the transcription factor BPH Ftz-F1. Moreover, it was noted that the nlu-miR-173 transcription was triggered by BR-C gene, indicating that it participates in molting function through its contribution to the 20E pathway. Targeting chitin synthase gene A (CHSA), represent a promising application of sRNAs in controlling N. lugens. The study of Li et al. (2017), have shown that two RNA silencing pathways, siRNAs and miRNAs, targeting CHSA in N. lugens via feeding affected the development of nymphs, reduced their chitin content, led to lethal phenotypes and adversely affected the egg hatchability in treated females.
The transmission of plant viruses mostly depends on vector insects, and vsiRNAs can target both viral and host transcripts due to viral infection. Rice viruses, such as RSV, RBSDV, and SRBSDV, are persistent-propagative viruses transmitted by different planthopper species and can cause severe diseases in rice. Therefore, controlling planthopper insects is a practical approach for controlling rice viruses. For instance, long noncoding RNAs (lncRNAs) have been shown to inhibit viral replication in host cells. The lncRNAs regulatory mechanisms are studied in the insect immune system to respond to RSV infection by predicting genes within SBPH transcriptomes (Chen et al., 2019). The transcriptomic analysis showed that the lncRNA genes were differently expressed in the ovary higher than salivary glands and viruliferous SBPH gut. The real-time quantitative PCR results also confirmed the differential expression of randomly selected lncRNAs from viruliferous and non-viruliferous SBPH. To study the regulatory role of vsiRNAs to gene expression in insect vectors, the transcriptomic profiles of sRNA and mRNA in RSV-infected SBPH were deeply analyzed (Yang et al., 2018). From the identified sRNAs, about 80,698 novel vsiRNAs were found only in SBPH. The downregulated genes were then identified in RSV-infected insects; among them, approximately 126 genes were reported as a vsiRNAs-potential targets. The analysis of gene function prediction indicated that these downregulated targets were enriched in the cuticle structure, oxidoreductase activity and serine-type endopeptidase activity. Similarly, 4,117 target genes that could be categorized into 45 functional groups were annotated and predicted for the 382 previously identified WBPH miRNAs (Chang et al., 2016). Eight upregulated and four downregulated miRNAs were predicted in the cells injected by SRBSDV compared to the non-viruliferous cells and it is suggested that miR-n98a and miR-14 may participate in the immune system response to viral infection. Moreover, the siRNA pathway is triggered by SRBSDV infection in cultured cells generated from SBPH and the intact insect midgut (Lan et al., 2016). When RNAi knocks down the Dicer-2, which is the core component of the siRNA pathway, the propagation ability of SRBSDV is highly increased in cultured cells and SBPH midgut. This mechanism also results in maintaining the viral titers in the midgut below a putative cytopathogenic threshold above which it might results in killing the insect instead of maintaining the persistent replication ensuring continuous viral infection.
There is an established link between the microbiota of planthoppers with miRNAs and the expression of their target genes. The influence of microbial symbionts on insect host physiology could be regulated through their effect of the sRNA. However, further investigations are needed to confirm such hypothesis. Previously, variation was observed in the expression of miRNAs in Wolbachia-infected and uninfected SBPH strains. Compared with the pure strain, Wolbachia infection upregulated 18 miRNAs and downregulated six miRNAs in SBPH males, while in females, 25 miRNAs were upregulated and 15 were downregulated (Liu et al., 2019).
While bacterial endosymbionts affect miRNA profiles in planthoppers, entomopathogenic microbes were also found to cause specific changes in miRNA profiles. In their investigation of the impact of the entomopathogenic fungus Metarhizium anisopliae against BPH (N. lugens), Xie et al. (2021) found that the high biocontrol potential of M. anisopliae was linked to specific changes in miRNA profiles, and target gene functions were predicted to include genes involved in energy metabolism, ubiquitin-mediated proteolysis, and mitogen-activated protein kinase signaling pathway. A previous study investigated the transcriptome of M. anisopliae-challenged insects and indicated that fungal infection caused specific changes in the expression of membrane-related genes and genes related to cellular immune response and humoral immune factors (Zhang et al., 2015). In addition to further investigations on post-transcriptional levels in rice planthoppers in response to associated microbes, these studies can provide insights useful in developing novel effective strategies by identifying new targets for RNAi.
There are many ways through which insect-associated microbes can influence host plant infestation by insects. Along with their effect on general fitness and adaptability of insects and facilitating the utilization of host plants by synthesizing essential nutrients or providing plant polymer-degrading enzymes, the associated microbiota also act as tools in plant-insect crosstalk through suppression of plant defense mechanisms or detoxification of plant-responsive phytochemicals and toxins (Shikano et al., 2017). The endosymbiont Wolbachia, which has been reported as the main microbe associated with rice planthopper, is known to play important roles in reproductive manipulation or inhibition of replication of several vector-borne pathogens in insects. It was also found to suppress defense-related genes in maize roots during western corn rootworm infestation (Barr et al., 2010). Although planthopper symbiotic microbes are generally used as weapons against rice plants, the presence of “double-agents” has also been reported, i.e., certain planthopper symbiotic microbiota that are secreted in honeydew on rice plants act as alert signals against their own host insect to stimulate rice to elicit defense against infestation (Wari et al., 2019b). In the same study, microbes in honeydew, that are derived from the insect gut, including members of the genera Staphylococcus, Acinetobacter, Pantoea, Microbacterium, Corynebacterium, and Serratia, were found to induce direct chemical defense by eliciting phytoalexins that serve as a direct defense against BPHs in rice, along with indirect defense by increasing release of volatile organic compounds (VOCs) to recruit natural enemies of herbivores (Aljbory and Chen, 2018; Wari et al., 2019b).
Plant microbiota can also influence insect behavior through plant defense and physiology. Plant pathogenic microbes can exert a significant influence on insect infestation through their effect on the host plant and consequently on the feeding behavior and distribution of insects (Shikano et al., 2017). This can be seen in the reportedly higher preference of the BPH (N. lugens) for rice plants that were infected with the bacterial blight pathogen Xanthomonas oryzae pv. oryzae. In contrast, Cyrtorhinus lividipennis, the major predator of BPH, showed a higher preference for healthy rice plants or infected plants that have been fed on by BPH. This can be explained by changes in VOCs, which were shown to be slightly increased with bacterial infection of rice plants and by herbivore-induced volatiles in plants fed on by BPH (Sun et al., 2016). This complex co-adaptation represents an image of evolution seen in the adaptation of BHP in targeting plants weakened by bacterial infection and the adaptation of BPH predators in targeting their prey by identifying plant volatiles signals emitted after the feeding activity of BPH. Moreover, plant-associated beneficial microbes have also been found to alter plant VOCs by utilizing such compounds as carbon sources or by emission of their own VOCs, which can influence insect feeding, mating, and oviposition behaviors (Aljbory and Chen, 2018; Davis et al., 2013). However, there is still a need to further investigate such microbiota-dependent VOC-mediated interactions with regard to rice-planthopper infestation, which can provide potential alternative control methods based on a detailed understanding of this network. An endophytic strain of Bacillus valensis was used in the root drenching treatment of rice seedlings, resulting in systemic acquired resistance against BPH. The transcriptome analysis in that study indicated that the acquired resistance was mediated by salicylic acid- and jasmonic acid-dependent pathways, and the main mechanisms were plant cell wall strengthening by increase in lignin and cellulose contents as well as chemical changes in secondary metabolites (Harun-Or-Rashid et al., 2018).
The effect of host plants on the planthopper microbial community has also been investigated. The gut microbes in two BPH groups feeding on susceptible and resistant rice plants were studied using 16S RNA amplicon sequencing (Wang et al., 2021). The results indicated that Proteobacteria was the most abundant phylum, while Acinetobacter were the most abundant genus. The bacterial richness in BPH feeding on resistant rice was significantly higher than in insects feeding on the susceptible plants. Although there were no significant differences in the dominant bacteria at all taxonomic levels in the two BPH groups, the relative abundances of the phyla, Firmicutes and Bacteroidetes, and the classes, Bacteroidia and Clostridia were notably different. Interestingly, Yan et al. (2021) illustrated species-specific changes in the three rice planthopper species, BPH, WBPH, and SBPH, with respect to different plants (rice and wheat). Although the three planthoppers were found to harbor distant gut microbes, there was no significant variation in the relative abundance or the bacterial diversity in planthoppers feeding on different plant hosts.
The roles of sRNAs in the regulation of growth, development, and reproduction of planthoppers may directly contribute to the infestation process in several ways (Sattar and Thompson, 2016). Moreover, there is a possibility that endosymbionts-derived sRNAs may be linked to regulation of processes not only in the microbe, but also in the host insect. The endosymbiont Wolbachia was found to utilize sRNAs for gene regulation and cross-kingdom gene regulation in different host insects, confirming that the functions of such sRNAs may be conserved across species and strains (Mayoral et al., 2014). SRNAs are also the main players during planthopper infestation of rice plants. In a study investigating miRNA and mRNA expression profiles of BPH resistant and wild-type rice varieties, 217 miRNAs, and 7,874 mRNAs were found to be differentially expressed following invasion in the resistant plants (Tan et al., 2020). Specific miRNAs were identified to be involved in rice resistance to BHP, and transcriptome profiling indicated enrichment in metabolic and defense-related physiological pathways, including cell wall organization, hormone transport, and synthesis of specific metabolites. Another study provided more insights into the molecular mechanism of OsmiR396 as a responsive miRNA induced by BPH infestation (Dai et al., 2019). The study highlighted the roles of flavonoids (which were suggested to be related to activation of the salicylic acid signaling pathway of resistance) in imparting rice resistance against BHP. Flavonoids were found to be tightly negatively regulated by OsmiR396. Understanding the role of OsmiR396 in the regulation of flavonoids and consequently in the resistance of rice to BHP paves the way for exploiting it as potential target for generation of OsmiR396-sequestered transgenic plants with improved BPH resistance.
Plant and insect microbiota and sRNAs are also important elements during the interaction involving planthopper transmitted viruses. In general, upon viral infection, vsiRNAs target both host and viral transcripts as most plant viruses depend on vector insects for transmission (Yang et al., 2018). The underlying mechanism of the pathogenicity of viral infection related to the plant-pathogen interactions is well studied (Xu et al., 2014b). The critical factor in the transmission process is that certain viruses are transmitted in a persistent, circulative-propagative manner by rice planthoppers (Wei and Li, 2016). The mechanism of virus transmission and the interactions between plant and insect microbiome and sRNA are illustrated in Fig. 4.
Considerable research has been devoted to investigating the structure of microbial communities and sRNAs and their versatile roles in planthoppers and rice plants. RNA molecules and their functional pathways play a vital role in plant-pathogen-herbivore interactions through plant responses against herbivore and insect attacks. The primary role of sRNAs is to induce pattern-triggered immune responses against biotic stressors, functioning as critical fine-tuning regulators of plant hormone pathways. To achieve sustainable food production, they can be used as a novel defense strategy to inhibit pathogenesis and control virulence without chemical pesticides. However, further research is needed to identify nucleotide sequence specificity that regulates the abundance of sRNA molecules in plants and other eukaryotic organisms. It is also important to further investigate the roles of sRNAs as a tool to manifest the influence of planthoppers-associated microbes and the potential intervention at that level for innovative pest management. In addition, recent research has used advanced analytical tools to focus on the interaction between host microbiota, sRNAs, and viral infection in planthoppers and rice. To date, it is unclear whether the composition and abundance of symbionts inside BPH are affected by viral infections, and further studies are needed to elucidate this interaction. Future studies should also investigate the functions of sRNAs and microbiota in the context of rice-planthopper-virus interactions. Understanding the microbiota and sRNAs that regulate planthopper-rice interactions may facilitate the development of novel, effective, and environment-friendly strategies for minimizing the damage of planthopper to rice crops.
Acknowledgments
This work was carried out with the support of “Cooperative Research Program for Agriculture Science and Technology Development (Project No. PJ01574901)” from Rural Development Administration, Republic of Korea.
Fig. 1
Structure of rice microbiome, including beneficial plant-associated microbes, the symbiome, and pathobiome. Pathogens produce virulence factors through secretory systems and adhesions to plants to suppress plant defense. Endophytes can produce toxins and antibiotics, regulate their behavior with quorum sensing molecules, and produce several phytohormones, biofilms, polysaccharides, elicitors of plant defense and metabolic products (Kim and Lee, 2020; Mannaa and Seo, 2021; Sánchez-Cañizares et al., 2017). AHLs, acylhomoserine lactones; HR, hypersensitive response; LPS, lipopolysaccharide; ROS, reactive oxygen species.

