 |
 |
| Plant Pathol J > Volume 38(5); 2022 > Article |
|
Abstract
Gummy stem blight (GSB), a common and serious disease in cucurbits worldwide, is caused by three genetically distinct species: Stagonosporopsis cucurbitacearum (syn. Didymella bryoniae), S. citrulli, and S. caricae. In Korea, however, the three species of Stagonosporopsis have been barely characterized. In this study, 21 Stagonosporopsis isolates were recovered from watermelon (Citrullus lanatus) and muskmelon (Cucumis melo) leaves and stem showing blight symptoms collected from 43 fields in Korea. Sequence analysis performed with an internal transcribed spacer region was not competent to differentiate the Stagonosporopsis isolates. On the contrary, analysis of β-tubulin (TUB) genes and three microsatellite markers, Db01, Db05, and Db06, successfully differentiated Stagonosporopsis isolates. Further sequence analysis identified two Stagonosporopsis species, S. citrulli and S. caricae, and one previously unknown species of Stagonosporopsis. Representative isolates from three species caused dark water-soaked lesions on the detached watermelon and muskmelon leaves with no significant differences in the aggressiveness. Our results indicate that the S. citrulli, S. caricae, and unknown Stagonosporopsis sp. are all causal agents of GSB for both watermelon and muskmelon. This is the first report of a new species and the population structure of Stagonosporopsis species causing GSB in Korea.
Watermelon (Citrullus lanatus Thunb.) and muskmelon (Cucumis melo L.), members of the Cucurbitaceae family (cucurbits), are economically important fruit crops, and their annual production in 2019 was estimated more than 120 million tons worldwide (Food and Agriculture Organization of the United Nations, 2019). It is widely grown in the most parts of Africa, the Southern United States, and Southeast Asia (Ma et al., 2021).
Gummy stem blight (GSB), also known as black rot when affecting fruits, is a devasting disease and a major production constraint of the Cucurbitaceae family (Wolukau et al., 2007). This disease was first observed on Cucumis sativus (cucumber) in France and on watermelon in the United States in 1892 (Gusmini et al., 2017) and now assumed to be distributed worldwide. Disease symptoms include foliar brown discoloration and necrotic lesions on the infected stems and fruits (Basim et al., 2016; Li et al., 2015). Fungicide applications are the main method to control GSB on cucurbits (Keinath, 2021). Therefore, identification of causal agents of the GSB is essential for choosing effective chemicals and setting up disease-management programs.
Didymella bryoniae was initially reported as a fungal pathogen causing the GSB on cucurbits worldwide including China, the United States, and Korea (Jensen et al., 2011; Koike, 1997; Kwon et al., 1997; Ling et al., 2010; Sudisha et al., 2004). Later, Stagonosporopsis cucurbitacearum was described as a synonym of D. bryoniae causing GSB (Aveskamp et al., 2010). However, a multi-locus sequencing study of diverse isolates of Stagonosporopsis species showed that GSB is caused by three morphologically similar, but genetically distinct Stagonosporopsis spp., S. cucurbitacearum, S. citrulli, and S. caricae (Brewer et al., 2015; Stewart et al., 2015). During the reclassification of the Stagonosporopsis spp., some of phylogenetically distinct isolates, previously classified as S. cucurbitacearum, were reassigned to the new species, S. citrulli (Stewart et al., 2015). One of the species, S. caricae, known to affect only papaya (Carica papaya) (Boerema et al., 2004; Brewer et al., 2015) was reported to be also aggressive on cucurbits, displaying a broader host range, and became a causal agent of GSB (Stewart et al., 2015). Previous population studies have shown that they vary in their geographic distribution worldwide (Garampalli et al., 2016; Stewart et al., 2015). S. citrulli was the most abundant species with the widest geographic distribution, and S. caricae was also widely distributed. However, S. cucurbitacearum has been only found in temperate regions (Garampalli et al., 2016; Stewart et al., 2015). To consistently compare GSB populations, 18 polymorphic microsatellite markers were developed, and three of them—Db01, Db05, and Db06—were successfully applied for polymerase chain reaction (PCR)-based markers for distinguishing Stagonosporopsis spp. (Brewer et al., 2015).
Causal agents of GSB have been commonly misidentified. For example, because S. caricae was described as infecting only papaya (Boerema et al., 2004; Brewer et al., 2015), some isolates of S. caricae infecting cucurbits had been misidentified as S. cucurbitacearum (Brewer et al., 2015). The host species of origin and similar morphology make many isolates of S. citrulli also misidentified as S. cucurbitacearum. Therefore, isolates previously identified as S. cucurbitacearum should be reidentified on the basis of its phylogenetic diversity. In addition, population structure of the GSB pathogens and diversity within and among the populations at the field level need to be determined to better understand the Stagonosporopsis spp. and set up GSB management strategies.
To our knowledge, there are only a few studies carrying out a molecular identification of Stagonosporopsis spp. causing GSB worldwide, and very few has been reported in Korea. Here, we describe the population structure of Stagonosporopsis spp. including one previously unknown species associated with GSB in Korea.
From 2019 to 2022, a total of 43 fields from 14 cities among the watermelon and muskmelon production area in Korea were selected for the survey of causal agents for GSB. Leaves and stems of watermelon and muskmelon showing GSB were harvested. Small pieces of tissues (10 × 10 mm) were removed from the margins between the healthy and symptomatic tissues and their surface were disinfected with 70% ethanol for 1 min, followed by rinsing with sterilized dH2O three times. The pieces were then placed on potato dextrose agar (PDA) (MBcell, Seoul, Korea) plates and incubated at 28°C in the light for 7 days. Multiple fungal colonies were obtained from each symptomatic tissue and only those with Stagonosporopsis morphologies were selected as isolates. The isolates were sub-cultured onto fresh PDA media for single spore purification.
The genomic DNAs were extracted following the protocol described previously (Chen and Ronald, 1999). For the extraction, Stagonosporopsis mycelia were scrapped from the surface of 7-day cultures grown on PDA and placed in a 2 ml microcentrifuge tube containing 700 μl cetyltrimethl-ammonium bromide (CTAB) extraction buffer (3% (w/v) CTAB, 20 mM EDTA pH 8.0, 1.42 M NaCl, 100 mM Tris-HCl pH 8.0, 2% (w/v) polyvinylptrrolidone, and 0.2% β-mercaptoethanol). After incubating for 10 min at 65°C, adding 570 μl of chloroform:isoamyl alcohol (24:1) to the tube, and centrifuging for 10 min at 13,000 ×g, genomic DNA was precipitated with 0.7 volumes of isopropanol. After centrifuging for 10 min at 13,000 ×g, the genomic DNA pellet was washed with 70% ethanol and then dried at room temperature. The genomic DNA was dissolved in 30 μl of sterilized dH2O and stored at −20°C.
The primers for the internal transcribe spacer (ITS) region of ribosomal DNA, β-tubulin (TUB) gene, and three microsatellite markers (Db01, Db05, and Db06) were used for PCRs (Table 1). PCR was performed in a 20 μl reaction mixture containing 0.1 μl FIREPol DNA polymerase (0.5 units) (Solis BioDyne, Tartu, Estonia), 1 μl each primer (10 μM), 0.2 μl dNTP mix (200 μM), 2 μl 10× buffer, 1 μl template (approximately 100 ng), 4 μl 5× Q-solution, and 10.7 μl sterilized dH2O. Amplifications were performed with an initial denaturation step at 94°C for 3 min, followed by 35 cycles of 15 s at 94°C, 15 s at 53°C and 40 s at 72°C for ITS region, 35 cycles of 15 s at 94°C, 15 s at 54°C and 90 s at 72°C for TUB gene, 35 cycles of 15 s at 94°C, 15 s at 53°C and 30 s at 72°C for three microsatellite markers (Db01, Db05, and Db06) and final extension at 72°C for 7 min. Amplified fragments were loaded on an agarose gel (0.8% w/v or 1.5% w/v) stained with RedSafe Nucleic Acid Staining Solution (20,000×) (Intronbio, Seongnam, Korea) and visualized under UV light.
