 |
 |
| Plant Pathol J > Volume 38(5); 2022 > Article |
|
Abstract
Lilies (Lilium spp.) are one of the most important ornamental flower crops grown in Korea. Most viral diseases in lilies are transmitted by infected bulbs, which cause serious economic losses due to reduced yields. Various diagnostic techniques and high-throughput sequencing methods have been used to detect lily viruses. According to Oxford Nanopore Technologies (ONT), MinION is a compact and portable sequencing device. In this study, three plant viruses, lily mottle, lily symptomless, and plantago asiatica mosaic virus, were detected in lily samples using the ONT platform. As a result of genome assembly of reads obtained through ONT, 100% coverage and 90.3-93.4% identity were obtained. Thus, we show that the ONT platform is a promising tool for the diagnosis and characterization of viruses that infect crops.
Lilies (Lilium spp.) are one of the most important ornamental flower crops grown in South Korea and are highly regarded for their ornamental value, summer blooming characteristics, and petal color and fragrance. The genus Lilium contains 294 genera with 4,500 species, including three commercially important divisions of lilies: Easter lily (Lilium longiflorum) and the Asian and Oriental hybrids (Sharma et al., 2005). According to the Ministry of Agriculture, Food and Rural Affairs, lilies are mainly cultivated in Gangwon, Jeju, and Chungcheongnam-do, with a total cultivated area of 215 ha and have an annual value of 34 million dollars (Kang et al., 2013).
Lilies can be grown as seeds, bulbs, and scales, and most lily bulbs grown in Korea are imported from the Netherlands (Jang and Kim, 2016; Sharma et al., 2005). Almost all lilies are cultivated by vegetative propagation, and infected bulbs used for vegative propagation help in the propagation of the plant viruses from one generation to the next (Zhang et al., 2020). These viral diseases have caused serious economic losses to farmers, as they reduce the yield of lily bulbs by more than 50% (Zhang et al., 2015a). More than 10 plant viruses have been reported worldwide, causing quantitative and qualitative damage to lilies (Ryu et al., 2002; Wang et al., 2010). Several viruses have been reported that infect lilies, including the Cucumber mosaic virus (CMV), Lily mottle virus (LMoV), Lily symptomless virus (LSV), Arabis mosaic virus, Lily virus X, Plantago asiatica mosaic virus (PlAMV), Strawberry latent ringspot virus, Tobacco rattle virus, and Tomato ringspot virus (Asjes, 2000; Komatsu et al., 2008; Kwon et al., 2013; Sharma et al., 2005).
Lilies can be infected by viruses via aphids, which is known to occur in many cultivated areas worldwide (Asjes, 2000; Choi and Ryu, 2003). Infection with one or more plant virus can cause more severe symptoms, including vein clearing, leaf rolling, spotting, mosaic patterns, and yellow streaking (Kwon et al., 2013).
Various techniques have been developed to diagnose lily viruses, including multiplex reverse-transcription polymerase chain reaction (RT-PCR), multiplex Luminex bead array, triplex IC-RT-PCR, indirect enzyme-lined immunosorbent assay, immunocapture RT-PCR, RT loop-mediated isothermal amplification (RT-LAMP), immunocapture RT-LAMP, and immunochromatographic strip. These studies mostly focused on the identification of the three main viruses: LSV, LMoV, and CMV (He et al., 2016; Jo and Cho, 2018; Kim et al., 2012; Kwon et al., 2013; Lim et al., 2016; Niimi et al., 2003; Zhang et al., 2015a, 2015b, 2017, 2022; Zhao et al., 2018). Advances in these molecular technologies can help clarify plant viral diseases and more effectively determine their etiology; early diagnosis facilitates plant disease management and is essential for successful biosecurity responses (Aboul-Ata et al., 2011; van der Want and Dijkstra, 2006; Vazquez-Iglesias et al., 2022).
Metagenomic high-throughput sequencing (HTS) technology allows for the detection of viral diseases without information about the organism in the sample, and this approach has enhanced plant virus detection in diagnostic and surveillance applications (Barzon et al., 2011; Batovska et al., 2017; Temmam et al., 2014). A recent study identified six different viruses using virome analysis in lilies or detected a hitherto unrecorded potyvirus (tentative name: lily yellow mosaic virus) (Jo and Cho, 2018; Li et al., 2018). Second-generation sequencing using massively parallel sequencing platforms requires multiple sequencing cycles; therefore, the sequencing process typically takes between 0.5 and 3 days depending on the read length and data output requirements (Wang et al., 2020).