Fig. 2
Small RNAs regulate plant defense against biotic stress. Plants regulate the expression of endogenous genes through small RNAs (sRNAs)-trigged mRNA cleavage in response to different biotic stressors. The essential functions are modulating the disease resistance against pathogens and enhancing plant immune responses. Upon pathogen infection, small plant RNAs function in host cells, move into invasive enemies, and accumulate the sRNAs regulators (i.e., miR393). Small plant RNAs induce auxin receptors by modifying plant hormone networks (i.e., indole acetic acid, jasmonic acid, salicylic acid, and abscisic acid) to increase plant immune responses against different biotic stressors. AGO, Argonaute proteins; miRNA, micro RNA; sRNAs, small RNAs; vsiRNAs, virus-derived small interfering RNAs.
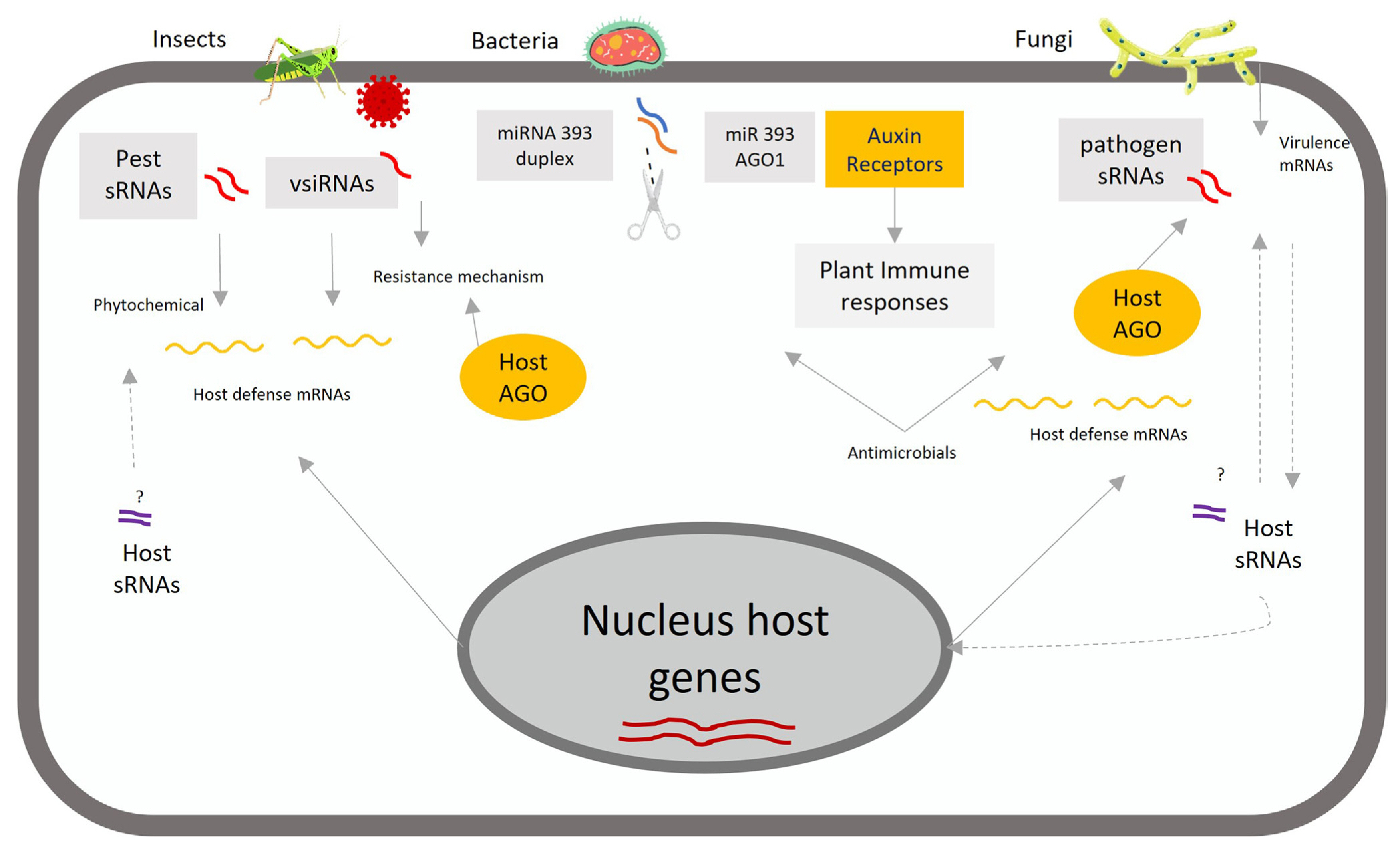
Fig. 3
The interaction among plants, microorganisms, insects, and the environment exhibits beneficial or harmful effects. PGPR, plant growth-promoting rhizobacteria.
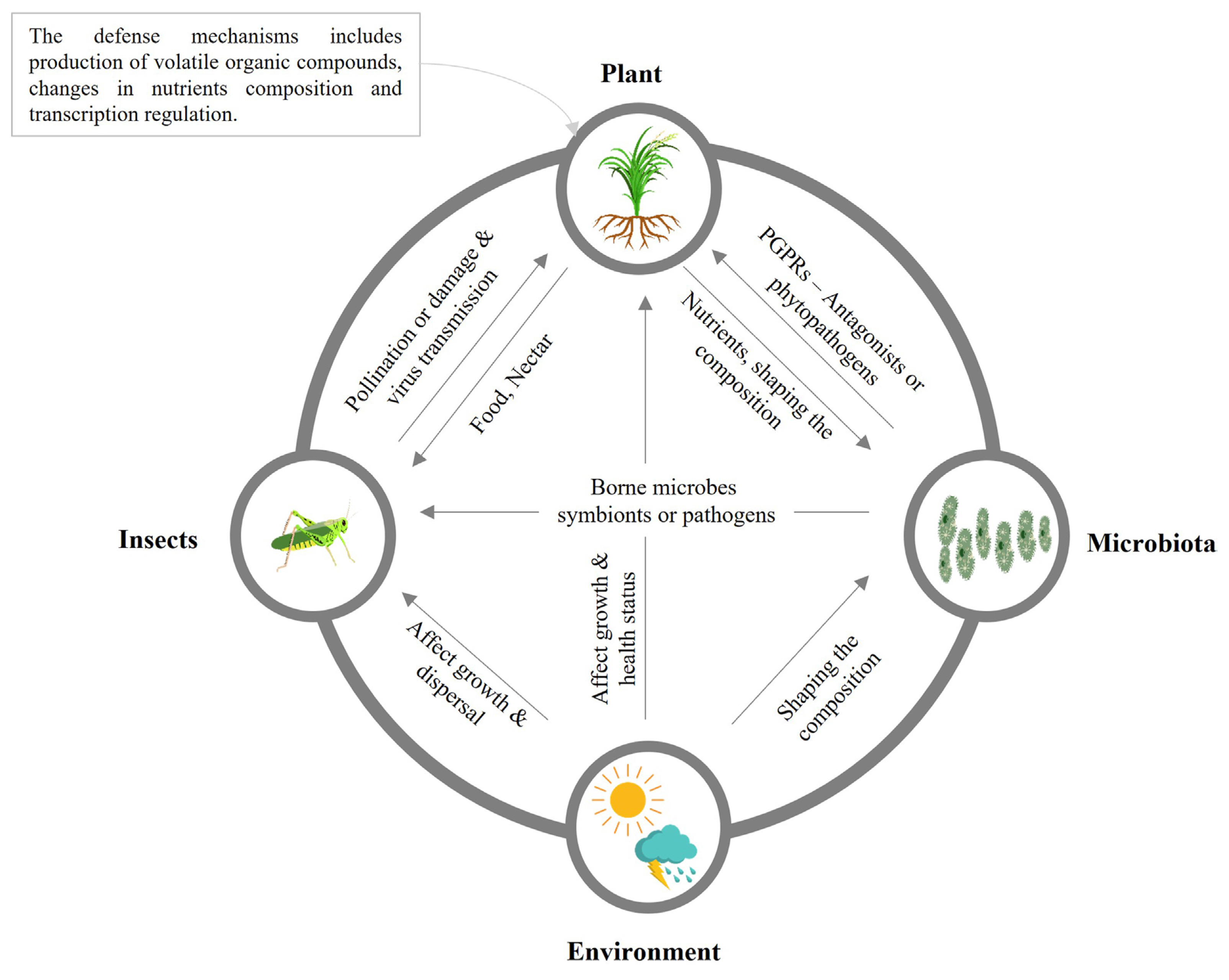
Fig. 4
Proposed mechanism of infection pathways of rice viruses in insect vectors and links with small RNA (sRNA) and plant microbes. Following virus ingestion by insect vectors from diseased rice plants, the viruses infect the epithelial tissues of the insect intestine and then the midgut, from where it can spread into the hemolymph, and finally to the salivary glands or the ovary of the female vector. miRNA, micro RNA; siRNA, small interfering RNA.