The morphology of Stagonosporopsis isolates was examined as previously described (Choi et al., 2010) with minor modification. For colony type, color, and margin, Stagonosporopsis isolates were incubated on PDA plates in a 28°C dark condition for 5 days. For spore observation, spores were recovered from watermelon cultivar ‘Seotaja’ (Lee et al., 2016) and muskmelon cultivar ‘Earls top one’ (Hassan et al., 2018) inoculated with Stagonosporopsis isolates, filtrated with sterilized gauze (Dong-A Health Care, Gwangju, Korea), and precipitated by centrifuging at 6,000 ×g for 1 min. Harvested spores were resuspended with 100 μl dH2O and examined using an inverted microscope Eclipse Ti (Nikon, Tokyo, Japan) fitted with an objective (200×).
The PCR amplification products of ITS region, TUB, and microsatellite marker Db05 were purified with FavorPrep GEL/PCR Purification Kit (Favorgen, Ping-Tung, Taiwan) and sequenced by Sanger sequencing (Macrogen, Seoul, Korea). Sequence similarity searches were performed using the nucleotide BLAST at National Center for Biotechnology Information (NCBI, https://blast.ncbi.nlm.nih.gov/Blast.cgi). DNA sequences of ITS region, TUB, and Db05 were aligned through ClustalW and refined manually using MEGA X (Kumar et al., 2016, 2018). Maximum likelihood based on the Jukes-Cantor model of ITS region, TUB, and Db05 was performed using MEGA X with 500 bootstrap replicates. Branches corresponding to partitions reproduced in less than 50%. The phylogenetic trees were viewed in MEGA X. All analyzed DNA sequences of ITS region and TUB from isolates were deposited in GenBank (https://www.ncbi.nlm.nih.gov/genbank/), and their accession numbers are listed in Table 2.
Stagonosporopsis isolates were inoculated according to the previously described detached-leaf inoculation method (Pryor and Michailides, 2002) with minor modification. Briefly, to prepare inoculum, Stagonosporopsis isolates were pure-cultured on PDA plates at 28°C in the dark for 7 days, and each mycelial plug was cut from the actively growing margin of the fungal colonies. Fully expanded leaves were collected from 6- to 7-week-old watermelon cultivar ‘Seotaja’ and muskmelon cultivar ‘Earls top one’ grown in a growth chamber. For the inoculation of each Stagonosporopsis isolate, three leaves from each cultivar were placed on papers soaked with distilled water in plastic boxes (25 × 19 × 5 cm [length × width × height]), and one mycelial plug was placed on each detached leaf. The plastic boxes were individually covered with transparent plastics to maintain 100% humidity and incubated at 25°C with a 12 h photoperiod per day for 7 days. Disease severity was calculated after each disease area was divided by whole leaf area using ImageJ. Stagonosporopsis isolates were re-isolated consistently from all the inoculated leaf to fulfill Koch’s postulates.
Leaves and stems of watermelon and muskmelon displaying GSB symptoms were collected from 43 farms distributed in 14 cities of Korea (Table 2, Fig. 1A). Multiple fungal colonies were obtained from each symptomatic tissue (Fig. 1B) and only those with Stagonosporopsis morphologies were selected as isolates and cultured to homogeneity.
ITS region of nuclear ribosomal RNA has been used to identify Stagonosporopsis spp. (Choi et al., 2010, 2015; Stewart et al., 2015; Zhang et al., 2019). The ITS regions of putative Stagonosporopsis isolates were amplified with primers, ITS1 and ITS4 (Table 1, Supplementary Fig. 1), and the sequences of each amplicon were compared with the NCBI database using the blastn algorithm. Blast results of the ITS sequences demonstrated that a total of 21 isolates belong to the Stagonosporopsis genus (Table 2). Phylogenetic analysis based on the ITS region was conducted with references retrieved from GenBank (5 from S. cucurbitacearum, 9 from S. citrulli, 3 from S. caricae, and 1 outgroup S. heliopsidis) by maximum likelihood based on the Jukes-Cantor model using MEGA X software (Supplementary Fig. 2). Four Korean Agriculture Culture Collection (KACC) strains previously identified as D. bryoniae, KACC 40937, KACC 40669, KACC 40900, KACC 40939 were included in the phylogenetic analysis. Although S. caricae references were grouped into one clade, all other references including S. cucurbitacearum and S. citrulli, were not successful in dividing into different clades. These results suggest that sequences from the ITS region alone cannot clearly discriminate between the Stagonosporopsis species.
The TUB gene has been also commonly used for inferring phylogenetic relationships among Stagonosporopsis spp. (Aveskamp et al., 2010; Stewart et al., 2015; Zhang et al., 2019). Therefore, after PCR amplification of TUB using a primer set, TUB-T1_F and TUB_T22_R (Table 1, Supplementary Fig. 3), TUB sequences from all isolates and four KACC strains and the references retrieved from GenBank were subjected to TUB-based phylogenetic analyses (Supplementary Fig. 3). All references in phylogenetic tree were placed within three different monophyletic clades with isolates together, indicating that the TUB gene is more useful than ITS region for assaying genetic diversity in the Stagonosporopsis spp.
Multi-locus analysis is often more useful for species classification and is successfully applied for many fungal pathogens (Aveskamp et al., 2010). Therefore, multi-locus phylogenetic analysis of ITS and TUB sequences from all Stagonosporopsis isolates and four KACC strains was conducted together with reference sequences retrieved from GenBank (Fig. 2). Twelve isolates, including YG211-10, YG211-5, GR213-3, GR214-1, GR213-2, GR212-29, GR212-27, CC203-17, CW211-2, CW211-3, CW211-11, and YP216-4, and four KACC strains previously identified as D. bryoniae, were located in one clade with reference S. citrulli. Six isolates, GC192-11, CW211-1, YG211-13, YP216-8, CA215-1, and CA217-1, were together with S. caricae. However, none of our isolates was located with S. cucurbitacearum. Interestingly, there was one clade consisting of three isolates, GJ201-3, YG211-4, and CW214-3, which did not belong to the three Stagonosporopsis spp. known to cause GSB, suggesting that they could be an unknown species of Stagonosporopsis causing GSB in Korea.
Additional multi-locus phylogenetic analysis of ITS and TUB sequences was conducted using all 21 species of the genus Stagonosporopsis available in GenBank (Aveskamp et al., 2010; Brewer et al., 2015). The unknown species was accurately placed within the Stagonosporopsis. However, it belonged to none of species in the Stagonosporopsis genus, suggesting that the unknown species is a new species of Stagonosporopsis (Supplementary Fig. 4).
