 |
 |
| Plant Pathol J > Volume 38(6); 2022 > Article |
|
Abstract
Colletotrichum species is known as the major causal pathogen of red pepper anthracnose in Korea and various groups of fungicides are registered for the management of the disease. However, the consistent use of fungicides has resulted in the development of resistance in many red pepper-growing areas of Korea. Effective management of the occurrence of fungicide resistance depends on constant monitoring and early detection. Thus, in this study, various methods such as agar dilution method (ADM), gene sequencing, allele-specific polymerase chain reaction (PCR), and polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) were applied for the detection of benzimidazole resistance among 24 isolates of Colletotrichum acutatum s. lat. and Colletotrichum gloeosporioides s. lat. The result of the ADM showed that C. gloeosporioides s. lat. was classified into sensitive and resistant isolates to benomyl while C. acutatum s. lat. was insensitive at ≥1 μg/ml of benomyl. The sequence analysis of the β-tubulin gene showed the presence of a single nucleotide mutation at the 198th amino acid position of five isolates (16CACY14, 16CAYY19, 15HN5, 15KJ1, and 16CAYY7) of C. gloeosporioides s. lat. Allele-specific PCR and PCR-RFLP were used to detect point mutation at 198th amino acid position and this was done within a day unlike ADM which usually takes more than one week and thus saving time and resources that are essential in the fungicide resistance management in the field. Therefore, the molecular techniques established in this study can warrant early detection of benzimidazole fungicide resistance for the adoption of management strategies that can prevent yield losses among farmers.
Benzimidazole fungicides are used for the management of diseases caused by Botrytis, Monilinia, Colletotrichum, Sclerotinia, Podosphaera, and so on (Bradley et al., 2006; Chen et al., 2014; Hwang et al., 2010; Sedláková and Lebeda, 2008). However, the use of the fungicide has resulted in point mutation that confers resistance of the pathogen to the fungicide (Albertini et al., 1999; Jones and Walker, 1976; Ross and Newberry, 1985). Resistance to benzimidazole-based fungicide was reported in New Zealand in 1975 on apple scab disease caused by Venturia inaequalis and it was also reported in 1977 in Japan on Venturia nashicola on pear (Hartill, 1986; Ishii and Yamaguch, 1977). In Korea, resistance to benzimidazole fungicide was reported on C. gloeosporioides s. lat. isolated on grapes and Botrytis cinerea isolated on cucumbers and ginseng (Hwang et al., 2010; Kim et al., 2009). Although fungicide resistance has been reported on various plant pathogenic fungi, the efficacy of the benzimidazole fungicides in the management of anthracnose in fruits and vegetables has resulted in its continuous use on some crops.
The benzimidazole fungicides were developed to specifically target the β-tubulin gene of the fungus and like all fungicides with a single site of action, a single nucleotide mutation can confer on the pathogen resistance to the fungicide (Staub, 1991). This fungicide binds with the β-tubulin gene in plant-fungal pathogen to prevent microtubule production, thus the pathogen cannot undergo cell division, and in extension, growth is suppressed (Davidse and Flach, 1977; Martin, 1997). This usually occurs due to mutation that happens because of frequent, prolonged, and/or inappropriate application of the fungicide. In this group of compounds, mutation takes place at the 198th amino acid position of the β-tubulin gene from glutamate to either alanine, glycine, or lysine and/or at the 200th amino acid position from phenylalanine to tyrosine and thus conferring resistance of the fungal pathogen to the fungicide (Fungicide Resistance Action Committee, 2022).
Various conventional assays and molecular techniques were validated and adopted for the evaluation and monitoring of resistance by the Fungicide Resistance Action Committee. Among these methods, an agar dilution method (ADM) is considered efficient, accurate, and reliable but it is time and resource-consuming, laborious, and requires expertise. Therefore, Chung et al. (2010) suggested polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) as an alternative tool for monitoring resistance in C. gloeosporioides s. lat. isolated from diseased fruits because it is a rapid and reliable method. Also, Yan et al. (2007) suggested the application of allele-specific polymerase chain reaction (PCR) for the detection of resistance in Cladosporium fulvum using primers designed based on allele substitution at the point of mutation because it is a simple and rapid method for the identification of resistant isolates. A similar allele-specific PCR technique was also used in the identification of resistant or sensitive isolates of C. acutatum s. lat. to strobilurin fungicides (Kim et al., 2019).