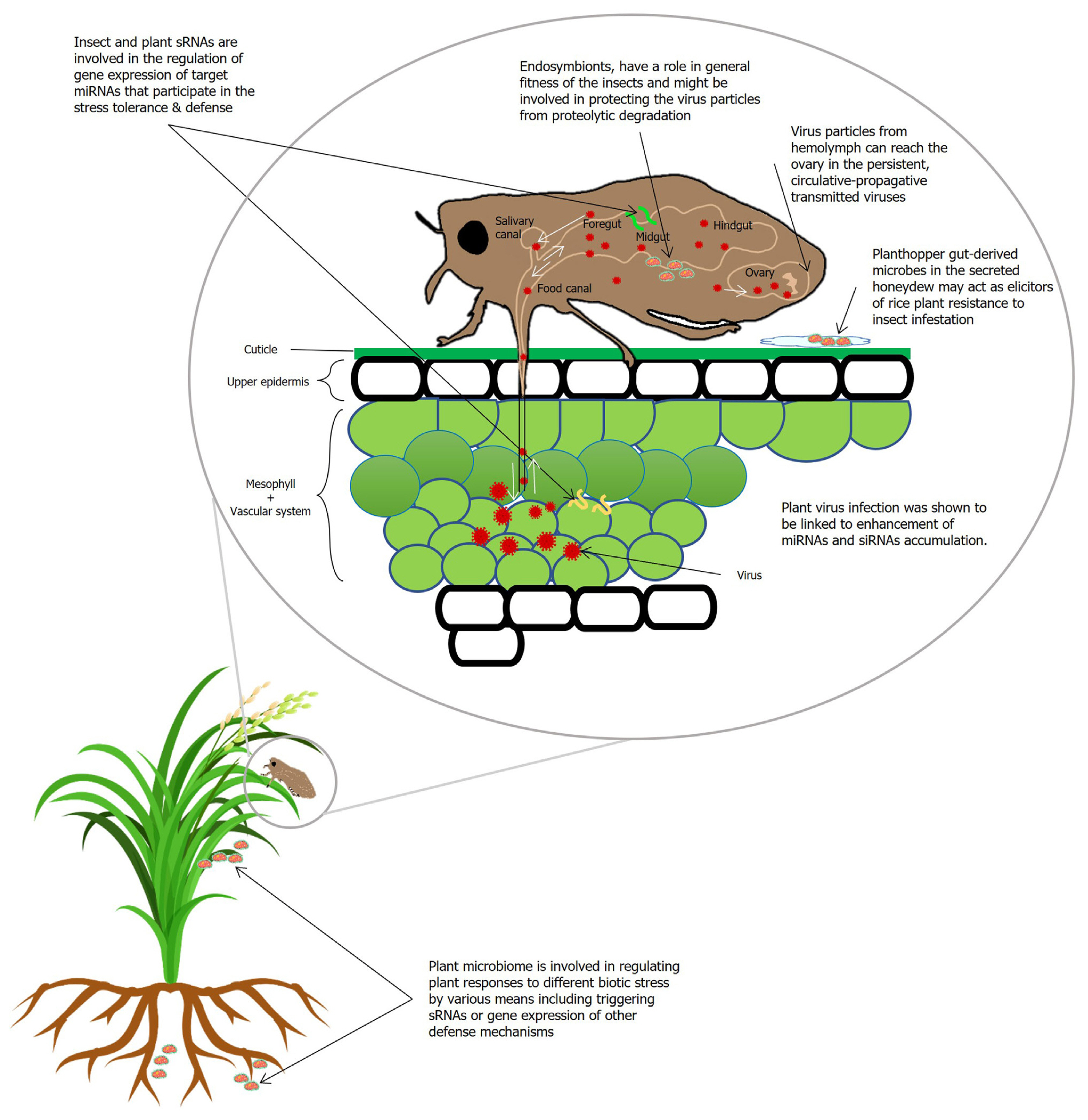
Table 1
Summary of previous studies demonstrating target genes and sRNAs of rice expressed during different biotic stressors
| sRNAs | Target genes | Fungal pathogen/insects | Reference |
|---|---|---|---|
| miR166k-5p | OsEIN2.1; OsEIN2.2 | Fusarium fujikuroi | Kettles et al. (2013) |
| miR396 | OsGRFs | Brown planthopper | Salvador-Guirao et al. (2018) |
| miR167 | OsARFs | Magnaporthe oryzae | Dai et al. (2019) |
| miR319 | OsTCP21; OsGAmyb | Rice ragged stunt virus Magnaporthe oryzae | Zhang et al. (2018a), Zhao et al. (2020) |
| miR169 | OsNF-YAs | Xanthomonas oryzae pv. oryzae | Zhang et al. (2016) |
| miR164a | OsNAC11; OsNAC60 | Magnaporthe oryzae Phytophthora infestans | Yu et al. (2018) |
| miR171b | OsSCL6-IIs | Rice stripe virus | Wang et al. (2018) |
| siR109944 | OsFBL55 | Rhizoctonia solani | Tong et al. (2017) |
| miR168 | OsAGO1 | Rice stripe virus | Qiao et al. (2020) |
| miR-14 | CsSpo; CsEcR | Rice stem borer | Wu et al. (2015) |
References
Akami, M, Ren, X-M, Qi, X, Mansour, A, Gao, B, Cao, S and Niu, C-Y 2019. Symbiotic bacteria motivate the foraging decision and promote fecundity and survival of Bactrocera dorsalis (Diptera: Tephritidae). BMC Microbiol 19:229.