Oxford Nanopore Technologies (ONT) is a compact, portable device for real-time single-molecule DNA and RNA sequencing that directly detects changes in the electric currents, which are generated when DNA and RNA molecules pass through nanopore proteins embedded in the device’s polymer membrane, thereby enabling the detection of biological molecules (Deamer et al., 2016; Eid et al., 2009; Phannareth et al., 2020). These electrical signals are converted into nucleotide sequences by base calling, which is a computational algorithm (Theuns et al., 2018). Until recently, HTS was limited to the laboratory because of the size of the sequencers available; however, with the release of MinION from ONT, sequencing in the field is now possible (Batovska et al., 2017). Moreover, additional time is required for analysis because the sequencing data cannot be applied to further analysis until the entire sequencing process is complete. In terms of diagnostics, the MinION sequencer and Flongle (flow cell dongle) are ideal for single or small numbers of samples in diagnostic settings, providing low sample throughput with data generated in a short time of 24-48 h at a cost suitable for diagnostic laboratories (Vazquez-Iglesias et al., 2022).
In recent years, ONT has demonstrated success across a wide variety of plant viruses by generating long-read single-molecule sequence data in real time with an innovation in third-generation sequencing technology (Jain et al., 2015; Lewandowski et al., 2019; Zhu et al., 2020). ONT has been used to detect plant viruses in a variety of plants, including peaches (Bronzato Badial et al., 2018), tomatoes (Chalupowicz et al., 2019), yams (Filloux et al., 2018), potatoes (Della Bartola et al., 2020), wheat (Fellers et al., 2019), cowpea (Naito et al., 2019), cassava (Boykin et al., 2019), Medicago arborea (Ben Chehida et al., 2021), Jasminum officinale (Amoia et al., 2022), grapevine (Javaran et al., 2021), roses (Vazquez-Iglesias et al., 2022), and lamon bean (Tarquini et al., 2022).
In this study, plant viruses were identified and characterized by virome analysis in lilies using MinION and flongle flow cells provided by ONT. In addition, we discuss the potential variant applications of MinION sequencing for the diagnosis of viruses affecting lilies.
In June 2021, lily (oriental hybrid “Siberia” cultivar) leaf samples with leaf curling and mosaic pattern symptoms (Fig. 1) were collected from Suncheon, Korea, and stored at −80°C.
Total RNAs was extracted using a Clear-S Total RNA extraction kit (InVirusTech Co., Gwangju, Korea), following the manufacturer’s instructions. DNA was removed from the extracted total RNA using a RapidOut DNA Removal Kit (Thermo Fisher Scientific, Waltham, MA, USA) for 30 min at 37°C. The quality and quantification of the extracted total RNA were evaluated using a spectrophotometer (BioDrop, Biochrom Ltd., Cambridge, UK) and a Qubit 4 Fluorometer with a Qubit RNA HS Assay Kit (Thermo Fisher Scientific). To enrich for viral nucleic acids present in plant samples, plant host rRNAs (ribodepletion) were removed using QIAseq FastSelect-rRNA Plant Kits (Qiagen, Hilden, Germany). The ribodepletized RNA was then purified with the AMPure XP Reagent (Beckman Coulter, Brea, CA, USA), quantified with a Qubit 4 Fluorometer with a Qubit RNA HS Assay Kit, and stored at −80°C before use for Nanopore library construction.
The library was prepared using a direct cDNA sequencing kit (SQK-DCS109) with a Flongle Starter pack (Oxford Nanopore Technologies, Oxford, UK), following the manufacturer’s instructions (Liefting et al., 2021). Briefly, cDNA was generated using the Maxima H Minus Double-Stranded cDNA Synthesis Kit (Thermo Fisher Scientific), then purified using GeneJET PCR Purification Kit (Thermo Fisher Scientific), and synthesized double-stranded cDNA was quantified using a Qubit 4 Fluorometer with a Qubit 1× dsDNA HS Assay Kit (Thermo Fisher Scientific). Then, the purified double-stranded cDNA was end-repaired and dA-tailing was performed using a NEBNext Ultra II End Repair/dA-Tailing Module (New England BioLabs, Ipswich, MA, USA) for 5 min at 20°C and 5 min at 65°C, purified with the AMPure XP Reagent, adapter ligation using Blunt/TA Ligase Master Mix (New England BioLabs) with Adapter Mix (SQK-DCS109, Oxford Nanopore Technologies) for 10 min at room temperature, purified with AMPure XP Reagent, and quantified with a Qubit 4 Fluorometer with a Qubit 1× dsDNA HS Assay Kit to prepare a nanopore library with a concentration of 3.54 ng/μl.