To further validate the species identification performed by multi-locus phylogenetic analysis, three PCR-based microsatellite markers, Db01, Db05, and Db06 (Brewer et al., 2015), were applied to all Stagonosporopsis isolates and four KACC strains (Fig. 3). In a previous report, S. citrulli displays three PCR amplicons of approximately 220, 280, and 360 bp produced by Db05, Db06, and Db01, respectively (Brewer et al., 2015). S. cucurbitacearum produces two PCR amplicons of approximately 220 and 270 bp Db05 and Db06, respectively. In S. caricae, one PCR amplicon is amplified approximately 220 bp by Db05. In this study, all KACC strains and 12 isolates, including YG211-10, YG211-5, GR213-3, GR214-1, GR213-2, GR212-29, GR212-27, CC203-17, CW211-2, CW211-3, CW211-11, and YP216-4 produced three PCR amplicons, which is typical of S. citrulli. In six isolates, GC192-11, CW211-1, YG211-13, YP216-8, CA215-1, and CA217-1, single amplicon of without additional bands was amplified, confirming them as S. caricae. Three isolates, GJ201-3, YG211-4, and CW214-3, not grouped with three Stagonosporopsis spp., known to GSB causal agents, produced two amplicons.
To gain insight into variation within and among the three Stagonosporopsis spp. and further investigate the unknown species, we sequenced Db05 amplicons, which are only PCR products obtained from all isolates and KACC strains, and then compared them with references (Fig. 4). DB05 sequences of the references were retrieved from GenBank (Brewer et al., 2015). Similar to multi-locus phylogenetic analysis (Fig. 2), twelve and six isolates were together with S. citrulli and S. caricae, respectively. Again, three unknown species isolates, GJ201-3, YG211-4, and CW214-3, were clustered into one clade, while S. cucurbitacearum references form distinct clades of different evolutionary origin. In the Db05 alignment, there was no variation within S. caricae isolates and S. caricae reference RG3, and within S. citrulli isolates and S. citrulli reference 09-090-1 (Supplementary Fig. 5). There was only one insertion/deletion (indel) (4 bp) and one single nucleotide polymorphism (SNP) within unknown species isolates. However, the unknown species isolate (CW214-3) differed from S. cucurbitacearum (reference RT3) by 33 nucleotide polymorphisms (SNPs) and three indels (5, 7, and 6 bp). This large diversity suggests that the unknown species is evolutionarily different from S. cucurbitacearum.
Morphological characteristics of the 21 Stagonosporopsis isolates, including 12 S. citrulli, 6 S. caricae, and 3 unknown species isolates, were examined on PDA plates. Light-white aerial mycelium was observed on top and white to olivaceous on the bottom in all isolates (Fig. 5A). There was no obvious difference among each isolate except that isolates of S. citrulli presented slightly less intense mycelial growth compared to S. caricae and unknown species. Conidia obtained from each isolate were observed under a light microscope, and three representatives—CC203-17 (S. citrulli), GC192-11 (S. caricae), and GJ201-3 (unknown)—were further analyzed (Fig. 5B). The conidia were round-ended, cylindrical, monoseptate, and hyaline, and their length and diameter ranged from approximately 3.5 to 11.1 and 1.2 to 3.9 μm, respectively.
The pathogenicity of isolates was evaluated on watermelon cultivar ‘Seotaja’ and muskmelon cultivar ‘Earls top one’ and disease symptoms caused by 14 representative isolates were presented (Supplementary Fig. 6). Visible symptoms of the leaves started 3 days after inoculation as dark brown lesions. The symptoms around the inoculation sites increased as blackish-brown water-soaked lesions. One week after inoculation, 100% of inoculated detached leaves of watermelon and muskmelon exhibited rapidly growing necrotic lesions with gray mycelium growth. The symptomatic tissues were cultured again on PDA, and the same pathogens were re-isolated confirming Koch’s postulates. Mock-inoculated leaves remained symptomless. To determine if the three species differed in aggressiveness, disease severity caused by three representative isolates—CC203-17 (S. citrulli), GC192-11 (S. caricae), and GJ201-3 (unknown)—from different species was quantified using ImageJ (Fig. 6). At 7 days after inoculation, there were no significant differences in aggressiveness of the three isolates on both watermelon and muskmelon.
The genus Stagonosporopsis causing GSB in cucurbits has been characterized relatively recently (Brewer et al., 2015; Stewart et al., 2015). S. citrulli was the most abundant species with the widest geographic distribution, and S. caricae was also distributed worldwide, whereas S. cucurbitacearum has been only found in temperate regions (Stewart et al., 2015). Given that causal agents of GSB have been reported as D. bryoniae until recently (Basim et al., 2016; Choi et al., 2010; Lee et al., 2016) and often misidentified as S. cucurbitacearum (Brewer et al., 2015), a careful study about the diversity and population structure of different GSB pathogens is necessary.
In this study, a multi-locus phylogenetic analysis of ITS and TUB sequences revealed that S. citrulli is the dominant species for the GSB and S. caricae is also relatively common in Korea. However, S. cucurbitacearum was not identified; instead, there was one clade consisting of three isolates, GJ201-3, YG211-4, and CW214-3, that did not belong to Stagonosporopsis spp., known to cause GSB. Additional multi-locus analysis conducted using all 19 species of the genus Stagonosporopsis available in GenBank displayed that although the three isolates accurately placed within the genus Stagonosporopsis, they were distinct from any previously known species of Stagonosporopsis. Our results were further confirmed using PCR-based microsatellite markers, Db01, DB05, and DB06. In addition, after sequence analysis of Db05 amplicons obtained from all isolates and references, we found that there is great diversity between the three isolates and other previously known GSB causal Stagonosporopsis spp. Therefore, we concluded that the isolates are the previously unknown Stagonosporopsis species causing GSB on both watermelon and muskmelon.
It was previously reported that although differences in pathogenicity among three species—S. citrulli, S. cucurbitacearum, and S. caricae—to papaya was detected, there were no differences in the aggressiveness of the species on cucurbit hosts including watermelon, Cucurbita moschata (pumpkin), and cucumber (Stewart et al., 2015). We were also unable to determine the differences in aggressiveness among S. citrulli, S. caricae, and unknown Stagonosporopsis species, to watermelon cultivar ‘Seotaja’ and muskmelon cultivar ‘Earls top one’. However, additional further experiments with various potential hosts need to be performed, because they could vary in pathogenicity to other species and different cultivars.
In the study, we described Stagonosporopsis spp. infecting watermelon and muskmelon in Korea from 2019 to 2022. Pathogenicity tests with isolates of S. citrulli and S. caricae as well as unknown Stagonosporopsis species confirmed that all three species were able to cause GSB on watermelon and muskmelon, thereby completing Koch’s postulates. Correct identification of these pathogens will facilitate the management strategies to prevent damage caused by GSB in cucurbits.
Acknowledgments
This study was carried out with the support of the “Cooperative Research Program for Agriculture Science & Technology Development (Project No. PJ01421303)” Rural Development Administration, Republic of Korea. We thank Marin Talbot Brewer (University of Georgia, Athens, USA) for sharing the sequences of Db05 amplicons obtained from Stagonosporopsis spp.
Electronic Supplementary Material
Supplementary materials are available at The Plant Pathology Journal website (http://www.ppjonline.org/).
Fig. 1
Watermelon and muskmelon farm locations and representative gummy stem blight (GSB) symptoms on leaves and stems. (A) Map showing 14 cities and counties in Korea. (B) Typical GSB displaying brown discoloration and necrotic lesions on watermelon and muskmelon leaves and stems. Arrowheads indicate typical symptoms of GSB. GSB causal agents were not isolated from a, b, d, f, g, and l.