Aljbory, Z and Chen, M-S 2018. Indirect plant defense against insect herbivores: a review. Insect Sci 25:2-23.



Arnold, AE, Mejía, LC, Kyllo, D, Rojas, EI, Maynard, Z, Robbins, N and Herre, EA 2003. Fungal endophytes limit pathogen damage in a tropical tree. Proc Natl. Acad. Sci. U. S. A 100:15649-15654.



Axtell, MJ 2013. Classification and comparison of small RNAs from plants. Annu. Rev. Plant Biol 64:137-159.


Baltrus, DA 2017. Adaptation, specialization, and coevolution within phytobiomes. Curr. Opin. Plant Biol 38:109-116.


Barr, KL, Hearne, LB, Briesacher, S, Clark, TL and Davis, GE 2010. Microbial symbionts in insects influence down-regulation of defense genes in maize. PLoS ONE 5:e11339.



Berlec, A 2012. Novel techniques and findings in the study of plant microbiota: search for plant probiotics. Plant Sci 193-194:96-102.

Bing, X-L, Zhao, D-S, Peng, C-W, Huang, H-J and Hong, X-Y 2020. Similarities and spatial variations of bacterial and fungal communities in field rice planthopper (Hemiptera: Delphacidae) populations. Insect Sci 27:947-963.