The classification system of Colletotrichum, which is known to cause anthracnose in various crops including pepper, has undergone several changes. In the past, the identification of Colletotrichum spp. causing red pepper anthracnose was based on characteristics such as the spore shape, the presence or absence of sexual generation, the presence of setae and sclerotia, and the pathogenicity to pepper fruits in Korea (Park and Kim, 1992). However, the morphological characteristics and pathogenicity of the fungal species may differ depending on the cultivation conditions or status of the pathogen, so there are limitations in such identification and classification (Damm et al., 2012). Thus, the use of molecular genetic markers provides more accurate identification. One of the genes that have been widely used for the identification of fungal pathogens is the internal transcribed spacer (ITS) region. This has made the identification and classification of C. acutatum s. lat. and C. gloeosporioides s. lat. easier. C. gloeosporioides s. lat. is sensitive or resistant to benzimidazole-based fungicide, but C. acutatum s. lat. showed insensitivity to benzimidazole-based fungicide, and even when treated with high concentrations of the fungicide, the pathogen growth is not completely suppressed (Chung et al., 2006; Kim et al., 2007). In Korea, it has been presented that the predominant causal pathogen of red pepper anthracnose has changed from C. gloeosporioides s. lat. to C. acutatum s. lat. by using the mycological characteristics and ITS gene’s sequence (Kim et al., 2008). C. gloeosporioides s. lat. has been known as a major anthracnose pathogen in crops such as apples, persimmons, grapes, and jujubes (Isa et al., 2021). In addition, both isolates of C. gloeosporioides s. lat. sensitive and resistant to benomyl are still detected in the field cultivating crops.
Therefore, in this study, both C. acutatum s. lat. and C. gloeosporioides s. lat. were isolated by collecting fruits with typical symptoms of red pepper anthracnose, and their resistance to benomyl was evaluated using an ADM. In addition, in order to evaluate a resistance to benomyl accurately and quickly, benomyl-resistant C. gloeosporioides s. lat. was detected by using allele-specific PCR and PCR-RFLP.
Colletotrichum species isolated from pepper in 2015/2016 using a single spore isolation method and stored in the plant-fungal disease laboratory, department of plant medicine, Chungbuk National University were used for the experiment as shown in Table 1. These isolates were also earlier identified as C. acutatum s. lat. and C. gloeosporioides s. lat. using species-specific primers (unpublished data).
The response of the Colletotrichum isolates was investigated using the ADM with benomyl (a.i. 50%, WP). The benomyl was serially diluted in sterile distilled water (SDW) to a pre-determined concentration (100, 10, 1, 0.1, and 0.01 μg/ml) and used to amend the potato dextrose agar (PDA; Becton, Dickinson and Company, Difco, Franklin Lakes, NJ, USA). A 3 mm mycelial disc taken from the edge of a 7-day-old growing culture of the isolates was transferred onto the PDA plates amended with each of the above concentrations. The mycelium discs were inoculated upside down on a fresh PDA medium with the mycelium facing down. The plates were incubated for 7-days at 25°C. PDA medium without the fungicide served as a standard check and three replicated were maintained for each isolate. The colony diameter for each isolate at various concentrations was measured and recorded. The mycelial growth inhibitory effect of each fungicide concentration was calculated by comparing the diameter of the colony grown on benomyl-amended PDA with the unamended control using the formula below as adopted from Isa et al. (2021).
Total genomic DNA was extracted from mycelia obtained from PDA culture of Colletotrichum spp. grown at 25°C for 7 days. Aerial mycelia were harvested from culture plates using a sterile transfer needle and placed in a sterile 1.5 ml microcentrifuge tube containing 300 μl of extraction buffer (0.2 M Tris-HCl, 0.25 M NaCl, 25 mM EDTA, and 2 ml sodium dodecyl sulfate, pH 8.5). Uncapped tubes were placed in a boiling water bath for 5 min and then cooled to 25°C. Two hundred microliter (200 μl) of phenol that was calibrated with extraction buffer (v/v) and 200 μl of chloroform was added. The tubes were vortexed for 4 min and then centrifuged at 13,000 rpm for 15 min. The supernatant was extracted with 200 μl of isopropanol and centrifuged at 13,000 rpm for 15 min. The nucleic acid pellet, after washing with 70% ethanol, followed by air-drying for 15 min, was re-suspended in 50 μl of TE buffer (10 mM Tris-HCl and 0.1 mM EDTA, pH 8.5). DNA was finally treated with ribonuclease A.
The β-tubulin gene was amplified using the primer TB2L (GYT TCC AGA TYA CCC ACT CC) and TB2R (TGA GCT CAG GAA CRC TGA CG) (Peres et al., 2004). A 20 μl PCR mixture containing 4 μl master mix (EzPCR 5× PCR master mix, Elpis Biotech, Daejeon, Korea), 1 μl reverse and forward primer each, 2 μl gDNA, and 12 μl SDW was prepared. The gene was amplified using the PCR kit (Bioneer Inc., Daejeon, Korea) in a thermal cycler (i-cycler version 3.021, Bio-Rad, Hercules, CA, USA). The amplification was conducted as follows: initial denaturation at 95°C for 2 min, followed by 40 cycles of denaturation at 95°C for 1 min, annealing at 55°C for 1 min, and extension at 72°C for 1 min, and lastly, a final extension at 72°C for 5 min. The β-tubulin gene sequences were aligned and blasted on the NCBI database.