Fig. 2
Multi-locus phylogenetic analysis based on the internal transcribed space (ITS) and β-tubulin (TUB) genes. Twenty-one Stagonosporopsis isolates, four Dydimella bryoniae Korean Agriculture Culture Collection (KACC) strains, and 17 Stagonosporopsis reference (nine S. citrulli, five S. cucurbitacearum, and three S. caricae) were analyzed. All Stagonosporopsis reference sequences were retrieved from GenBank, and their GenBank accession numbers are shown after the name of each reference (ITS + TUB). Black circles indicate Stagonosporopsis isolates isolated in this study. Red circles indicate four D. bryoniae strains from KACC. Others without circles are references for S. citrulli, S. cucurbitacearum, and S. caricae. S. heliopsidis was used as outgroup. For each branch, bootstrap support values obtained by maximum likelihood are indicated based on 500 replicates. Bootstrap support values less than 50% are not shown.

Fig. 3
Analysis of Stagonosporopsis isolates using microsatellite markers. Gel electrophoresis analysis after polymerase chain reaction using microsatellite markers, Db01, Db05, and Db06 (Table 1). Twenty-one Stagonosporopsis isolates and four Dydimella bryoniae Korean Agriculture Culture Collection (KACC) strains were analyzed. Alternaria alternata was used as a negative control.