Bottrell, DG and Schoenly, KG 2012. Resurrecting the ghost of green revolutions past: the brown planthopper as a recurring threat to highly-yielding rice production in tropical Asia. J. Asia-Pac. Entomol 15:122-140.
Brader, G, Compant, S, Vescio, K, Mitter, B, Trognitz, F, Ma, L-J and Sessitsch, A 2017. Ecology and genomic insights into plant-pathogenic and plant-nonpathogenic endophytes. Annu. Rev. Phytopathol 55:61-83.


Breidenbach, B, Pump, J and Dumont, MG 2016. Microbial community structure in the rhizosphere of rice plants. Front. Microbiol 6:1537.



Buée, M, de Boer, W, Martin, F, van Overbeek, L and Jurkevitch, E 2009. The rhizosphere zoo: an overview of plant-associated communities of microorganisms, including phages, bacteria, archaea, and fungi, and of some of their structuring factors. Plant Soil 321:189-212.


Carvalho, FP 2006. Agriculture, pesticides, food security and food safety. Environ. Sci. Policy 9:685-692.

Chang, Z-X, Akinyemi, IA, Guo, D-Y and Wu, Q 2018. Characterization and comparative analysis of microRNAs in the rice pest Sogatella furcifera
. PLoS ONE 13:e0204517.



Chang, Z-X, Tang, N, Wang, L, Zhang, L-Q, Akinyemi, IA and Wu, Q-F 2016. Identification and characterization of microRNAs in the white-backed planthopper, Sogatella furcifera
. Insect Sci 23:452-468.


Chaudhari, PR, Ahire, DV, Ahire, VD, Chkravarty, M and Maity, S 2013. Soil bulk density as related to soil texture, organic matter content and available total nutrients of coimbatore soil. Int. J. Sci. Res. Publ 3:1-8.
Chen, C-J, Liu, Q, Zhang, Y-C, Qu, L-H, Chen, Y-Q and Gautheret, D 2011. Genome-wide discovery and analysis of microRNAs and other small RNAs from rice embryogenic callus. RNA Biol 8:538-547.


Chen, J, Li, TC, Pang, R, Yue, XZ, Hu, J and Zhang, WQ 2018. Genome-wide screening and functional analysis reveal that the specific microRNA nlu-miR-173 regulates molting by targeting Ftz-F1 in Nilaparvata lugens
. Front. Physiol 9:1854.



Chen, J, Liang, Z, Liang, Y, Pang, R and Zhang, W 2013. Conserved microRNAs miR-8-5p and miR-2a-3p modulate chitin biosynthesis in response to 20-hydroxyecdysone signaling in the brown planthopper, Nilaparvata lugens
. Insect Biochem. Mol. Biol 43:839-848.


Chen, M-Y, Ye, W-Y, Xiao, H-M, Li, M-Z, Cao, Z-H, Ye, X-H, Zhao, X-X, He, K and Li, F 2019. LncRNAs are potentially involved in the immune interaction between small brown planthopper and rice stripe virus. J. Integr. Agric 18:2814-2822.

Chen, Q, Lu, L, Hua, H, Zhou, F, Lu, L and Lin, Y 2012. Characterization and comparative analysis of small RNAs in three small RNA libraries of the brown planthopper (Nilaparvata lugens). PLoS ONE 7:e32860.



Cook, DE, Mesarich, CH and Thomma, BPHJ 2015. Understanding plant immunity as a surveillance system to detect invasion. Annu. Rev. Phytopathol 53:541-563.


Dai, Z, Tan, J, Zhou, C, Yang, X, Yang, F, Zhang, S, Sun, S, Miao, X and Shi, Z 2019. The OsmiR396-OsGRF8-OsF3H-flavonoid pathway mediates resistance to the brown planthopper in rice (Oryza sativa). Plant Biotechnol. J 17:1657-1669.




Davis, TS, Crippen, TL, Hofstetter, RW and Tomberlin, JK 2013. Microbial volatile emissions as insect semiochemicals. J. Chem. Ecol 39:840-859.



De Costa, DM, Pinto, MTC, Geethanjalee, HDN and Dissanayake, N 2006. Suppression of rice pathogens by phyllosphere associated microflora of different rice varieties in Sri Lanka. Trop. Sci 46:97-104.

Ding, L-J, Cui, H-L, Nie, S-A, Long, X-E, Duan, G-L and Zhu, Y-G 2019. Microbiomes inhabiting rice roots and rhizosphere. FEMS Microbiol. Ecol 95:fiz040.


Engel, P and Moran, NA 2013. The gut microbiota of insects: diversity in structure and function. FEMS Microbiol. Rev 37:699-735.


Feng, Q, Li, Y, Zhao, Z-X and Wang, W-M 2021. Contribution of small RNA pathway to interactions of rice with pathogens and insect pests. Rice 14:15.




Franche, C, Lindström, K and Elmerich, C 2009. Nitrogen-fixing bacteria associated with leguminous and non-leguminous plants. Plant Soil 321:35-59.


Gordon, J, Knowlton, N, Relman, DA, Rohwer, F and Youle, M 2013. Superorganisms and holobionts. Microbe 8:152-153.

Guo, W, Wu, G, Yan, F, Lu, Y, Zheng, H, Lin, L, Chen, H and Chen, J 2012. Identification of novel Oryza sativa miRNAs in deep sequencing-based small RNA libraries of rice infected with Rice stripe virus. PLoS ONE 7:e46443.



Griffiths, RI, Thomson, BC, James, P, Bell, T, Bailey, M and Whiteley, AS 2011. The bacterial biogeography of British soils. Environ. Microbiol 13:1642-1654.


Hardoim, PR, van Overbeek, LS and van Elsas, JD 2008. Properties of bacterial endophytes and their proposed role in plant growth. Trends Microbiol 16:463-471.


Harsonowati, W, Astuti, RI and Wahyudi, AT 2017. Leaf blast disease reduction by rice-phyllosphere actinomycetes producing bioactive compounds. J. Gen. Plant Pathol 83:98-108.


Harun-Or-Rashid, M, Kim, H-J, Yeom, S-I, Yu, H-A, Manir, MM, Moon, S-S, Kang, YJ and Chung, YR 2018.
Bacillus velezensis YC7010 enhances plant defenses against brown planthopper through transcriptomic and metabolic changes in rice. Front. Plant Sci 9:1904.



Huang, C-Y, Wang, H, Hu, P, Hamby, R and Jin, H 2019. Small RNAs: big players in plant-microbe interactions. Cell Host Microbe 26:173-182.


Hudzik, C, Hou, Y, Ma, W and Axtell, MJ 2020. Exchange of small regulatory RNAs between plants and their pests. Plant Physiol 182:51-62.



Hussain, Q, Liu, Y, Zhang, A, Pan, G, Li, L, Zhang, X, Song, X, Cui, L and Jin, Z 2011. Variation of bacterial and fungal community structures in the rhizosphere of hybrid and standard rice cultivars and linkage to CO2 flux. FEMS Microbiol. Ecol 78:116-128.