The Flongle flow cell (FLO-FLG001, R9.4.1, Oxford Nanopore Technologies) was primed with 200 μl of a mixture containing 117 μl of flush buffer and 3 μl of flush ether (FLT). Then, a total volume of 30 μl of the sequencing mix, including 6 μl of eluted library and 14 μl of sequencing buffer II (SQBII) and loading beads II (LBII), was loaded into the Flongle flow cell (R9.4.1) port of the MinION (Oxford Nanopore Technologies).
Sequencing runs were operated for 24 h using the MinKNOW software (version 21.11.7), and base-calling and quality assessment on the generated sequencing data were performed in real-time using the high-accuracy mode (quality score of 7) in the Guppy (version 5.1.12) with MinKNOW softwares (version 21.11.7, Oxford Nanopore Technologies).
The base-called FASTQ files were converted to FASTA files using Geneious Prime (version 21.1.1), and the adapter sequences were removed from the trim ends. The FASTQ ‘What’s in my Pot?’ (WIMP) workflow on the EPI2ME Agent (version 3.4.2) and a BLASTn search (-outfmt 6) of the sequences against the National Center for Biotechnology Information (NCBI) Viral Genome data (Viral RefSeq Database; VirDB) (version 21.11.4) were used to confirm the sequences of plant viruses. The sequencing reads were mapped to plant virus genomes identified with a high BLAST scores (top hits) in NCBI BLASTn searches using the ‘Map to Reference’ tool in Geneious Prime with default settings (Medium Sensitivity/Fast) and complete genome consensus sequences were obtained.
For phylogenetic analysis, the plant virus complete genome sequences obtained in this study and the genome sequences retrieved from NCBI BLASTn were aligned using the BioEdit Sequence Alignment Editor (version 7.0.5.3) with ClustalW Multiple alignment, and maximum-likelihood phylogenetic trees were inferred using MEGAX (version 10.2.4) with the nucleotide substitution selected Tamura-Nei model (Kumar et al., 2018; Tamura et al., 2004). The reliability of the phylogenetic tree was evaluated by calculating bootstrap values from 1000 replicates.
RT-PCR was used to confirm the presence of the identified plant viruses by ONT sequencing, such as LMoV, LSV, and PlAMV, with primers listed in Supplementary Table 1. Total RNA was amplified using the Suprimescript RT-PCR premix (Genet Bio, Daejeon, Korea) in accordance with the manufacturer’s instructions. All RT-PCR products were sequenced using Sanger sequencing, and the sequences were validated using a BLASTn search.
The FASTQ files generated in this study were deposited in the NCBI Sequence Read Archive (SRA) repository under accession number SRR18587584 in the NCBI BioProject database as PRJNA822696, and the plant virus sequences were deposited in GenBank with their individual accession numbers.
MinKNOW was run on lily leaf samples showing symptoms of leaf curling and mosaic pattern symptoms (Fig. 1), and the data generated a total of 566,812 reads and an average read length of 581.4 by 83 pores of the Flongle flow cell. Of these, 387,588 reads were obtained in the adapter trimming and quality filtering steps (>Q7) and the average length of the trimmed reads was 164.1 (Table 1).
The WIMP workflow was used to rapidly identify and quantify species in metagenomic samples, and of the 566,812 reads, which 54% were classified as bacteria, 27% as viruses, 19% as eukaryotes, and less than 1% as archaea (data not shown). To identify viruses infecting lilies, trimmed reads obtained from ONT sequencing were BLASTed from the NCBI VirDB to detect three RNA viruses, including LMoV, LSV, and PlAMV (Table 2). The same virus list was analyzed using the WIMP workflow (data not shown).
Genome assembly was performed on the reference genomes of LMoV, LSV, and PlAMV isolated from lily samples to obtain 100% complete sequences (Fig. 2). For LMoV, 7,049 reads (1.81% of the total reads) were mapped with 93.4% identity. For LSV, 27,334 reads (7.05% of the total reads) were mapped with 91.9% identity. For PlAMV, 15,569 reads (4.02% of the total reads) were mapped with 90.3% identity (Table 2, Fig. 3A). The complete genome consensus sequences of each virus were deposited in GenBank (LMoV, LC704533; LSV, LC720794; and PlAMV, LC720795) (Table 2). As for the length of the reads mapped to each reference genome, the maximum length of LMoV was 3,801 bp (Table 2, Fig. 3B), the maximum length of LSV was 3,237 bp (Table 2, Fig. 3C), and the maximum length of PlAMV was 2,591 bp (Table 2, Fig. 3D).