Fig. 4
Phylogenetic tree analysis based on Db05 sequence of 21 Stagonosporopsis isolates, four Korean Agriculture Culture Collection (KACC) strains, and 17 Stagonosporopsis reference (nine S. citrulli, five S. cucurbitacearum, and three S. caricae). Sequences of Db05 amplicons from Stagonosporopsis references were obtained from a previous study (Brewer et al., 2015). Black circles indicate Stagonosporopsis isolates isolated in this study. Red circles indicate KACC strains. Others are references for S. citrulli, S. cucurbitacearum, and S. caricae. For each branch, bootstrap support values obtained by maximum likelihood are indicated based on 500 replicates. Bootstrap support values less than 50% are not shown.
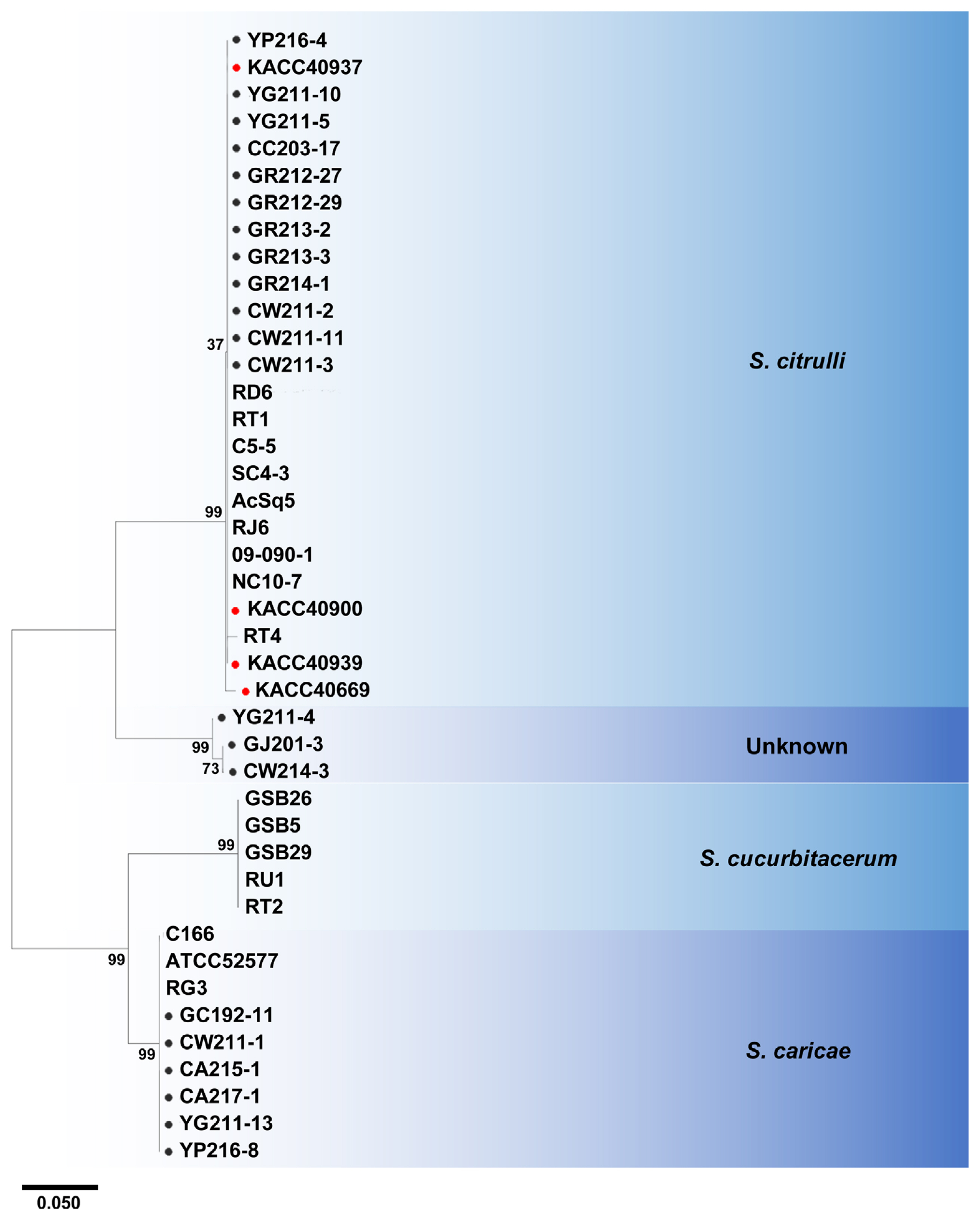
Fig. 5
Colonies of Stagonosporopsis isolates. (A) Pure cultures and morphological characteristics of all Stagonosporopsis isolates on potato dextrose agar (PDA) (front and reverse) 7 days after incubation at 28°C under continuous light condition. (B) Conidial morphology of S. citrulli GR213-3, S. caricae CW211-1, and unknown GJ214-3 on PDA media. Scale bar = 50 μm.
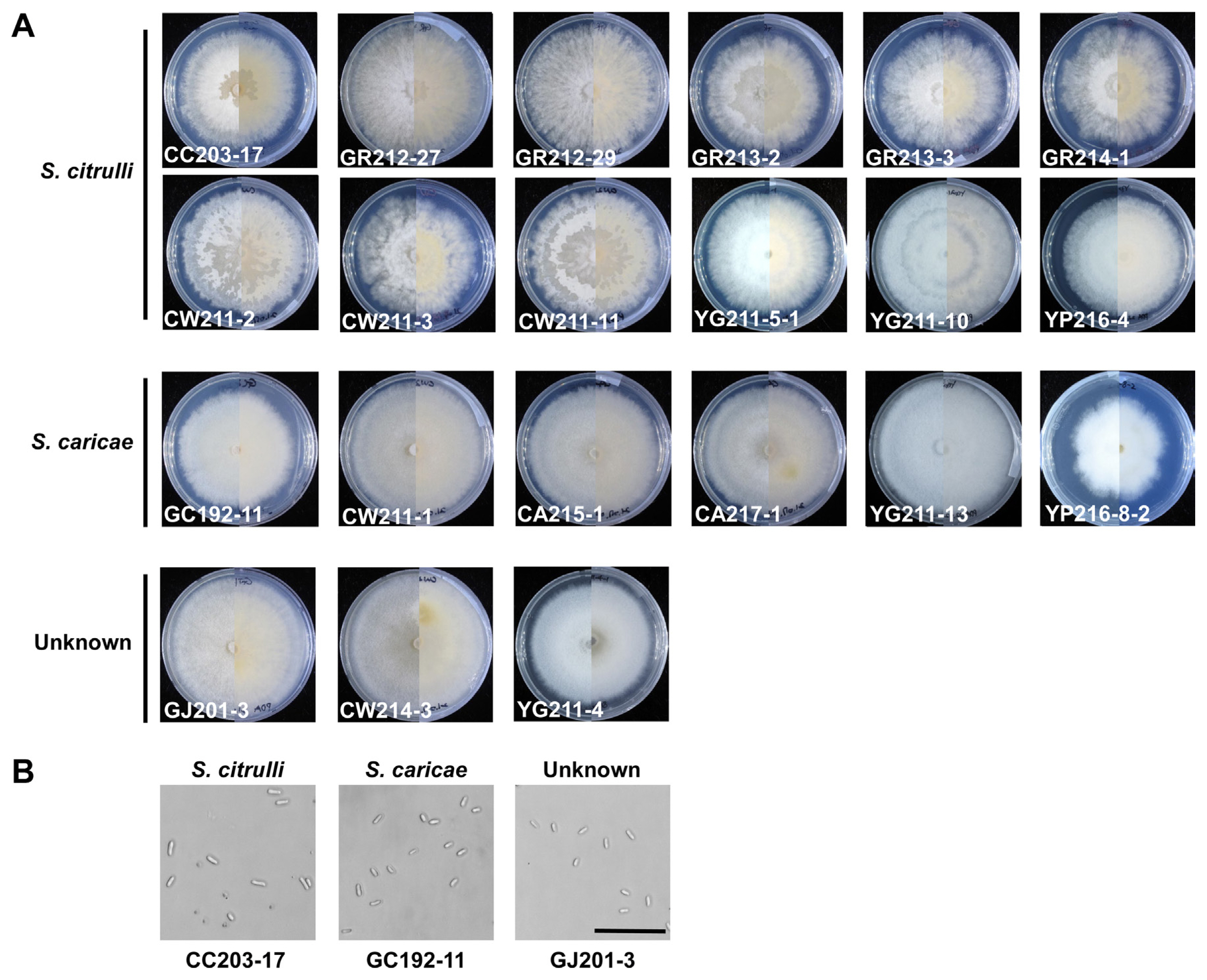
Fig. 6
Disease severity of three representative isolates of three Stagonosporopsis species on detached watermelon or muskmelon leaves. (A) One agar disk growing each Stagonosporopsis species was placed on the detached of watermelon cultivar ‘Seotaja’ and muskmelon cultivar ‘Earls top one’. Three leaves from each cultivar were inoculated, and images were obtained 7 days after inoculation. Scale bars = 3.0 cm. (B) Disease severity was calculated after each disease area was divided by whole leaf area using ImageJ. The data are the mean ± SE of at least three independent replicates. The values with the same letter are not significantly different at P<0.05.