Hussain, Q, Pan, GX, Liu, YZ, Zhang, A, Li, LQ, Zhang, XH and Jin, ZJ 2012. Microbial community dynamics and function associatedwith rhizosphere over periods of rice growth. Plant Soil Environ 58:55-61.

Jiang, L, Bonkowski, M, Luo, L, Kardol, P, Zhang, Y, Chen, X, Li, D, Xiao, Z, Hu, F and Liu, M 2020. Combined addition of chemical and organic amendments enhances plant resistance to aboveground herbivores through increasing microbial abundance and diversity. Biol. Fertil. Soils 56:1007-1022.


Jiang, Y, Liang, Y, Li, C, Wang, F, Sui, Y, Suvannang, N, Zhou, J and Sun, B 2016. Crop rotations alter bacterial and fungal diversity in paddy soils across East Asia. Soil Biol. Biochem 95:250-261.

Jones, JDG, Vance, RE and Dangl, JL 2016. Intracellular innate immune surveillance devices in plants and animals. Science 354:aaf6395.


Juarez, MT, Kui, JS, Thomas, J, Heller, BA and Timmermans, MCP 2004. microRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature 428:84-88.



Kettles, GJ, Drurey, C, Schoonbeek, H-J, Maule, AJ and Hogenhout, SA 2013. Resistance of A rabidopsis thaliana to the green peach aphid, Myzus persicae, involves camalexin and is regulated by microRNAs. New Phytol 198:1178-1190.




Kim, H and Lee, Y-H 2020. The rice microbiome: a model platform for crop holobiome. Phytobiomes J 4:5-18.

Knief, C, Delmotte, N, Chaffron, S, Stark, M, Innerebner, G, Wassmann, R, von Mering, C and Vorholt, JA 2012. Metaproteogenomic analysis of microbial communities in the phyllosphere and rhizosphere of rice. ISME J 6:1378-1390.




Lan, H, Wang, H, Chen, Q, Chen, H, Jia, D, Mao, Q and Wei, T 2016. Small interfering RNA pathway modulates persistent infection of a plant virus in its insect vector. Sci. Rep 6:20699.




Lee, HJ, Jeong, SE, Kim, PJ, Madsen, EL and Jeon, CO 2015. High resolution depth distribution of Bacteria, Archaea, methanotrophs, and methanogens in the bulk and rhizosphere soils of a flooded rice paddy. Front. Microbiol 6:639.



Li, F, Hua, H, Ali, A and Hou, M 2019. Characterization of a bacterial symbiont Asaia sp. in the white-backed planthopper, Sogatella furcifera, and its effects on host fitness. Front. Microbiol 10:2179.



Li, T, Chen, J, Fan, X, Chen, W and Zhang, W 2017. MicroRNA and dsRNA targeting chitin synthase A reveal a great potential for pest management of the hemipteran insect Nilaparvata lugens
. Pest Manag. Sci 73:1529-1537.



Li, T-P, Zha, S-S, Zhou, C-Y, Gong, J-T, Zhu, Y-X, Zhang, X, Xi, Z and Hong, X-Y 2020. Newly introduced Cardinium endosymbiont reduces microbial diversity in the rice brown planthopper Nilaparvata lugens
. FEMS Microbiol. Ecol 96:fiaa194.



Lindow, SE and Andersen, GL 1996. Influence of immigration on epiphytic bacterial populations on navel orange leaves. Appl. Environ. Microbiol 62:2978-2987.




Liu, L, Zhang, K-J, Rong, X, Li, Y-Y and Liu, H 2019. Identification of Wolbachia-responsive miRNAs in the small brown planthopper, Laodelphax striatellus
. Front. Physiol 10:928.



Liu, W, Zhang, X, Wu, N, Ren, Y and Wang, X 2020. High diversity and functional complementation of alimentary canal microbiota ensure small brown planthopper to adapt different biogeographic environments. Front. Microbiol 10:2953.



Lugtenberg, B and Kamilova, F 2009. Plant-growth-promoting rhizobacteria. Annu. Rev. Microbiol 63:541-556.


Mannaa, M and Seo, Y-S 2021. Plants under the attack of allies: moving towards the plant pathobiome paradigm. Plants 10:125.



Mayoral, JG, Hussain, M, Joubert, DA, Iturbe-Ormaetxe, I, O’Neill, SL and Asgari, S 2014.
Wolbachia small noncoding RNAs and their role in cross-kingdom communications. Proc Natl. Acad. Sci. U. S. A 111:18721-18726.



Mendes, R, Garbeva, P and Raaijmakers, JM 2013. The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev 37:634-663.


Miller, DL, Parish, AJ and Newton, ILG 2019. Transitions and transmission: behavior and physiology as drivers of honey bee-associated microbial communities. Curr. Opin. Microbiol 50:1-7.


Miura, K, Ikeda, M, Matsubara, A, Song, X-J, Ito, M, Asano, K, Matsuoka, M, Kitano, H and Ashikari, M 2010.
OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat. Genet 42:545-549.



Müller, T and Ruppel, S 2014. Progress in cultivation-independent phyllosphere microbiology. FEMS Microbiol. Ecol 87:2-17.



Navarro, L, Dunoyer, P, Jay, F, Arnold, B, Dharmasiri, N, Estelle, M, Voinnet, O and Jones, JDG 2006. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312:436-439.


Nevita, T, Sharma, GD and Pandey, P 2018. Differences in rice rhizosphere bacterial community structure by application of lignocellulolytic plant-probiotic bacteria with rapid composting traits. Ecol. Eng 120:209-221.

Niu, D, Lii, YE, Chellappan, P, Lei, L, Peralta, K, Jiang, C, Guo, J, Coaker, G and Jin, H 2016. miRNA863-3p sequentially targets negative immune regulator ARLPKs and positive regulator SERRATE upon bacterial infection. Nat. Commun 7:11324.




Pathak, MD and Khan, ZR 1994. Insect pests of rice. International Rice Research Institute, Manila, Philippines. pp. 89.
Peng, T, Lv, Q, Zhang, J, Li, J, Du, Y and Zhao, Q 2011. Differential expression of the microRNAs in superior and inferior spikelets in rice (Oryza sativa). J. Exp. Bot 62:4943-4954.


Qiao, L, Zheng, L, Sheng, C, Zhao, H, Jin, H and Niu, D 2020. Rice siR109944 suppresses plant immunity to sheath blight and impacts multiple agronomic traits by affecting auxin homeostasis. Plant J 102:948-964.



Raaijmakers, JM, Paulitz, TC, Steinberg, C, Alabouvette, C and Moënne-Loccoz, Y 2009. The rhizosphere: a playground and battlefield for soilborne pathogens and beneficial microorganisms. Plant Soil 321:341-361.

Ren, G-D, Zhu, J-G and Jia, Z-J 2014. Contrasting response patterns of rice phyllosphere bacterial taxa to elevated CO2
. Pedosphere 24:544-552.

Rincon-Florez, VA, Carvalhais, LC and Schenk, PM 2013. Culture-independent molecular tools for soil and rhizosphere microbiology. Diversity 5:581-612.

Salvador-Guirao, R, Hsing, Y-I and San Segundo, B 2018. The polycistronic miR166k-166h positively regulates rice immunity via post-transcriptional control of EIN2
. Front. Plant Sci 9:337.