Neighbor-joining phylogenetic trees estimated with the Tamura-Nei model were generated for each complete coat protein (CP) sequence of the virus genomes identified by ONT sequencing (Fig. 4). The phylogenetic tree based on the complete CP sequence of LMoV was divided into two clades, and was most closely related to 99.76% identity with the Japan isolate (AB053256) (Fig. 4A). The LSV was divided into two clades and was most closely related with 99.54% identity with the Chinese isolate (HM222522) from Lilium (Fig. 4B). PlAMV was also divided into two clades and was most closely related with 100% identity to the South Korean isolate (KY923700) from Rehmannia glutinosa and the Russian isolate (Z21647) (Fig. 4C).
To confirm the true presence of viruses identified from lily samples, RT-PCR was performed using primers known to be virus-specific. The results of RT-PCR were the same as those of viruses analyzed by ONT sequencing and were amplified according to the amplicon size of each specific primer (Fig. 5). The RT-PCR amplification products were sequenced by Sanger sequencing, and the obtained sequences were confirmed for the presence of each virus using a BLASTn search (data not shown).
The development of rapid detection and accurate diagnostic techniques is essential for preventing the spread of plant diseases caused by plant viruses. Immunoassays or PCR-based methods have limitations in the detection of viral pathogens because of the limited number of targets that can detect or process already known targets, and HTS (RNA-Seq, DNA-Seq) and Sanger sequencing are both costly and time consuming, delaying control actions (Boykin et al., 2018; Bronzato Badial et al., 2018). The ability to simultaneously detect multiple plant viruses in a single sample is an important feature of diagnostic tools, and the ONT platform has been successfully applied to identify viral pathogens (Mehetre et al., 2021). The ONT platform has recently proven to be a reliable, real-time, and portable technology that enhances the study and monitoring of various viral pathogens including human and other animal viruses (Fang and Ramasamy, 2015). However, to date, this technique has rarely been applied in plant virology (Filloux et al., 2018). Therefore, we evaluated the potential of the ONT platform to detect and characterize viruses infecting lily plants, and showed that nanopore sequencing is a promising alternative means of identifying viral pathogens in plants.
The ONT platform is sequenced using massive parallel sequencing technology, the ONT platform offers the potential for genomic studies of known or unknown viruses, but still provides low single-base accuracy (Cao et al., 2019; Mehetre et al., 2021). However, according to a recent study, the raw read accuracy was approximately 85% but due to recent improvements in nanopore equipment, reagents, and subsequent analytical algorithms, including base-calling, more than 99.9% consensus accuracy has been reported (Chang et al., 2020; Liefting et al., 2021). Flongle flow cells have much lower throughput than flow cells but are cost-effective and make them more attractive when they are not needed because of the large datasets of plant virus diagnostics (Liefting et al., 2021).
The library constructed using a direct cDNA sequencing kit yielded positive results when present at concentrations as low as 3.5 ng/μl. The reason for using a low-concentration library is to use a Flongle flow cell that generates sufficient yield data for diagnostic purposes and low cost. Because the protocol for the Flongle flow cell is currently not available for cDNA library preparation kits at the ONT, the assay was performed using a low-concentration library. This is because ONT library preparation kits for RNA samples are currently designed for polyadenylated (poly(A)) RNA, and plant viruses without a poly(A) tail are not sequenced in this manner. Therefore, in this study, the desired data were obtained by successfully loading a direct cDNA sequencing kit into the flogle flow cell by reducing the concentration of the library, according to a previous study by Liefting et al. (2021).