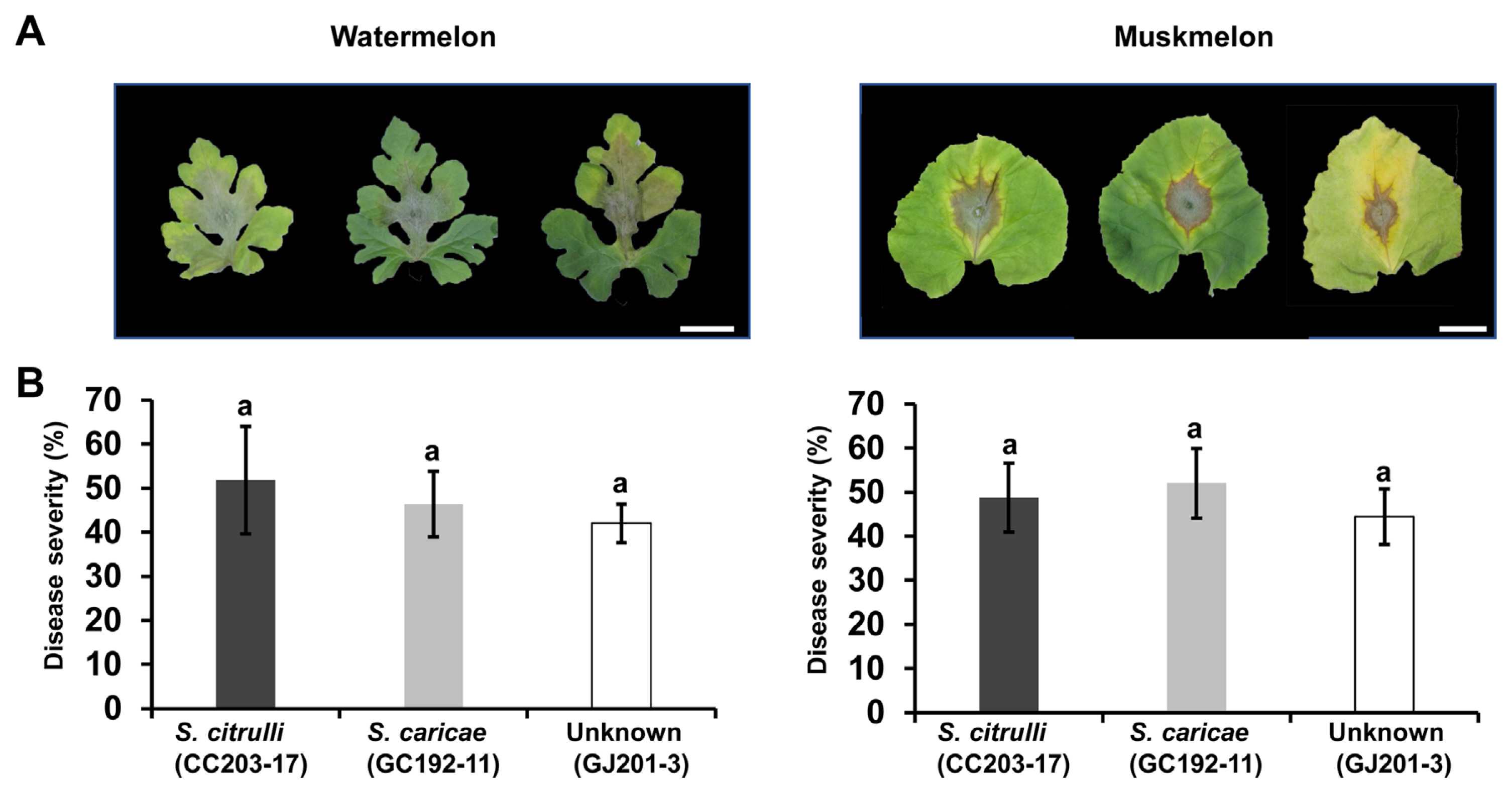
Table 1
Primers used in this study
| DNA region | Primer name | Primer sequence (5′-3′) | Reference |
|---|---|---|---|
| Universal primers | |||
| ITS | ITS1 | TCCGTAGGTGAACCTGCGG | White et al. (1990) |
| ITS4 | TCCTCCGCTTATTGATATGC | ||
| TUB | TUB_T1_F | TCTGGATGTTGTTGGGAATCC | O’Donnell and Cigelnik (1997) |
| TUB_T22_R | ACCCGCTGAACTTAAGC | ||
| Primers for microsatellite markers | |||
| Db01 | Db01_F | CGGTCCGGTCAACCTACTAC | Brewer et al. (2015) |
| Db01_R | CACGCCAGCAAATCTCACTA | ||
| Db05 | Db05_F | TATGACGTTGGGCAAGTGAG | Brewer et al. (2015) |
| Db05_R | TTTGCTGGGATGGTGTTGTA | ||
| Db06 | Db06_F | GGTGACATCTTGCGTGAATG | Brewer et al. (2015) |
| Db06_R | TGGTTGTTTGGTTGTTTGGA | ||
Table 2
Origins of Stagonosporopsis isolates and GenBank accession numbers of ITS regions and TUB genes
| Origin (city/county) | Isolate | Host | Accession no. | Species | Locationa | |
|---|---|---|---|---|---|---|
|
|
||||||
| ITS | TUB | |||||
| Gochang | GC192-11 | Watermelon | ON055719 | ON093927 | Stagonosporopsis caricae | h |
| Chuncheon | CC203-17 | Muskmelon | ON055703 | ON093933 | Stagonosporopsis citrulli | j |
| Gwangju | GJ201-3 | Muskmelon | ON055716 | ON093945 | Stagonosporopsis sp. | c |
| Goryeong | GR212-27 | Muskmelon | ON055704 | ON093934 | Stagonosporopsis citrulli | m |
| Goryeong | GR212-29 | Muskmelon | ON055705 | ON093935 | Stagonosporopsis citrulli | m |