A reverse primer (ASR 1; TTG TCA ATG CAG AAG GTC G) designed to specifically amplify a single point mutation at 198 amino acid position by Yan et al. (2007) was used to amplify the resistant isolates of Colletotrichum species. A forward primer (HTI-1; CTA CCC AGG CTT CCG GC) specific to the β-tubulin gene of Colletotrichum spp. was designed based on the fourth intron of the aligned sequences of MT513083-MT513070 on the NCBI database. A 20 μl PCR mixture containing 4 μl master mix (EzPCR 5× PCR master mix, Elpis Biotech), 1 μl reverse and forward primer each, 2 μl gDNA, and 12 μl SDW was prepared. initial denaturation at 94°C for 4 min, followed by 30 cycles of denaturation at 98°C for 10 s, annealing at 61°C for 30 s, and extension at 72°C for 1 min, and a final extension at 72°C for 7 min. To confirm the amplification of the resistant isolates by PCR, 1.5% agarose gel was made with 0.5× TBE buffer, and safe view classic was added according to the recommended usage. The PCR products were separated by electrophoresis for 40 min at 100 volts and the gel was viewed in a UV-transilluminator.
The BstUI restriction enzyme that restricts 5′-CG/CG-3′ and 3′-GC/GC-5′ was used for the experiment. The PCR was conducted using the primer TB2L and TB2R. Furthermore, a 30 μl volume reaction consisting of 17 μl SDW, 10 μl PCR product, 10× buffer, and 1 μl BstUI restriction enzyme was prepared. The mixture was incubated in a thermal cycler at 37°C for 1 h, followed by inactivation at 65°C for 20 min. The products were separated by electrophoresis for 40 min at 100 V and the gel was viewed in a UV-transilluminator.
Colletotrichum species isolated from red pepper in Korea showed different responses to benomyl using the ADM. The Colletotrichum isolates used in this study can be grouped into three types based on their response to benomyl, viz: sensitive Cg (C. gloeosporioides s. lat.) type, resistant Cg type, and insensitive Ca (C. acutatum s. lat.) type. Isolates with a mycelial growth inhibition rate of 80% or more at 0.8 μg/ml were grouped as the sensitive Cg type, isolates with less than 30% inhibition were grouped as the resistant Cg type, and lastly, isolates with 30-80% inhibition rates at 0.16 and 0.8 μg/ml treatment were grouped as the insensitive Ca type as shown in Fig. 1A and B. The result showed that 10 isolates of C. gloeosporioides s. lat. were sensitive to the benomyl, while five isolates were resistant to the fungicide. However, all nine isolates of C. acutatum s. lat. showed no clear response to the fungicide and were thus classified as the insensitive type as shown in Fig. 1B.
The primer TB2L/TB2R was used to amplify 414 bp of the β-tubulin gene of 24 isolates of Colletotrichum spp. isolated on red pepper fruits. Analysis of the aligned sequences showed that five isolates of C. gloeosporioides s. lat. possessed GCG (alanine) codon at the 198th amino acid position which is the site of action of the benzimidazole fungicide and thus, conferred resistance of the isolates to the fungicide. On the other hand, 10 isolates of C. gloeosporioides s. lat. possessed GAG (glutamate) at the same position as shown in Table 2. The result also showed that the isolates identified as C. acutatum s. lat. and grouped as insensitive in the ADM possessed the GAG codon as shown in Table 2 and Fig. 1. The analysis of the β-tubulin gene also showed that mutation was not observed at the 200th amino acid position on all the isolates used in the study as shown in Table 2.
The allele-specific PCR was performed to amplify the resistant isolates of Colletotrichum species used in this experiment. The result showed that the primer HTI-1 and ASR1 successfully amplified 460 bp of only the resistant isolates 16CACY14, 16CAYY19, 15HN5, 15KJ1, and 16CAYY7 that were earlier identified as members of the C. gloeosporioides s. lat. The gel electrophoresis result is as shown in Fig. 2. The sensitive isolates of C. gloeosporioides s. lat. and insensitive isolates of C. acutatum s. lat. was not amplified by the primer.
The primer TB2L/TB2R amplified 414 bp of Colletotrichum isolates used in this experiment. Furthermore, the restriction enzyme BstUI restricted the PCR products of the resistant C. gloeosporioides s. lat. to generate two amplicons of 262 and 152 bp as shown in Fig. 3. On the contrary, restriction did not occur on the PCR products of sensitive C. gloeosporioides s. lat. and insensitive C. acutatum s. lat. as shown in Fig. 3.
C. acutatum s. lat. and C. gloeosporioides s. lat., which belong to the genus of Colletotrichum are now being sub-divided into several species through multi-gene phylogenetic analysis using the ITS, actin (ACT), β-tubulin (TUB2), chitin synthase 1 (CHS-1), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and histone3 (HIS3) as reported by Damm et al. (2012) and Weir et al. (2012). However, although C. acutatum s. lat. and C. gloeosporioides s. lat. showed distinct differences between species in response to benomyl, the newly re-identified Colletotrichum species had no clear association between species and response to fungicides. Therefore, in this study, C. acutatum s. lat. and C. gloeosporioides s. lat. were used as they were, without identifying Colletotrichum as a new species through polygenic phylogenetic analysis.