The ONT platform successfully detected three plant viruses, including LMoV, LSV, and PlAMV, in lily samples. When using the WIMP workflow on raw data obtained using the ONT platform, cactus virus X (104 reads) and mint virus X (8 reads) were also identified (data not shown). When genome assembly was performed for these viruses, the complete sequence could not be obtained with a very short read, and was not identified in the BLASTn search. These results require additional updates because only the number of reads can be determined through the WIMP workflow, and it is not known where these reads are mapped to the genome or their length.
Similar to other HTS methods, benchtop sequencers are routinely used in laboratories because ONT-based studies such as MinION require sophisticated bioinformatics tools for data management and analysis (Marcolungo et al., 2022). A single run can detect and diagnose multiple virus species in real time and has great potential for a variety of applications (Ben Chehida et al., 2021). The ONT platform was introduced as a portable MinION sequencer that meets the minimum requirements for technical skills and bioinformatics knowledge and can be performed under field conditions and in small laboratories with a minimal package of laboratory equipment (Mehetre et al., 2021). The ease of point-of-care testing using MinION has been demonstrated and reported in several field studies (Faria et al., 2017; Maestri et al., 2019; Marcolungo et al., 2022; Quick et al., 2016). Therefore, these technologies will provide new avenues for point-of-care diagnostics for plant viral diseases.
In conclusion, this study demonstrated that the ONT platform is a promising tool for the identification and characterization of viral diseases in lily samples (Fig. 6). The widespread use of these ONT platforms will revolutionize plant disease diagnosis and enable early diagnosis of novel viruses in new crops. Addressing current limitations, such as error correction and base modification detection, is expected to have a tremendous impact not only on plant virus detection but also on RNA modification research, plant epigenomics, epitranscriptomics, and various aspects of plant biology and virology.
Acknowledgments
This work was carried out with the support of Cooperative Research Program for Agriculture Science and Technology Development (Project No. PJ014947032022) Rural Development Administration, Republic of Korea.
Electronic Supplementary Material
Supplementary materials are available at The Plant Pathology Journal website (http://www.ppjonline.org/).
Fig. 2
Genome assembly of identified viruses genome generated by nanopore sequencing. Genomic mapping of lily mottle virus (LMoV) (A), lily symptomless virus (LSV) (B), and plantago asiatica mosaic virus (PlAMV) (C) isolates identified in lilies. The blue graph is the mapping distribution of reads, and the black bars represent each read.
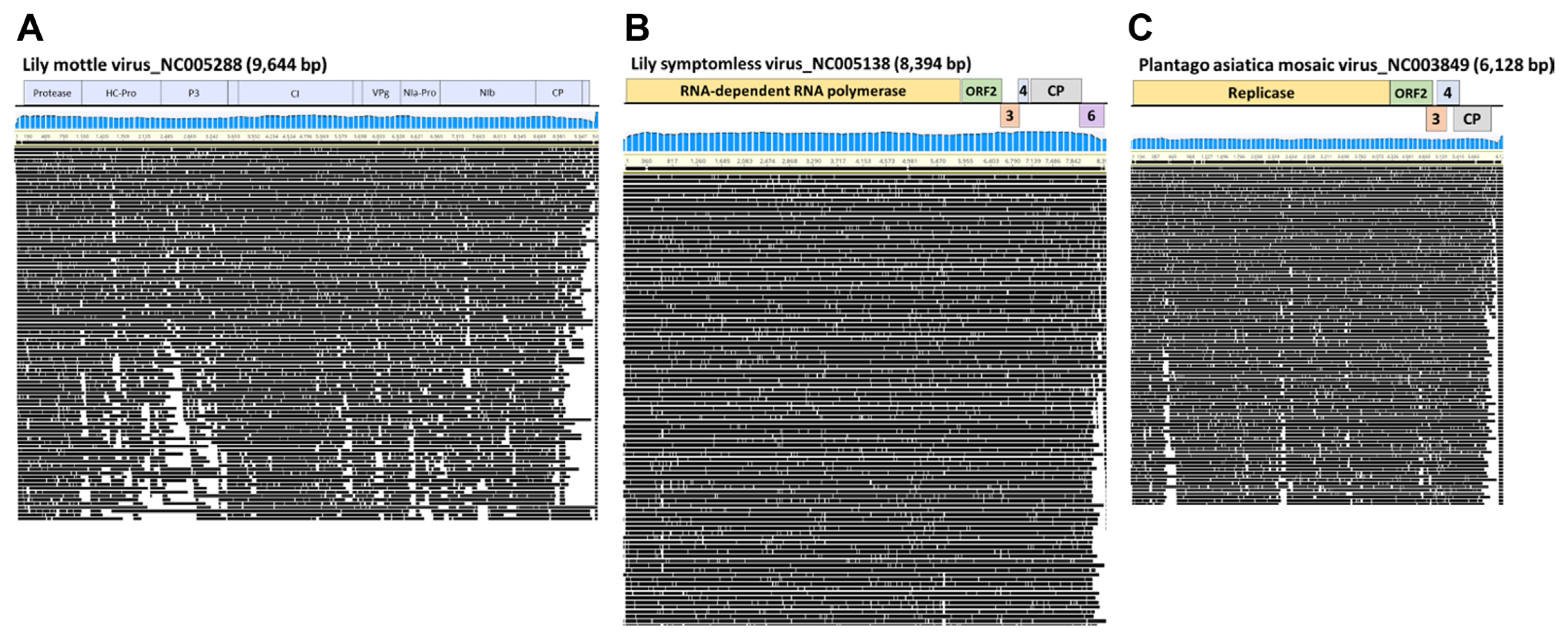
Fig. 3
The distribution of the read number and length of nanopore sequencing data. (A) Number of reads associated with each virus. Length distribution of reads associated with lily mottle virus (LMoV) (B), lily symptomless virus (LSV) (C), and plantago asiatica mosaic virus (PlAMV) (D).
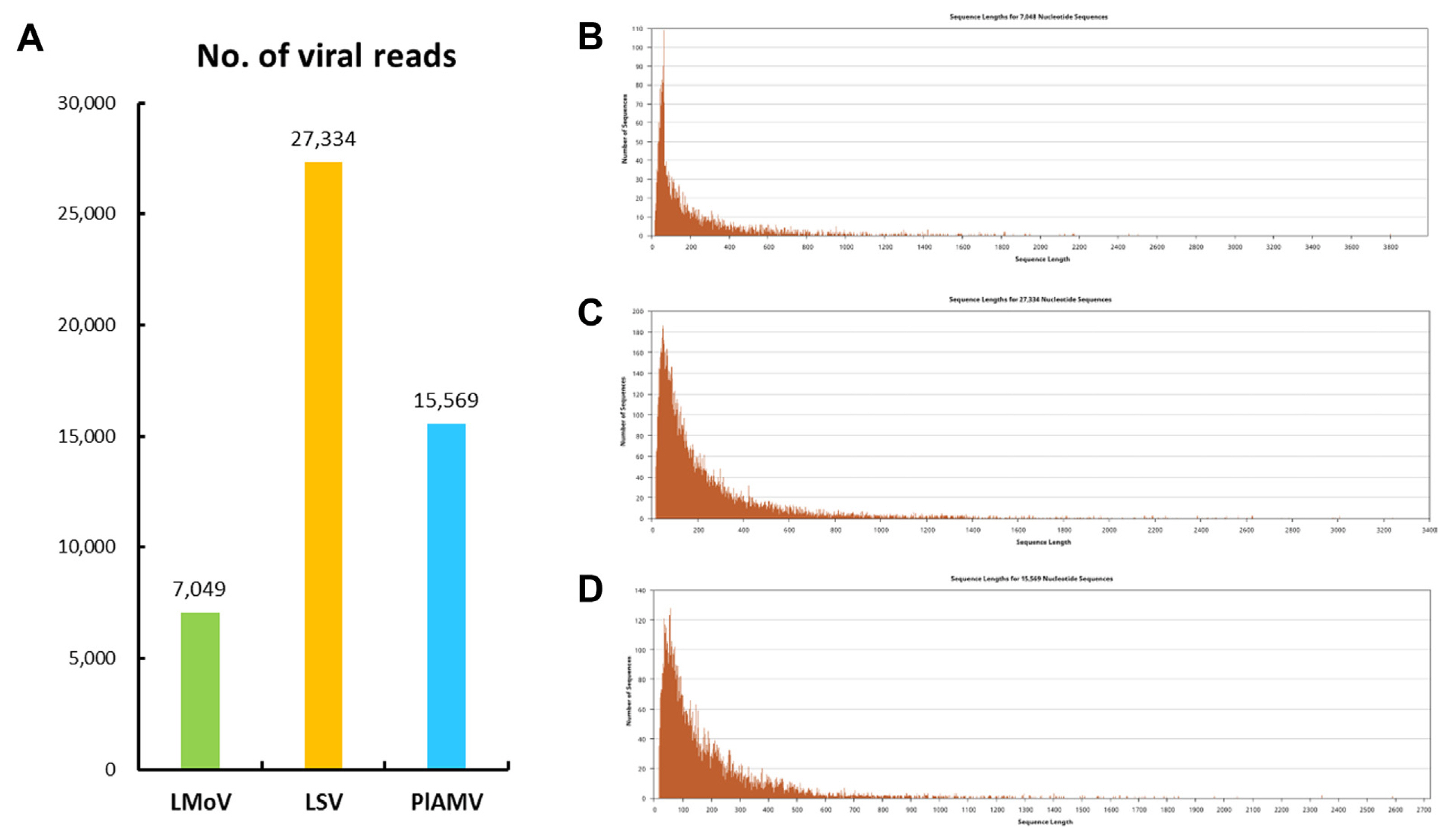
Fig. 4
Maximum-likelihood phylogenetic trees calculated from the coat protein sequences of lily mottle virus (LMoV) (A), lily symptomless virus (LSV) (B), and plantago asiatica mosaic virus (PlAMV) (C). The genomes of the viral isolates identified in this study are indicated by black circles.
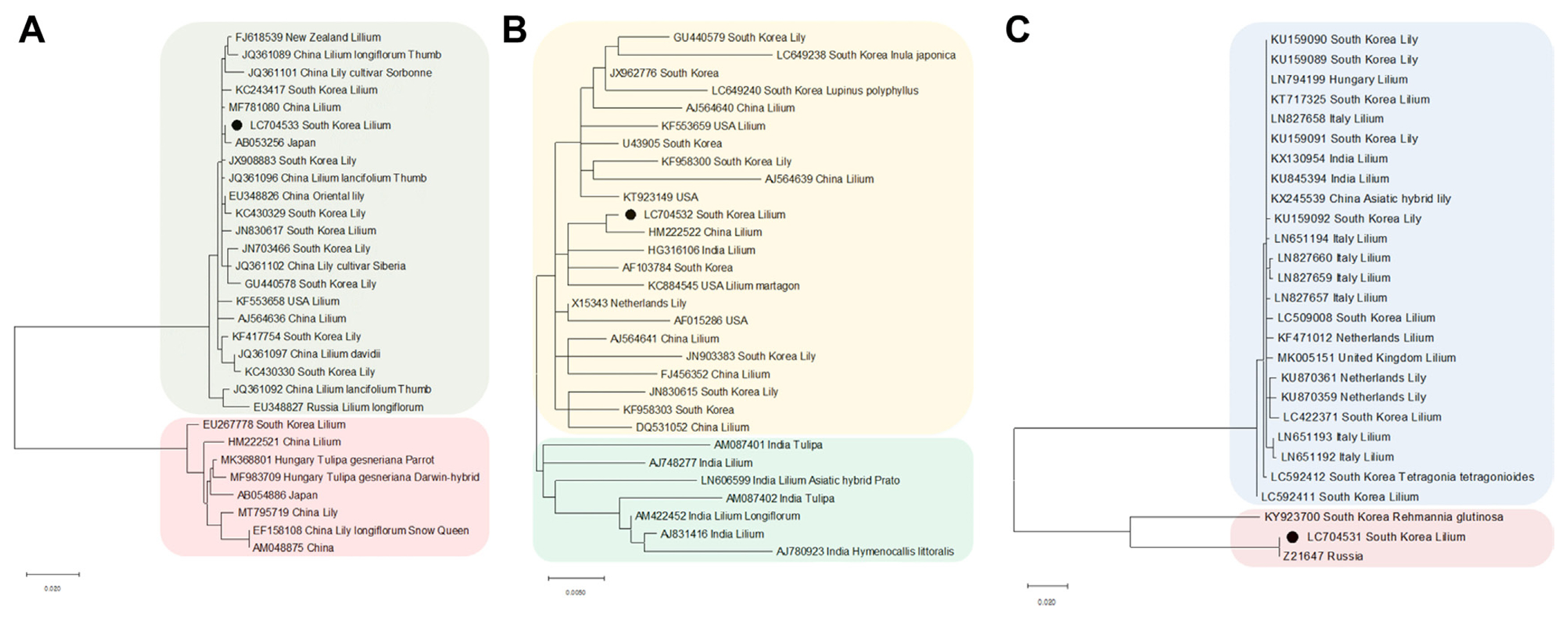
Fig. 5
Confirmation of the presence of the virus identified in the lily sample by reverse-transcription polymerase chain reaction. M, 100 bp ladder; LMoV, lily mottle virus; LSV, lily symptomless virus; PlAMV, plantago asiatica mosaic virus.
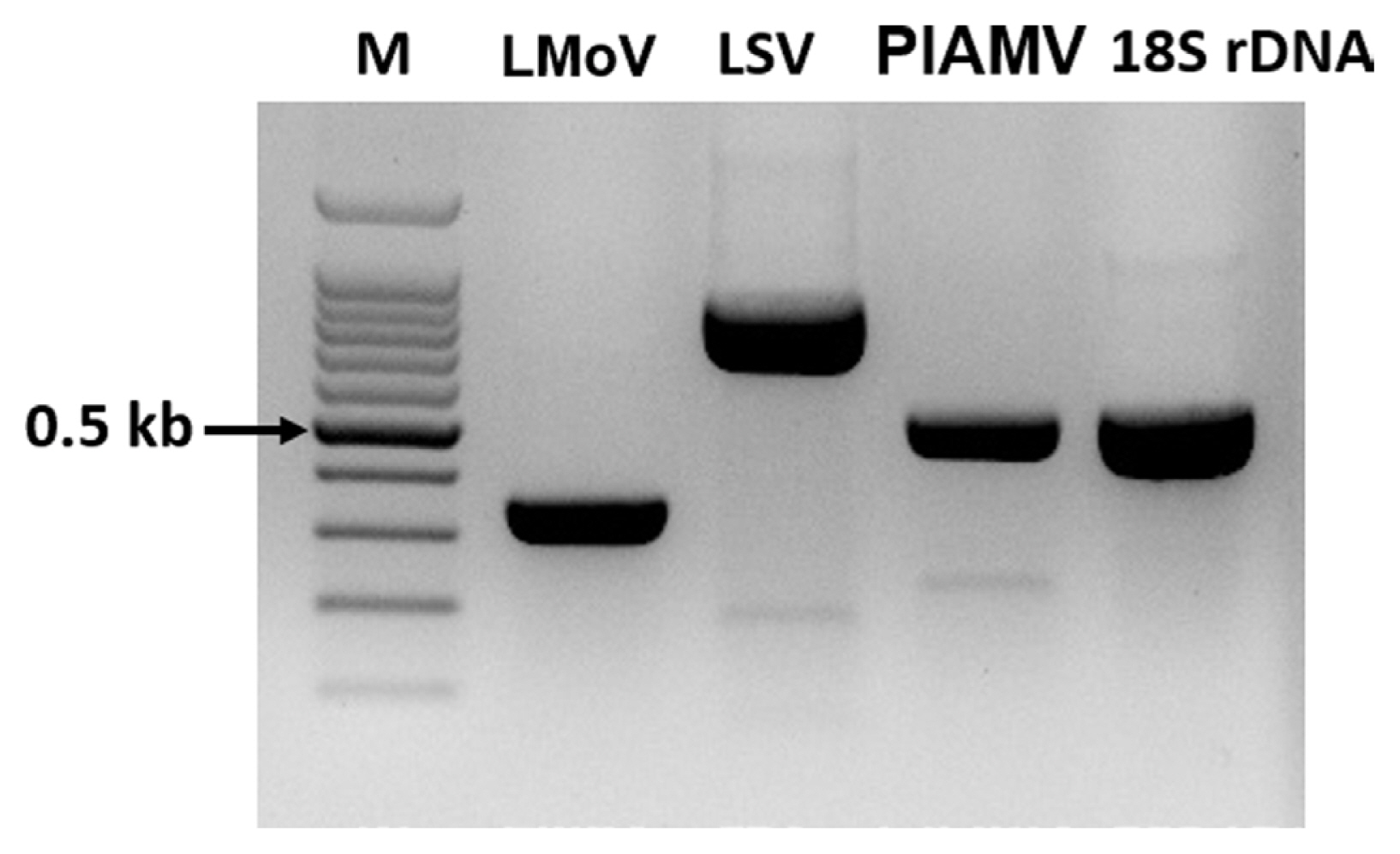
Fig. 6
Workflow for preparation Oxford Nanopore Technologies (ONT) sequencing for MinION. Created with Biorender (biorender.com).
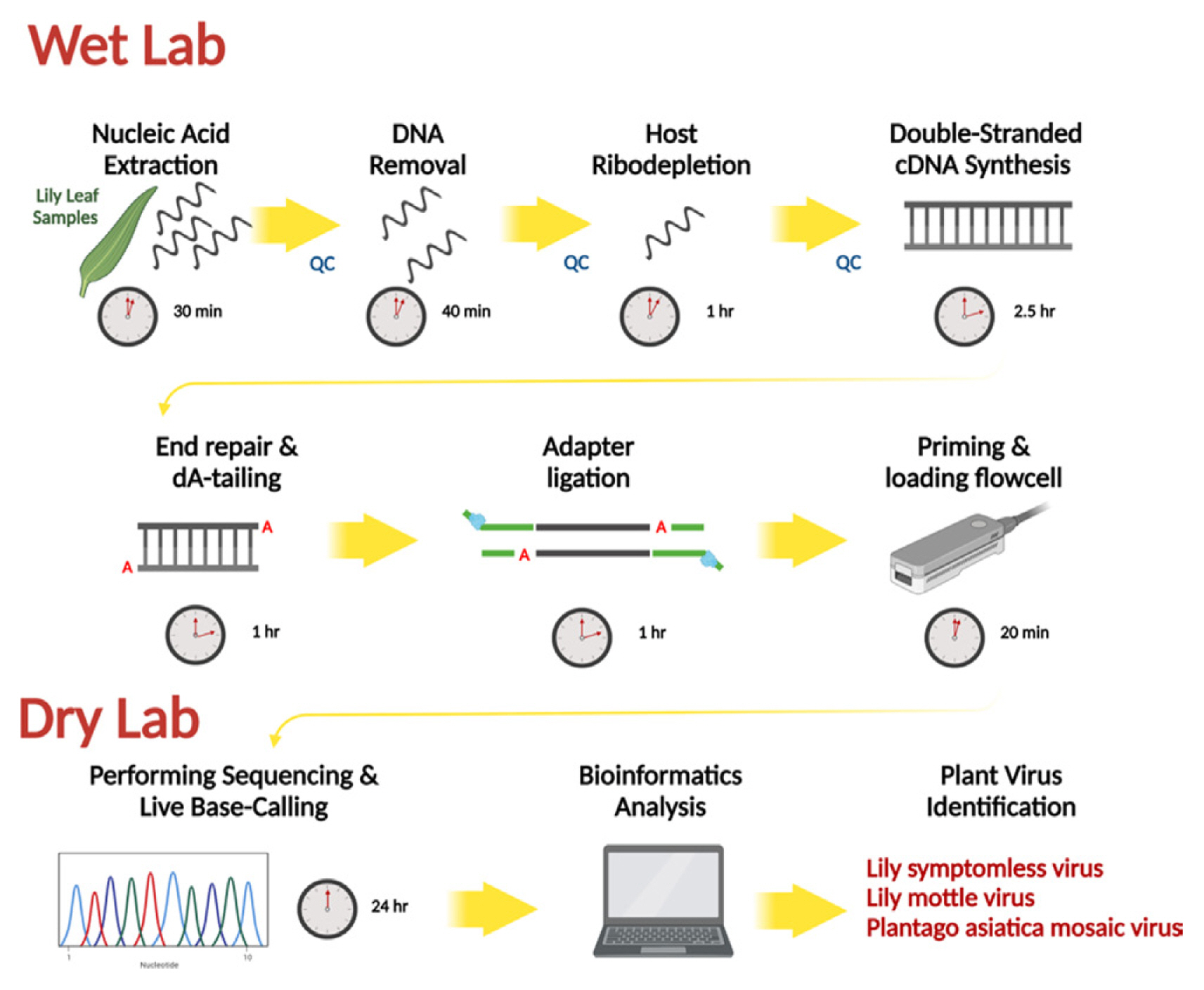
Table 1
Summary of ONT sequencing data
Table 2
List of identified viruses in lily samples
References
Aboul-Ata, AA, Mazyad, H, El-Attar, AK, Soliman, AM, Anfoka, G, Zeidan, M, Gorovits, R, Sobol, I and Czosnek, H 2011. Diagnosis and control of cereal viruses in the Middle East. Adv. Virus Res 81:33-61.