| Goryeong | GR213-2 | Muskmelon | ON055706 | ON093936 | Stagonosporopsis citrulli | m |
| Goryeong | GR213-3 | Muskmelon | ON055707 | ON093937 | Stagonosporopsis citrulli | m |
| Goryeong | GR214-1 | Muskmelon | ON055708 | ON093938 | Stagonosporopsis citrulli | m |
| Changwon | CW211-1 | Muskmelon | ON055720 | ON093928 | Stagonosporopsis caricae | n |
| Changwon | CW211-2 | Muskmelon | ON055709 | ON093939 | Stagonosporopsis citrulli | n |
| Changwon | CW211-3 | Muskmelon | ON055710 | ON093940 | Stagonosporopsis citrulli | n |
| Changwon | CW211-11 | Muskmelon | ON055711 | ON093941 | Stagonosporopsis citrulli | n |
| Changwon | CW214-3 | Watermelon | ON055717 | ON093946 | Stagonosporopsis sp. | n |
| Chunan | CA215-1 | Muskmelon | ON055721 | ON093929 | Stagonosporopsis caricae | e |
| Chunan | CA217-1 | Muskmelon | ON055722 | ON093930 | Stagonosporopsis caricae | e |
| Yanggu | YG211-4 | Watermelon | ON055718 | ON093947 | Stagonosporopsis sp. | i |
| Yanggu | YG211-5 | Watermelon | ON055712 | ON093942 | Stagonosporopsis citrulli | i |
| Yanggu | YG211-10 | Watermelon | ON055713 | ON093943 | Stagonosporopsis citrulli | i |
| Yanggu | YG211-13 | Watermelon | ON055723 | ON093931 | Stagonosporopsis caricae | i |
| Yangpyeong | YP216-4 | Watermelon | ON055714 | ON093944 | Stagonosporopsis citrulli | k |
| Yangpyeong | YP216-8 | Watermelon | ON055724 | ON093932 | Stagonosporopsis caricae | k |
References
Aveskamp, MM, de Gruyter, J, Woudenberg, JH, Verkley, GJ and Crous, PW 2010. Highlights of the Didymellaceae: a polyphasic approach to characterise Phoma and related pleosporalean genera. Stud. Mycol 65:1-60.