Sánchez-Cañizares, C, Jorrín, B, Poole, PS and Tkacz, A 2017. Understanding the holobiont: the interdependence of plants and their microbiome. Curr. Opin. Microbiol 38:188-196.


Sattar, S and Thompson, GA 2016. Small RNA regulators of plant-hemipteran interactions: micromanagers with versatile roles. Front. Plant Sci 7:1241.



Sazama, EJ, Ouellette, SP and Wesner, JS 2019. Bacterial endosymbionts are common among, but not necessarily within, insect species. Environ. Entomol 48:127-133.


Shah, AZ, Ma, C, Zhang, Y, Zhang, Q, Xu, G and Yang, G 2022. Decoyinine induced resistance in rice against small brown planthopper Laodelphax striatellus
. Insects 13:104.



Shikano, I, Rosa, C, Tan, C-W and Felton, GW 2017. Tritrophic interactions: microbe-mediated plant effects on insect herbivores. Annu. Rev. Phytopathol 55:313-331.


Song, X, Wang, D, Ma, L, Chen, Z, Li, P, Cui, X, Liu, C, Cao, S, Chu, C, Tao, Y and Cao, X 2012. Rice RNA-dependent RNA polymerase 6 acts in small RNA biogenesis and spikelet development. Plant J 71:378-389.


Song, Y, Shi, J, Xiong, Z, Shentu, X and Yu, X 2021. Three antimicrobials alter gut microbial communities and causing different mortality of brown planthopper, Nilaparvata lugens Stål. Pestic. Biochem. Physiol 174:104806.


Sun, W, Xu, XH, Li, Y, Xie, L, He, Y, Li, W, Lu, X, Sun, H and Xie, X 2020. OsmiR530 acts downstream of OsPIL15 to regulate grain yield in rice. New Phytol 226:823-837.




Sun, Z, Liu, Z, Zhou, W, Jin, H, Liu, H, Zhou, A, Zhang, A and Wang, M-Q 2016. Temporal interactions of plant-insect-predator after infection of bacterial pathogen on rice plants. Sci. Rep 6:26043.




Sunkar, R, Zhou, X, Zheng, Y, Zhang, W and Zhu, J-K 2008. Identification of novel and candidate miRNAs in rice by high throughput sequencing. BMC Plant Biol 8:25.



Tan, J, Wu, Y, Guo, J, Li, H, Zhu, L, Chen, R, He, G and Du, B 2020. A combined microRNA and transcriptome analyses illuminates the resistance response of rice against brown planthopper. BMC Genomics 21:144.




Tang, T, Zhang, Y, Cai, T, Deng, X, Liu, C, Li, J, He, S, Li, J and Wan, H 2021. Antibiotics increased host insecticide susceptibility via collapsed bacterial symbionts reducing detoxification metabolism in the brown planthopper, Nilaparvata lugens
. J. Pest Sci 94:757-767.


Thapa, S, Ranjan, K, Ramakrishnan, B, Velmourougane, K and Prasanna, R 2018. Influence of fertilizers and rice cultivation methods on the abundance and diversity of phyllosphere microbiome. J. Basic Microbiol 58:172-186.



Tong, A, Yuan, Q, Wang, S, Peng, J, Lu, Y, Zheng, H, Lin, L, Chen, H, Gong, Y, Chen, J and Yan, F 2017. Altered accumulation of osa-miR171b contributes to rice stripe virus infection by regulating disease symptoms. J. Exp. Bot 68:4357-4367.



United Nations 2005. World population prospects. The 2004 revision. Highlights. Population Division, Department of Economicand Social Affairs. United Nations, New York, USA. pp. 91.
Urayama, S, Moriyama, H, Aoki, N, Nakazawa, Y, Okada, R, Kiyota, E, Miki, D, Shimamoto, K and Fukuhara, T 2010. Knock-down of OsDCL2 in rice negatively affects maintenance of the endogenous dsRNA virus, Oryza sativa endornavirus. Plant Cell Physiol 51:58-67.


Ursell, LK, Metcalf, JL, Parfrey, LW and Knight, R 2012. Defining the human microbiome. Nutr. Rev 70(Suppl 1):S38-S44.



Van Ex, F, Jacob, Y and Martienssen, RA 2011. Multiple roles for small RNAs during plant reproduction. Curr. Opin. Plant Biol 14:588-593.



Vijayakumar, MM, More, RP, Rangasamy, A, Gandhi, GR, Muthugounder, M, Thiruvengadam, V, Samaddar, S, Jalali, SK and Sa, T 2018. Gut bacterial diversity of insecticide-susceptible and -resistant nymphs of the brown planthopper Nilaparvata lugens Stål (Hemiptera: Delphacidae) and elucidation of their putative functional roles. J. Microbiol. Biotechnol 28:976-986.


Walker, TS, Bais, HP, Grotewold, E and Vivanco, JM 2016. Root exudation and rhizosphere biology. Plant Physiol 132:44-51.




Wang, J, Yao, W, Zhu, D, Xie, W and Zhang, Q 2015. Genetic basis of sRNA quantitative variation analyzed using an experimental population derived from an elite rice hybrid. eLife 4:e03913.




Wang, Z-L, Pan, H-B, Wu, W, Li, M-Y and Yu, X-P 2021. The gut bacterial flora associated with brown planthopper is affected by host rice varieties. Arch. Microbiol 203:325-333.



Wang, Z-L, Wang, T-Z, Zhu, H-F, Pan, H-B and Yu, X-P 2020. Diversity and dynamics of microbial communities in brown planthopper at different developmental stages revealed by high-throughput amplicon sequencing. Insect Sci 27:883-894.



Wang, Z, Xia, Y, Lin, S, Wang, Y, Guo, B, Song, X, Ding, S, Zheng, L, Feng, R, Chen, S, Bao, Y, Sheng, C, Zhang, X, Wu, J, Niu, D, Jin, H and Zhao, H 2018. Osa-miR164a targets OsNAC60 and negatively regulates rice immunity against the blast fungus Magnaporthe oryzae
. Plant J 95:584-597.


Wari, D, Alamgir, KM, Mujiono, K, Hojo, Y, Tani, A, Shinya, T, Nakatani, H and Galis, I 2019a. Brown planthopper honeydew-associated symbiotic microbes elicit momilactones in rice. Plant Signal. Behav 14:1655335.



Wari, D, Kabir, MA, Mujiono, K, Hojo, Y, Shinya, T, Tani, A, Nakatani, H and Galis, I 2019b. Honeydew-associated microbes elicit defense responses against brown planthopper in rice. J. Exp. Bot 70:1683-1696.



Wei, LQ, Yan, LF and Wang, T 2011. Deep sequencing on genome-wide scale reveals the unique composition and expression patterns of microRNAs in developing pollen of Oryza sativa
. Genome Biol 12:R53.




Wu, J, Yang, Z, Wang, Y, Zheng, L, Ye, R, Ji, Y, Zhao, S, Ji, S, Liu, R, Xu, L, Zheng, H, Zhou, Y, Zhang, X, Cao, X, Xie, L, Wu, Z, Qi, Y and Li, Y 2015. Viral-inducible Argonaute18 confers broad-spectrum virus resistance in rice by sequestering a host microRNA. eLife 4:e05733.