The C. gloeosporioides s. lat. isolates used in this study distinctly showed the difference in the mycelial growth between the benomyl sensitive and resistant phenotypes at concentrations ≥1 μg/ml. Peres et al. (2004) reported that C. gloeosporioides s. lat. resistance to benomyl was not affected by the fungicide at concentrations ≤10 μg/ml, while the mycelial growth of sensitive isolates was inhibited by more than 80% at 0.1 μg/ml. In contrast, C. acutatum s. lat. was moderately resistant (40-60% inhibition) at concentrations of ≥1 μg/ml and this is similar to the findings of Goes and Kimati (1998) that reported a mycelial growth inhibition ratio of 40% to 85% on C. acutatum s. lat. at concentrations of ≥1 μg/ml. Kim et al. (2007) also reported that C. acutatum s. lat. was insensitive to benzimidazoles in ADM and such response can be used to distinguish between C. acutatum s. lat. and C. gloeosporioides s. lat. Furthermore, the response of C. gloeosporioides s. lat. to the benzimidazole fungicide has been described as qualitative while the response of C. acutatum s. lat. is quantitative respectively.
The resistance of C. gloeosporioides s. lat. to the benomyl correlated with an amino acid substitution at the 198 (E198A) position on the β-tubulin gene. On the other hand, such mutation was not found in the sensitive and moderate isolates of C. gloeosporioides s. lat. and C. acutatum s. lat. respectively. The Fungicide Resistance Action Committee (2022) reported the mutation E198A as one of the reasons for the high resistance of fungal species to benzimidazole fungicides. Kim et al. (2007) also reported that highly resistant isolates of C. gloeosporioides s. lat. possessed alanine instead of glutamate at the 198th position of the -tubulin gene. On the other hand, there was no mutation at the same position on all C. acutatum s. lat. evaluated by Peres et al. (2004) and Moreira et al. (2019). The analysis of the -tubulin gene also revealed that there was no mutation at the 200th amino acid position (F200Y) in both the C. gloeosporioides s. lat. and C. acutatum s. lat. used in this study. This is contrary to the findings of Yan et al. (2007) that detected a change from phenylalanine to tyrosine at the 200th amino acid position on Cladosporium fulvum isolated from tomato.
The PCR-RFLP analysis of the β-tubulin gene showed there was no restriction cleavage on the sensitive and insensitive isolates of C. gloeosporioides s. lat. and C. acutatum s. lat. respectively. However, the restriction cut that occurred on the resistant isolates of C. gloeosporioides s. lat. corroborates the findings of Chung et al. (2010) that recommended PCR-RFLP as a potential tool for monitoring the resistance of C. gloeosporioides s. lat. to benomyl for effective cost and time management. The allele-specific PCR was used to detect the resistant isolates of C. gloeosporioides s. lat. because of the modification of a single allele mismatch at the 3′end of the primers (Yan et al., 2007). The use of allele-specific PCR has been described as a simple and rapid method for the identification of benzimidazole resistance. However, only a primer capable of detecting isolates resistant to benomyl was developed in this experiment, so for more accurate resistance monitoring for benomyl, it is necessary to develop a primer capable of detecting isolates sensitive to the fungicide in the future. The molecular monitoring method for detecting the benomyl resistance established in this study may seem less important because C. gloeosporioides s. lat. was not a major pathogen in red pepper. Based on the results of this study, it can be used as an assay that can quickly and accurately detect C. gloeosporioides s. lat. resistant to benomyl causing anthracnose in apples, jujube, and strawberries, where C. gloeosporioides s. lat. is still reported as a major pathogen.
Acknowledgments
This work was supported by the Korean Institute of Planning and Evaluation for Technology in Food, Agriculture, Forestry (IPET) through the Crop Viruses and Pests Response Industry Technology Development Program, funded by the Ministry of Agriculture, Food, and Rural Affairs (MAFRA; Grant No. 320042-5).
Fig. 1
Inhibitory effect of benomyl on the mycelial growth of Colletotrichum gloeosporioides s. lat. (A) and C. acutatum s. lat. (B). Inhibitory effect of benomyl was evaluated by using an agar dilution method. A mycelium disc with a diameter of 3 mm was cut off from the hyphal tip of the colony cultured for 7 days in potato dextrose agar (PDA) at 25°C for using as an inoculum. The inoculum was inoculated into PDA with benomyl and cultured at 25°C for 7 days, then the diameter of the colony was measured. The mycelial growth inhibitory effect of benomyl was investigated at each concentration by comparing the colony diameter of Colletotrichum spp. on PDA without benomyl with that of the PDA amended with benomyl.