Basim, E, Basim, H, Abdulai, M, Baki, D and Öztürk, N 2016. Identification and characterization of Didymella bryoniae causing gummy stem blight disease of watermelon (Citrullus lanatus) in Turkey. Crop Prot 90:150-156.

Boerema, GH, de Gruyter, J, Noordeloos, ME and Hamers, MEC 2004. Phoma identification manual: differentiation of specific and infra-specific taxa in culture. CABI Publishing, Wallingford, Oxfordshire, UK. pp. 470.
Brewer, MT, Rath, M and Li, H-X 2015. Genetic diversity and population structure of cucurbit gummy stem blight fungi based on microsatellite markers. Phytopathology 105:815-824.


Chen, D-H and Ronald, PC 1999. A rapid DNA minipreparation method suitable for AFLP and other PCR applications. Plant Mol. Biol. Rep 17:53-57.
Choi, IY, Choi, JN, Choi, DC, Sharma, PK and Lee, WH 2010. Identification and characterization of the causal organism of gummy stem blight in the muskmelon (Cucumis melo L.). Mycobiology 38:166-170.



Choi, IY, Kim, JH, Lee, WH, Park, JH and Shin, HD 2015. First report of black rot caused by Phoma cucurbitacearum on Momordica charantia in Korea. Plant Dis 99:727.

Food and Agriculture Organization of the United Nations 2019 FAOSTAT Online Database URL https://www.fao.org/faostat/en/#data
. 10 July 2022.
Garampalli, RH, Gapalkrishna, MK, Li, H-X and Brewer, MT 2016. Two Stagonosporopsis species identified as causal agents of gummy stem blight epidemics of gherkin cucumber (Cucumis sativus) in Karnataka, India. Eur. J. Plant Pathol 145:507-512.


Gusmini, G, Rivera-Burgos, LA and Wehner, TC 2017. Inheritance of resistance to gummy stem blight in watermelon. HortScience 52:1477-1482.

Hassan, MZ, Khan Robin, AH, Rahim, MA, Natarajan, S, Kim, H-T, Park, J-I and Nou, I-S 2018. Screening of melon genotypes identifies gummy stem blight resistance associated with Gsb1 resistant loci. J. Plant Biotechnol 45:217-227.

Jensen, BD, Massawe, A and Swai, IS 2011. First report of gummy stem blight caused by Didymella bryoniae on watermelon and confirmation of the disease on pumpkin in Tanzania. Plant Dis 95:768.

Keinath, PA 2021. Premix fungicides that reduce development of fruiting bodies but not leaf lesions by Stagonosporopsis citrulli on watermelon leaves in the field. Plant Dis 105:1415-1421.


Koike, ST 1997. First report of gummy stem blight, caused by Didymella bryoniae, on watermelon transplants in California. Plant Dis 81:1331.

Kumar, S, Stecher, G, Li, M, Knyaz, C and Tamura, K 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol 35:1547-1549.



Kumar, S, Stecher, G and Tamura, K 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol 33:1870-1874.




Kwon, MK, Hong, JR, Sun, HJ, Sung, KY, Cho, BH and Kim, KC 1997. Standardization of a mass-production technique for pycnidiospores of Didymella bryoniae, gummy stem blight fungus of cucurbits. Korean J. Plant Pathol 13:105-112 (in Korean).
Lee, JH, Jang, KS, Choi, YH, Kim, J-C and Choi, GJ 2016. Development of an efficient screening system for resistance of watermelon plants to Didymella bryoniae
. Res. Plant Dis 22:72-80 (in Korean).

Li, P-F, Ren, R-S, Yao, X-F, Xu, J-H, Babu, B, Paret, ML and Yang, X-P 2015. Identification and characterization of the causal agent of gummy stem blight from muskmelon and watermelon in East China. J. Phytopathol 163:314-319.


Ling, K-S, Wechter, WP, Somai, BM, Walcott, RR and Keinath, AP 2010. An improved real-time PCR system for broad-spectrum detection of Didymella bryoniae, the causal agent of gummy stem blight of cucurbits. Seed Sci. Technol 38:692-703.

Ma, G, Bao, S, Zhao, J, Sui, Y and Wu, X 2021. Morphological and molecular characterization of alternaria species causing leaf blight on watermelon in China. Plant Dis 105:60-70.


O’Donnell, K and Cigelnik, E 1997. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Mol. Phylogenet. Evol 7:103-116.


Pryor, BM and Michailides, TJ 2002. Morphological, pathogenic, and molecular characterization of alternaria isolates associated with alternaria late blight of pistachio. Phytopathology 92:406-416.


Stewart, JE, Turner, AN and Brewer, MT 2015. Evolutionary history and variation in host range of three Stagonosporopsis species causing gummy stem blight of cucurbits. Fungal Biol 119:370-382.


Sudisha, J, Kumar, TV, Niranjana, SR and Shetty, HS 2004. First report of gummy stem blight caused by Didymella bryoniae on muskmelon (Cucumis melo) in India. Plant Pathol 53:533.

White, TJ, Bruns, T, Lee, S and Taylor, J 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: PCR protocols: a guide to methods and applications, eds. by MA Innis, DH Gelfand, JJ Sninsky and TJ White, pp. 315-322. Academic Press, London, UK.

- TOOLS
-
METRICS

-
- 0 Crossref
- 0 Scopus
- 2,791 View
- 136 Download
- Related articles
-
Incidence of Alternaria Species Associated with Watermelon Leaf Blight in Korea2021 August;37(4)



 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Supplement
Supplement Print
Print