Amoia, SS, Minafra, A, Nicoloso, V, Loconsole, G and Chiumenti, MA 2022. A new jasmine virus C isolate identified by nanopore sequencing is associated to yellow mosaic symptoms of Jasminum officinale in Italy. Plants 11:309.



Asjes, CJ 2000. Control of aphid-borne Lily symptomless virus and Lily mottle virus in Lilium in the Netherlands. Virus Res 71:23-32.


Barzon, L, Lavezzo, E, Militello, V, Toppo, S and Palù, G 2011. Applications of next-generation sequencing technologies to diagnostic virology. Int. J. Mol. Sci 12:7861-7884.



Batovska, J, Lynch, SE, Rodoni, BC, Sawbridge, TI and Cogan, NO 2017. Metagenomic arbovirus detection using MinION nanopore sequencing. J. Virol. Methods 249:79-84.


Ben Chehida, S, Filloux, D, Fernandez, E, Moubset, O, Hoareau, M, Julian, C, Blondin, L, Lett, J-M, Roumagnac, P and Lefeuvre, P 2021. Nanopore sequencing is a credible alternative to recover complete genomes of geminiviruses. Microorganisms 9:903.



Boykin, LM, Ghalab, A, De Marchi, BR, Savill, A, Wainaina, JM, Kinene, T, Lamb, S, Rodrigues, M, Kehoe, M, Ndunguru, J, Tairo, F, Sseruwagi, P, Kayuki, C, Mark, D, Erasto, J, Bachwenkizi, H, Alicai, T, Okao-Okuja, G, Abidrabo, P, Osingada, JF, Akono, J, Ateka, E, Muga, B and Kiarie, S 2018. Real time portable genome sequencing for global food security. bioRxiv preprint
https://doi.org/10.1101/314526
.

Boykin, LM, Sseruwagi, P, Alicai, T, Ateka, E, Mohammed, IU, Stanton, JL, Kayuki, C, Mark, D, Fute, T, Erasto, J, Bachwenkizi, H, Muga, B, Mumo, N, Mwangi, J, Abidrabo, P, Okao-Okuja, G, Omuut, G, Akol, J, Apio, HB, Osingada, F, Kehoe, MA, Eccles, D, Savill, A, Lamb, S, Kinene, T, Rawle, CB, Muralidhar, A, Mayall, K, Tairo, F and Ndunguru, J 2019. Tree lab: portable genomics for early detection of plant viruses and pests in sub-saharan africa. Genes 10:632.



Bronzato Badial, A, Sherman, D, Stone, A, Gopakumar, A, Wilson, V, Schneider, W and King, J 2018. Nanopore sequencing as a surveillance tool for plant pathogens in plant and insect tissues. Plant Dis 102:1648-1652.


Cao, Y, Li, J, Chu, X, Liu, H, Liu, W and Liu, D 2019. Nanopore sequencing: a rapid solution for infectious disease epidemics. Sci. China Life Sci 62:1101-1103.




Chalupowicz, L, Dombrovsky, A, Gaba, V, Luria, N, Reuven, M, Beerman, A, Lachman, O, Dror, O, Nissan, G and Manulis-Sasson, S 2019. Diagnosis of plant diseases using the Nanopore sequencing platform. Plant Pathol 68:229-238.


Chang, JJM, Ip, YCA, Bauman, AG and Huang, D 2020. MinION-in-ARMS: nanopore sequencing to expedite barcoding of specimen-rich macrofaunal samples from autonomous reef monitoring structures. Front. Mar. Sci 7:448.

Choi, SA and Ryu, KH 2003. The complete nucleotide sequence of the genome RNA of Lily symptomless virus and its comparison with that of other carlaviruses. Arch. Virol 148:1943-1955.



Deamer, D, Akeson, M and Branton, D 2016. Three decades of nanopore sequencing. Nat. Biotechnol 34:518-524.




Della Bartola, M, Byrne, S and Mullins, E 2020. Characterization of potato virus y isolates and assessment of nanopore sequencing to detect and genotype potato viruses. Viruses 12:478.