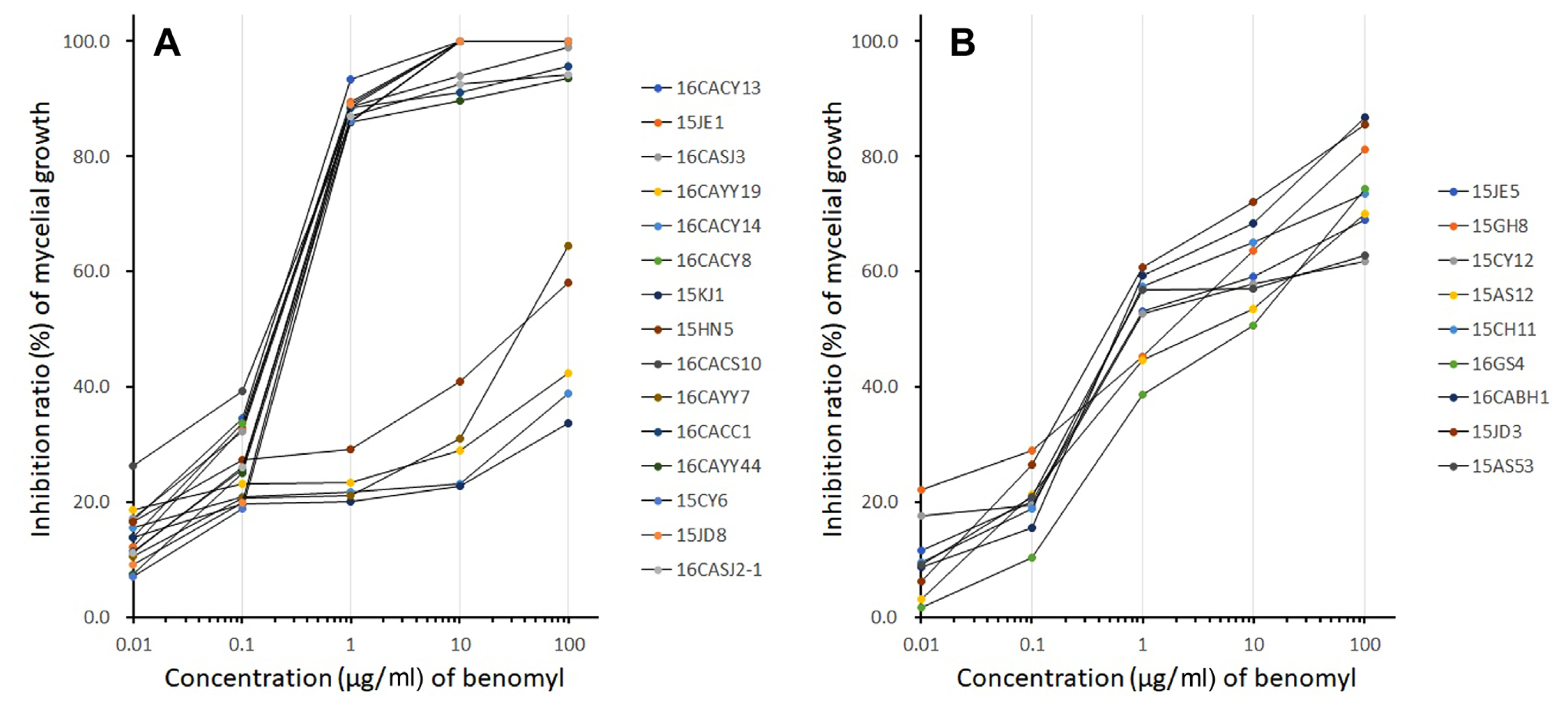
Fig. 2
Allele-specific polymerase chain reaction of Colletotrichum spp. with primer HTI-1 and ASR1. The primer HTI-1 and ASR1 successfully amplified 460 bp of only the resistant isolates. According to the results of the agar dilution method, the first 5 isolates (16CACY14, 16CAYY19, 15HN5, 15KJ1, and 16CAYY7) were Colletotrichum gloeosporioides s. lat. resistant to benomyl, and the rest were sensitive or insensitive. Sensitive isolates of C. gloeosporioides s. lat.; 15CY6, 16CACY13, 16CAYY44, 16CACS10, 16CASJ2-1, 16CACC1, 15JD8, 16CASJ3, 15JE1, and 16CACY8, insensitive isolates of C. acutatum s. lat.; 15CY12, 15GH8, 15AS12, 16CABH1, 15JD3, 15AS53, 16GS4, 15JE5, and 15CH11. M, DNA size marker; N, negative control.