Wu, L, Zhou, H, Zhang, Q, Zhang, J, Ni, F, Liu, C and Qi, Y 2010. DNA methylation mediated by a microRNA pathway. Mol. Cell 38:465-475.


Xia, X, Gurr, GM, Vasseur, L, Zheng, D, Zhong, H, Qin, B, Lin, J, Wang, Y, Song, F, Li, Y, Lin, H and You, M 2017. Metagenomic sequencing of diamondback moth gut microbiome unveils key holobiont adaptations for herbivory. Front. Microbiol 8:663.



Xie, J, Peng, Y and Xia, Y 2021. Genome-wide identification and analysis of Nilaparvata lugens microRNAs during challenge with the entomopathogenic fungus Metarhizium anisopliae
. J. Fungi 7:295.



Xie, Z, Johansen, LK, Gustafson, AM, Kasschau, KD, Lellis, AD, Zilberman, D, Jacobsen, SE and Carrington, JC 2004. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol 2:e104.



Xu, D, Mou, G, Wang, K and Zhou, G 2014a. MicroRNAs responding to southern rice black-streaked dwarf virus infection and their target genes associated with symptom development in rice. Virus Res 190:60-68.


Xu, H-X, Zheng, X-S, Yang, Y-J, Wang, X, Ye, G-Y and Lu, Z-X 2014b. Bacterial community in different populations of rice brown planthopper nilaparvata lugens (Stål). Rice Sci 21:59-64.

Xu, L, Zhang, J, Zhan, A, Wang, Y, Ma, X, Jie, W, Cao, Z, Omar, MAA, He, K and Li, F 2020. Identification and analysis of microRNAs associated with wing polyphenism in the brown planthopper, Nilaparvata lugens
. Int. J. Mol. Sci 21:9754.



Yan, X-T, Ye, Z-X, Wang, X, Zhang, C-X, Chen, J-P, Li, J-M and Huang, H-J 2021. Insight into different host range of three planthoppers by transcriptomic and microbiomic analysis. Insect Mol. Biol 30:287-296.



Yan, Y, Zhang, Y, Yang, K, Sun, Z, Fu, Y, Chen, X and Fang, R 2011. Small RNAs from MITE-derived stem-loop precursors regulate abscisic acid signaling and abiotic stress responses in rice. Plant J 65:820-828.


Yang, M, Xu, Z, Zhao, W, Liu, Q, Li, Q, Lu, L, Liu, R, Zhang, X and Cui, F 2018. Rice stripe virus-derived siRNAs play different regulatory roles in rice and in the insect vector Laodelphax striatellus
. BMC Plant Biol 18:219.




Ye, X, Xu, L, Li, X, He, K, Hua, H, Cao, Z, Xu, J, Ye, W, Zhang, J, Yuan, Z and Li, F 2019. MiR-34 modulates wing polyphenism in planthopper. PLoS Genet 15:e1008235.



Yu, C, Chen, Y, Cao, Y, Chen, H, Wang, J, Bi, Y-M, Tian, F, Yang, F, Rothstein, SJ, Zhou, X and He, C 2018. Overexpression of miR169o, an overlapping microRNA in response to both nitrogen limitation and bacterial infection, promotes nitrogen use efficiency and susceptibility to bacterial blight in rice. Plant Cell Physiol 59:1234-1247.


Yu, H, Ji, R, Ye, W, Chen, H, Lai, W, Fu, Q and Lou, Y 2014. Transcriptome analysis of fat bodies from two brown planthopper (Nilaparvata lugens) populations with different virulence levels in rice. PLoS ONE 9:e88528.



Yuan, C, Zhang, L, Hu, H, Wang, J, Shen, J and He, J 2018. The biogeography of fungal communities in paddy soils is mainly driven by geographic distance. J. Soils Sediments 18:1795-1805.


Zhang, C, Ding, Z, Wu, K, Yang, L, Li, Y, Yang, Z, Shi, S, Liu, X, Zhao, S, Yang, Z, Wang, Y, Zheng, L, Wei, J, Du, Z, Zhang, A, Miao, H, Li, Y, Wu, Z and Wu, J 2016. Suppression of jasmonic acid-mediated defense by viral-inducible microRNA319 facilitates virus infection in rice. Mol. Plant 9:1302-1314.


Zhang, J-H, Yu, N, Xu, X-X and Liu, Z-W 2019. Community structure, dispersal ability and functional profiling of microbiome existing in fat body and ovary of the brown planthopper, Nilaparvata lugens
. Insect Sci 26:683-694.



Zhang, W, Chen, J, Keyhani, NO, Zhang, Z, Li, S and Xia, Y 2015. Comparative transcriptomic analysis of immune responses of the migratory locust, Locusta migratoria, to challenge by the fungal insect pathogen, Metarhizium acridum
. BMC Genomics 16:867.



Zhang, X, Bao, Y, Shan, D, Wang, Z, Song, X, Wang, Z, Wang, J, He, L, Wu, L, Zhang, Z, Niu, D, Jin, H and Zhao, H 2018a.
Magnaporthe oryzae induces the expression of a microRNA to suppress the immune response in rice. Plant Physiol 177:352-368.



Zhang, X, Li, T-P, Zhou, C-Y, Zhao, D-S, Zhu, Y-X, Bing, X-L, Huang, H-J and Hong, X-Y 2020. Antibiotic exposure perturbs the bacterial community in the small brown planthopper Laodelphax striatellus
. Insect Sci 27:895-907.



Zhang, X, Zhao, H, Gao, S, Wang, W-C, Katiyar-Agarwal, S, Huang, H-D, Raikhel, N and Jin, H 2011.
Arabidopsis Argonaute 2 regulates innate immunity via miRNA393*-mediated silencing of a Golgi-localized SNARE gene, MEMB12
. Mol. Cell 42:356-366.



Zhang, Y, Tang, T, Li, W, Cai, T, Li, J and Wan, H 2018b. Functional profiling of the gut microbiomes in two different populations of the brown planthopper, Nilaparvata lugens
. J. Asia-Pac. Entomol 21:1309-1314.

Zhang, Y-C, Yu, Y, Wang, C-Y, Li, Z-Y, Liu, Q, Xu, J, Liao, J-Y, Wang, X-J, Qu, L-H, Chen, F, Xin, P, Yan, C, Chu, J, Li, H-Q and Chen, Y-Q 2013. Overexpression of microRNA OsmiR397 improves rice yield by increasing grain size and promoting panicle branching. Nat. Biotechnol 31:848-852.



Zhao, Z-X, Feng, Q, Cao, X-L, Zhu, Y, Wang, H, Chandran, V, Fan, J, Zhao, J-Q, Pu, M, Li, Y and Wang, W-M 2020.
Osa-miR167d facilitates infection of Magnaporthe oryzae in rice. J. Integr. Plant Biol 62:702-715.



- TOOLS
-
METRICS

-
- 1 Crossref
- 0 Scopus
- 3,288 View
- 159 Download
- Related articles



 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Print
Print