Eid, J, Fehr, A, Gray, J, Luong, K, Lyle, J, Otto, G, Peluso, P, Rank, D, Baybayan, P, Bettman, B, Bibillo, A, Bjornson, K, Chaudhuri, B, Christians, F, Cicero, R, Clark, S, Dalal, R, Dewinter, A, Dixon, J, Foquet, M, Gaertner, A, Hardenbol, P, Heiner, C, Hester, K, Holden, D, Kearns, G, Kong, X, Kuse, R, Lacroix, Y, Lin, S, Lundquist, P, Ma, C, Marks, P, Maxham, M, Murphy, D, Park, I, Pham, T, Phillips, M, Roy, J, Sebra, R, Shen, G, Sorenson, J, Tomaney, A, Travers, K, Trulson, M, Vieceli, J, Wegener, J, Wu, D, Yang, A, Zaccarin, D, Zhao, P, Zhong, F, Korlach, J and Turner, S 2009. Real-time DNA sequencing from single polymerase molecules. Science 323:133-138.


Fang, Y and Ramasamy, RP 2015. Current and prospective methods for plant disease detection. Biosensors 5:537-561.



Faria, NR, Quick, J, Claro, IM, Thézé, J, de Jesus, JG, Giovanetti, M, Kraemer, M, Hill, SC, Black, A, da Costa, AC, Franco, LC, Silva, SP, Wu, CH, Raghwani, J, Cauchemez, S, du Plessis, L, Verotti, MP, de Oliveira, WK, Carmo, EH, Coelho, GE, Santelli, ACFS, Vinhal, LC, Henriques, CM, Simpson, JT, Loose, M, Andersen, KG, Grubaugh, ND, Somasekar, S, Chiu, CY, Muñoz-Medina, JE, Gonzalez-Bonilla, CR, Arias, CF, Lewis-Ximenez, LL, Baylis, SA, Chieppe, AO, Aguiar, SF, Fernandes, CA, Lemos, PS, Nascimento, BLS, Monteiro, HAO, Siqueira, IC, de Queiroz, MG, de Souza, TR, Bezerra, JF, Lemos, MR, Pereira, GF, Loudal, D, Moura, LC, Dhalia, R, França, RF, Magalhães, T, Marques, ET Jr, Jaenisch, T, Wallau, GL, de Lima, MC, Nascimento, V, de Cerqueira, EM, de Lima, MM, Mascarenhas, DL, Neto, JPM, Levin, AS, Tozetto-Mendoza, TR, Fonseca, SN, Mendes-Correa, MC, Milagres, FP, Segurado, A, Holmes, EC, Rambaut, A, Bedford, T, Nunes, MRT, Sabino, EC, Alcantara, LCJ, Loman, NJ and Pybus, OG 2017. Establishment and cryptic transmission of Zika virus in Brazil and the Americas. Nature 546:406-410.




Fellers, JP, Webb, C, Fellers, MC, Shoup Rupp, J and De Wolf, E 2019. Wheat virus identification within infected tissue using nanopore sequencing technolog. Plant Dis 103:2199-2203.

Filloux, D, Fernandez, E, Loire, E, Claude, L, Galzi, S, Candresse, T, Winter, S, Jeeva, ML, Makeshkumar, T, Martin, DP and Roumagnac, P 2018. Nanopore-based detection and characterization of yam viruses. Sci. Rep 8:17879.




He, X, Xue, F, Xu, S and Wang, W 2016. Rapid and sensitive detection of Lily symptomless virus by reverse transcription loop-mediated isothermal amplification. J. Virol. Methods 238:38-41.


Jain, M, Fiddes, IT, Miga, KH, Olsen, HE, Paten, B and Akeson, M 2015. Improved data analysis for the MinION nanopore sequencer. Nat. Methods 12:351-356.




Jang, H and Kim, S 2016. Analysis of the economic value of the production of lily bulbs in Korea. Korean J. Agric. Sci 43:481-495 (in Korean).

Javaran, VJ, Moffett, P, Lemoyne, P, Xu, D, Adkar-Purushothama, CR and Fall, ML 2021. Grapevine virology in the third-generation sequencing era: from virus detection to viral epitranscriptomics. Plants 10:2355.



Jo, Y and Cho, WK 2018. RNA viromes of the oriental hybrid lily cultivar “Sorbonne”. BMC Genomics 19:748.




Kang, Y-I, Joung, HY, Goo, DH, Choi, YJ, Choi, MP, An, HR, Ko, J-Y, Choi, K-J, Lee, KH and Hong, KW 2013. A survey on cut flower cultivar trends and horticultural status of lilies (Lilium hybrids) in South Korea. HortTechnology 23:629-634.

Kim, JH, Yoo, HN, Bae, EH and Jung, Y-T 2012. Development of an indirect ELISA and immunocapture rt-PCR for lily virus detection. J. Microbiol. Biotechnol 22:1776-1781.


Komatsu, K, Yamaji, Y, Ozeki, J, Hashimoto, M, Kagiwada, S, Takahashi, S and Namba, S 2008. Nucleotide sequence analysis of seven Japanese isolates of Plantago asiatica mosaic virus (PlAMV): a unique potexvirus with significantly high genomic and biological variability within the species. Arch. Virol 153:193-198.



Kumar, S, Stecher, G, Li, M, Knyaz, C and Tamura, K 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol 35:1547-1549.



Kwon, JY, Ryu, KH and Choi, SH 2013. Reverse transcription polymerase chain reaction-based system for simultaneous detection of multiple lily-infecting viruse. Plant Pathol. J 29:338-343.


Lewandowski, K, Xu, Y, Pullan, ST, Lumley, SF, Foster, D, Sanderson, N, Vaughan, A, Morgan, M, Bright, N, Kavanagh, J, Vipond, R, Carroll, M, Marriott, AC, Gooch, KE, Andersson, M, Jeffery, K, Peto, T, Crook, DW, Walker, AS and Matthews, PC 2019. Metagenomic nanopore sequencing of influenza virus direct from clinical respiratory samples. J. Clin. Microbiol 58:e00963-e00919.



Li, Y, Jia, A, Qiao, Y, Xiang, J, Zhang, Y and Wang, W 2018. Virome analysis of lily plants reveals a new potyvirus. Arch. Virol 163:1079-1082.



Liefting, LW, Waite, DW and Thompson, JR 2021. Application of oxford nanopore technology to plant virus detection. Viruses 13:1424.



Lim, MS, Kim, SM and Choi, SH 2016. Simultaneous detection of three lily-infecting viruses using a multiplex Luminex bead array. J. Virol. Methods 231:34-37.


Maestri, S, Cosentino, E, Paterno, M, Freitag, H, Garces, JM, Marcolungo, L, Alfano, M, Njunjić, I, Schilthuizen, M, Slik, F, Menegon, M, Rossato, M and Delledonne, M 2019. A rapid and accurate MinION-based workflow for tracking species biodiversity in the field. Genes 10:468.



Marcolungo, L, Passera, A, Maestri, S, Segala, E, Alfano, M, Gaffuri, F, Marturano, G, Casati, P, Bianco, PA and Delledonne, M 2022. Real-time on-site diagnosis of quarantine pathogens in plant tissues by nanopore-based sequencing. Pathogens 11:199.