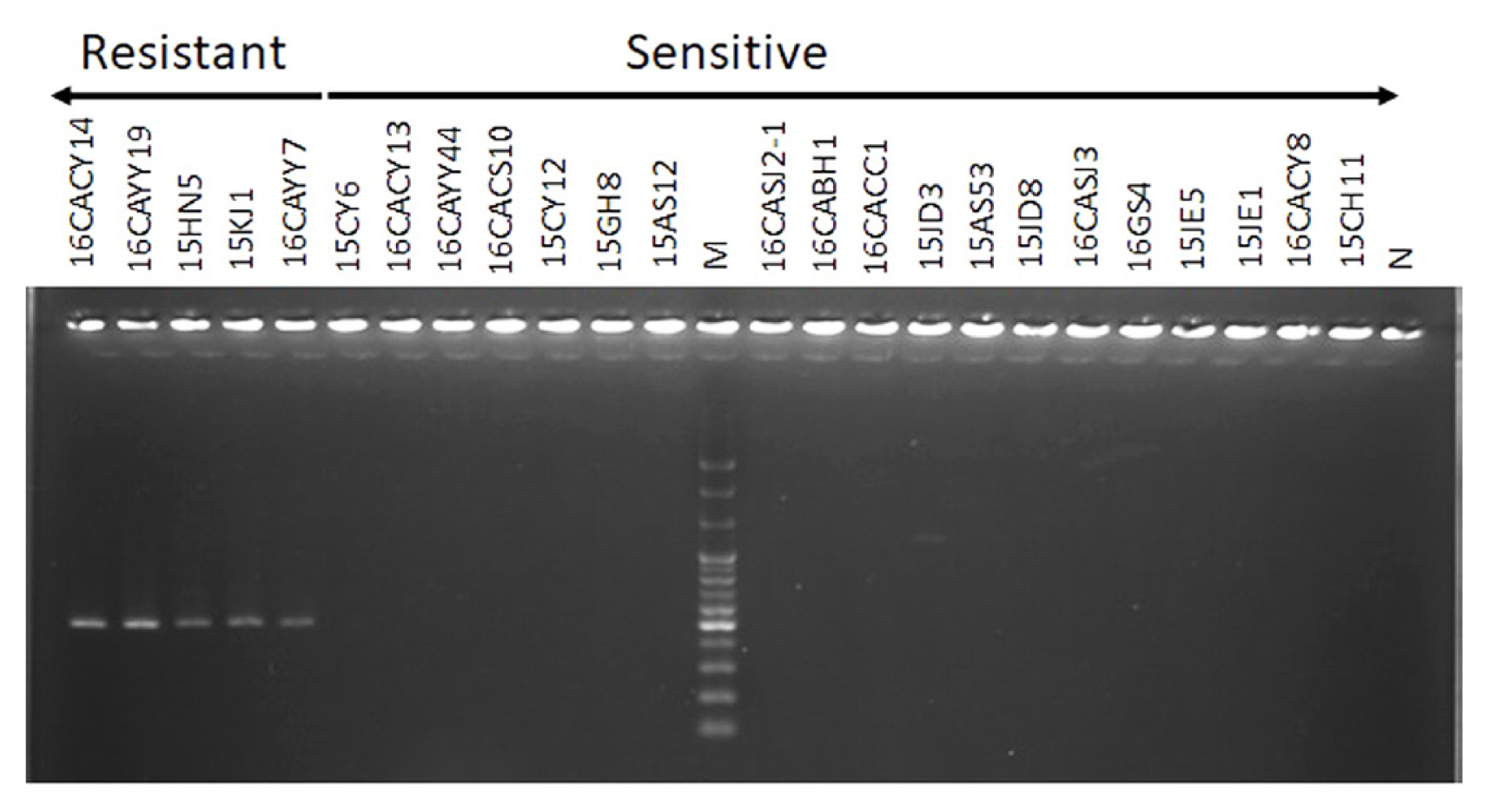
Fig. 3
Polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) of β-tubulin gene of Colletotrichum species treated with restriction enzyme BstUI. Restriction enzyme BstUI was used to conduct PCR-RFLP of the β-tubulin gene product amplified using primer TB2L/TB2R. (A) The amplified β-tubulin gene without restriction enzyme treatment. (B) The polymerase chain reaction-restriction product after the restriction enzyme treatment on the amplified β-tubulin gene. Some isolates of Colletotrichum spp. with different responses to benomyl were used for PCR-RFLP. Resistant isolates of C. gloeosporioides s. lat.; 16CACY14, 16CAYY19, 15HN5, 15KJ1, and 16CAYY7, sensitive isolates of C. gloeosporioides s. lat.; 15CY6, 16CACY13, 16CAYY44, and 16CACS10, and insensitive isolates of C. acutatum s. lat.; 15CY12, 15GH8 and 15AS12. N, negative control; M, DNA size marker.
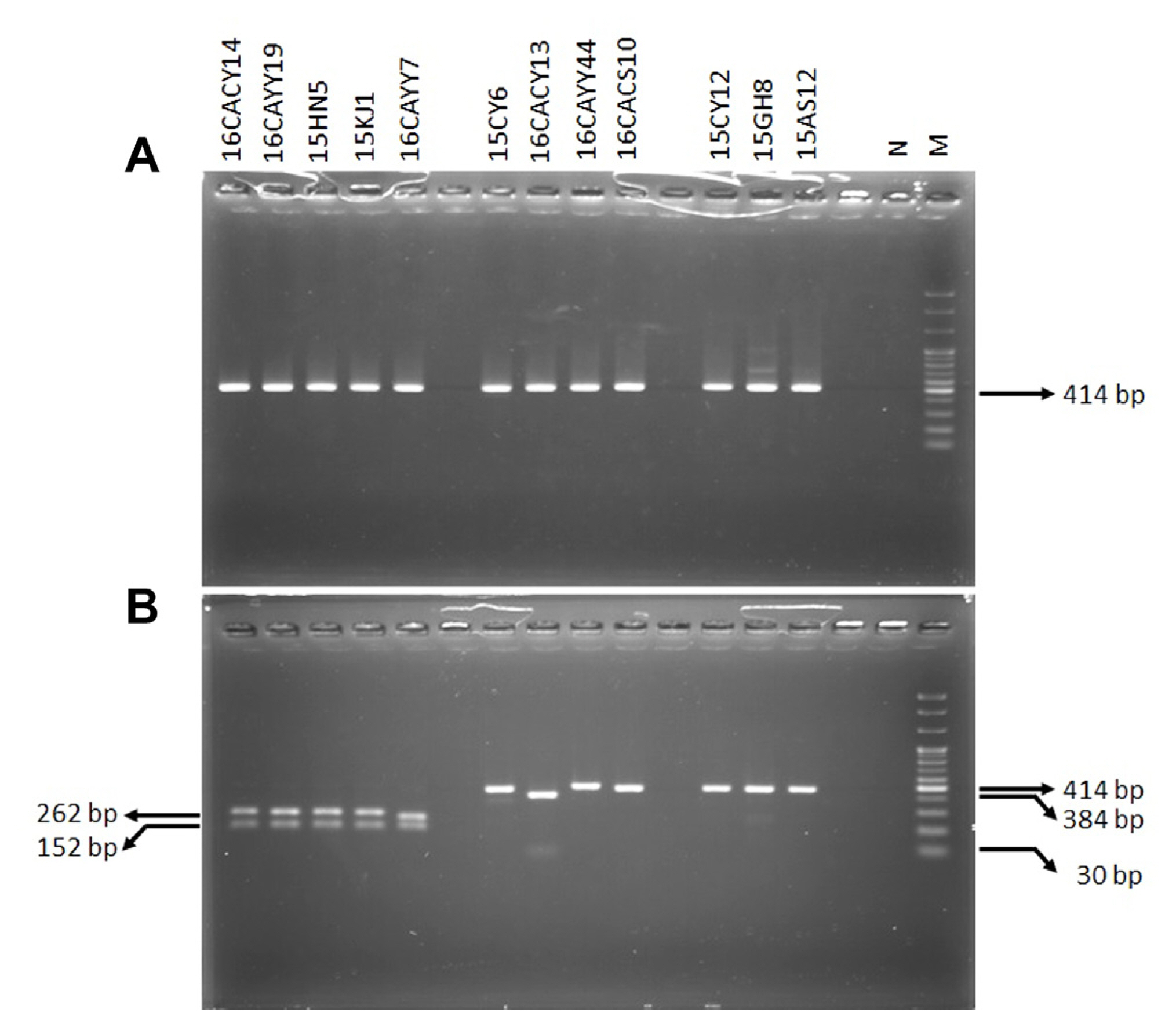
Table 1
List of Colletotrichum species isolates used in this study
Table 2
Benomyl resistance of Colletotrichum gloeosporioides isolated from diseased pepper fruits investigated using several methodsa
| Isolates | Identification | ADM | β-Tubulin | Amino acid sequenceb | PCR-RFLP | Allele-specific PCR | ||
|---|---|---|---|---|---|---|---|---|
|
|
||||||||
| 198 | 200 | R/S | ||||||
| 16CACY14 | C. gloeosporioides s. lat. | Rc | +d | GCG(A) | TTC(F) | R | R | R |
| 16CAYY19 | C. gloeosporioides s. lat. | R | + | GCG(A) | TTC(F) | R | R | R |
| 15HN5 | C. gloeosporioides s. lat. | R | + | GCG(A) | TTC(F) | R | R | R |
| 15KJ1 | C. gloeosporioides s. lat. | R | + | GCG(A) | TTC(F) | R | R | R |
| 16CAYY7 | C. gloeosporioides s. lat. | R | + | GCG(A) | TTC(F) | R | R | R |
| 15JE1 | C. gloeosporioides s. lat. | S | + | GAG(E) | TTC(F) | S | S | S |
| 16CACY13 | C. gloeosporioides s. lat. | S | + | GAG(E) | TTC(F) | S | S | S |
| 16CACY8 | C. gloeosporioides s. lat. | S | + | GAG(E) | TTC(F) | S | S | S |
| 16CASJ3 | C. gloeosporioides s. lat. | S | + | GAG(E) | TTC(F) | S | S | S |
| 16CAYY44 | C. gloeosporioides s. lat. | S | + | GAG(E) | TTC(F) | S | S | S |
| 15CY6 | C. gloeosporioides s. lat. | S | + | GAG(E) | TTC(F) | S | S | S |
| 15JD8 | C. gloeosporioides s. lat. | S | + | GAG(E) | TTC(F) | S | S | S |
| 16CACC1 | C. gloeosporioides s. lat. | S | + | GAG(E) | TTC(F) | S | S | S |
| 16CACS10 | C. gloeosporioides s. lat. | S | + | GAG(E) | TTC(F) | S | S | S |
| 16CASJ2-1 | C. gloeosporioides s. lat. | S | + | GAG(E) | TTC(F) | S | S | S |
| 15AS12 | C. acutatum s. lat. | IS | + | GAG(E) | TTC(F) | S | S | S |
| 15AS53 | C. acutatum s. lat. | IS | + | GAG(E) | TTC(F) | S | S | S |
| 15CH11 | C. acutatum s. lat. | IS | + | GAG(E) | TTC(F) | S | S | S |
| 15CY12 | C. acutatum s. lat. | IS | + | GAG(E) | TTC(F) | S | S | S |
| 16CABH1 | C. acutatum s. lat. | IS | + | GAG(E) | TTC(F) | S | S | S |
| 15JE5 | C. acutatum s. lat. | IS | + | GAG(E) | TTC(F) | S | S | S |
| 15JD3 | C. acutatum s. lat. | IS | + | GAG(E) | TTC(F) | S | S | S |
| 15GH8 | C. acutatum s. lat. | IS | + | GAG(E) | TTC(F) | S | S | S |
| 16GS4 | C. acutatum s. lat. | IS | + | GAG(E) | TTC(F) | S | S | S |
a The methods used for the benomyl resistance assay of C. gloeosporioides were the agar dilution method (ADM), β-tubulin gene sequencing, polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP), and allele-specific polymerase chain reaction (PCR).