Mehetre, GT, Leo, VV, Singh, G, Sorokan, A, Maksimov, I, Yadav, MK, Upadhyaya, K, Hashem, A, Alsaleh, AN, Dawoud, TM, Almaary, KS and Singh, BP 2021. Current developments and challenges in plant viral diagnostics: a systematic review. Viruses 13:412.



Naito, F, Melo, FL, Fonseca, M, Santos, C, Chanes, CR, Ribeiro, BM, Gilbertson, RL, Boiteux, LS and de Cássia Pereira-Carvalho, R 2019. Nanopore sequencing of a novel bipartite New World begomovirus infecting cowpea. Arch. Virol 164:1907-1910.



Niimi, Y, Han, D-S, Mori, S and Kobayashi, H 2003. Detection of cucumber mosaic virus, lily symptomless virus and lily mottle virus in Lilium species by RT-PCR technique. Sci. Hortic 97:57-63.

Phannareth, T, Nunziata, SO, Stulberg, MJ, Galvez, ME and Rivera, Y 2020. Comparison of nanopore sequencing protocols and real-time analysis for phytopathogen diagnostics. Plant Health Prog 22:31-36.

Quick, J, Loman, NJ, Duraffour, S, Simpson, JT, Severi, E, Cowley, L, Bore, JA, Koundouno, R, Dudas, G, Mikhail, A, Ouédraogo, N, Afrough, B, Bah, A, Baum, JH, Becker-Ziaja, B, Boettcher, JP, Cabeza-Cabrerizo, M, Camino-Sanchez, A, Carter, LL, Doerrbecker, J, Enkirch, T, Dorival, IGG, Hetzelt, N, Hinzmann, J, Holm, T, Kafetzopoulou, LE, Koropogui, M, Kosgey, A, Kuisma, E, Logue, CH, Mazzarelli, A, Meisel, S, Mertens, M, Michel, J, Ngabo, D, Nitzsche, K, Pallash, E, Patrono, LV, Portmann, J, Repits, JG, Rickett, NY, Sachse, A, Singethan, K, Vitoriano, I, Yemanaberhan, RL, Zekeng, EG, Trina, R, Bello, A, Sall, AA, Faye, O, Faye, O, Magassouba, N, Williams, CV, Amburgey, V, Winona, L, Davis, E, Gerlach, J, Washington, F, Monteil, V, Jourdain, M, Bererd, M, Camara, A, Somlare, H, Camara, A, Gerard, M, Bado, G, Baillet, B, Delaune, D, Nebie, KY, Diarra, A, Savane, Y, Pallawo, RB, Gutierrez, GJ, Milhano, N, Roger, I, Williams, CJ, Yattara, F, Lewandowski, K, Taylor, J, Rachwal, P, Turner, D, Pollakis, G, Hiscox, JA, Matthews, DA, O’Shea, MK, Johnston, AM, Wilson, D, Hutley, E, Smit, E, Di Caro, A, Woelfel, R, Stoecker, K, Fleischmann, E, Gabriel, M, Weller, SA, Koivogui, L, Diallo, B, Keita, S, Rambaut, A, Formenty, P, Gunther, S and Carroll, MW 2016. Real-time, portable genome sequencing for Ebola surveillance. Nature 530:228-232.




Ryu, KH, Park, HW and Park, JK 2002. Characterization and sequence analysis of a lily isolate of Cucumber mosaic virus from Lithium tsingtauense
. Plant Pathol J 18:85-92.

Sharma, A, Mahinghara, BK, Singh, AK, Kulshrestha, S, Raikhy, G, Singh, L, Verma, N, Hallan, V, Ram, R and Zaidi, AA 2005. Identification, detection and frequency of lily viruses in Northern India. Sci. Hortic 106:213-227.

Tamura, K, Nei, M and Kumar, S 2004. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. U. S. A 101:11030-11035.



Tarquini, G, Martini, M, Maestri, S, Firrao, G and Ermacora, P 2022. The virome of ‘Lamon Bean’: application of MinION sequencing to investigate the virus population associated with symptomatic beans in the Lamon Area, Italy. Plants 11:779.



Temmam, S, Davoust, B, Berenger, JM, Raoult, D and Desnues, C 2014. Viral metagenomics on animals as a tool for the detection of zoonoses prior to human infection? Int. J. Mol. Sci 15:10377-10397.



Theuns, S, Vanmechelen, B, Bernaert, Q, Deboutte, W, Vandenhole, M, Beller, L, Matthijnssens, J, Maes, P and Nauwynck, HJ 2018. Nanopore sequencing as a revolutionary diagnostic tool for porcine viral enteric disease complexes identifies porcine kobuvirus as an important enteric virus. Sci. Rep 8:9830.




Vazquez-Iglesias, I, McGreig, S, Pufal, H, Robinson, R, Clover, GRG, Fox, A, Boonham, N and Adams, IP 2022. A novel high-throughput sequencing approach reveals the presence of a new virus infecting Rosa: rosa ilarvirus-1 (RIV-1). J. Virol. Methods 300:114417.


Wang, M, Fu, A, Hu, B, Tong, Y, Liu, R, Liu, Z, Gu, J, Xiang, B, Liu, J, Jiang, W, Shen, G, Zhao, W, Men, D, Deng, Z, Yu, L, Wei, W, Li, Y and Liu, T 2020. Nanopore targeted sequencing for the accurate and comprehensive detection of SARS-CoV-2 and other respiratory viruses. Small 16:e2002169.




Wang, R, Wang, G, Zhao, Q, Zhu, Y, Zhan, J, Xie, Z, An, L and Wang, Y 2010. Molecular and cytopathologic evidences for a mixed infection of multiple viruses on Lanzhou lily (Lilium davidii var. unicolor) in Northwestern China. J. Plant Dis. Prot 117:145-149.


Zhang, Y, Wang, Y, Meng, J, Xie, Z, Wang, R, Kutcher, HR and Guo, Z 2015a. Development of an immunochromatographic strip test for rapid detection of lily symptomless virus. J. Virol. Methods 220:13-17.


Zhang, Y, Wang, Y, Xie, Z, Wang, R, Guo, Z and He, Y 2020. Rapid detection of Lily mottle virus and Arabis mosaic virus infecting lily (Lilium spp.) using reverse transcription loop-mediated isothermal amplification. Plant Pathol. J 36:170-178.




Zhang, Y, Wang, Y, Xie, Z, Yang, G, Guo, Z and Wang, L 2017. Simultaneous detection of three lily viruses using triplex IC-RT-PCR. J. Virol. Methods 249:69-75.


Zhang, Y, Wang, Y, Yang, W, Xie, Z, Wang, R, Kutcher, HR and Guo, Z 2015b. A rapid immunochromatographic test to detect the lily mottle virus. J. Virol. Methods 220:43-48.


Zhang, Y, Xie, Z, Wang, Y, Wang, R, Guo, Z, Qiu, Y and He, Y 2022. Development of immunocapture reverse transcription loop-mediated isothermal amplification assays to detect both lily symptomless virus and cucumber mosaic virus. Ann. Appl. Biol 180:132-139.


- TOOLS
-
METRICS

- ORCID iDs
-
Rae-Dong Jeong

https://orcid.org/0000-0002-6370-8909 - Related articles




 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Supplement
Supplement Print
Print