References
Albertini, C., Gredt, M. and Leroux, P. 1999. Mutations of the β-tubulin gene associated with different phenotypes of benzimidazole resistance in the cereal eyespot fungi Tapesia yallundae and Tapesia acuformis
. Pestic. Biochem. Physiol 64:17-31.

Bradley, C.A., Lamey, H.A., Endres, G.J., Henson, R.A., Hanson, B.K., McKay, K.R., Halvorson, M., LeGare, D.G. and Porter, P.M. 2006. Efficacy of fungicides for control of Sclerotinia stem rot of canola. Plant Dis 90:1129-1134.


Chen, S.N., Shang, Y., Wang, Y., Schnabel, G., Lin, Y., Yin, L.F. and Luo, C.X. 2014. Sensitivity of Monilinia fructicola from peach farms in China to four fungicides and characterization of isolates resistant to carbendazim and azoxystrobin. Plant Dis 98:1555-1560.


Chung, W.-H., Chung, W.-C., Peng, M.-T., Yang, H.-R. and Huang, J.-W. 2010. Specific detection of benzimidazole resistance in Colletotrichum gloeosporioides from fruit crops by PCR-RFLP. N. Biotechnol 27:17-24.


Chung, W.-H., Ishii, H., Nishimura, K., Fukaya, M., Yano, K. and Kajitani, Y. 2006. Fungicide sensitivity and phylogenetic relationship of anthracnose fungi isolated from various fruit crops in Japan. Plant Dis 90:506-512.


Damm, U., Cannon, P.F., Woudenberg, J.H.C. and Crous, P.W. 2012. The Colletotrichum acutatum species complex. Stud. Mycol 73:37-113.



Davidse, L.C. and Flach, W. 1977. Differential binding of methyl benzimidazol-2-yl carbamate to fungal tubulin as a mechanism of resistance to this antimitotic agent in mutant strains of Aspergillus nidulans
. J. Cell Biol 72:174-193.



Fungicide Resistance Action Committee 2022 FRAC code list 2022: fungal control agents sorted by cross-resistance pattern and mode of action (including coding for FRAC Groups on product labels) URL https://www.frac.info/docs/default-source/publications/frac-code-list/frac-code-list-2022--final.pdf?sfvrsn=b6024e9a_2 on 24/04/2022. [2 June 2022].
Goes, A. and Kimati, H. 1998.
Colletotrichum acutatum, agente causal da queda prematura dos frutos citricos: resistente ou insensivel a benomyl? Summa Phytopathol 24:246-253.
Hartill, W.F.T. 1986. Resistance of plant pathogens to fungicides in New Zealand. N. Z. J. Exp. Agric 14:239-245.

Hwang, S., Kim, H.-R., Kim, J., Park, J.-H., Lee, S.-B., Cheong, S.-R. and Kim, H.T. 2010. Sensitivity of Colletotrichum spp. isolated from grapes in Korea to carbendazim and the mixture of carbendazim plus diethofencarb. Plant Pathol. J 26:49-56.

Isa, D.A., Min, J.Y. and Kim, H.-T. 2021. Responses to carbendazim and analysis of field populations of Colletotrichum spp. isolated from several host plants. Korean J. Pestic. Sci 25:73-81 (in Korean).

Ishii, H. and Yamaguchi, A. 1977. Tolerance of Venturia nashicola to thiophanate-methyl and benomyl in Japan. Ann. Phytopathol. Soc. Jpn 43:557-561.

Jones, A.L. and Walker, R.J. 1976. Tolerance of Venturia inaequalis to dodine and benzimidazole fungicides in Michigan. Plant Dis. Rep 60:40-44.
Kim, J., Min, J.Y., Bae, Y.S. and Kim, H.T. 2009. Molecular analysis of Botrytis cinerea causing ginseng grey mold resistant to carbendazim and the mixture of carbendazim plus diethofencarb. Plant Pathol. J 25:322-327.
Kim, J.T., Park, S.-Y., Choi, W., Lee, Y.-H. and Kim, H.T. 2008. Characterization of Colletotrichum isolates causing anthracnose of pepper in Korea. Plant Pathol. J 24:17-23.

Kim, S., Min, J. and Kim, H.T. 2019. Occurrence and mechanism of fungicide resistance in Colletotrichum acutatum causing pepper anthracnose against pyraclostrobin. Korean J. Pestic. Sci 23:202-211 (in Korean).

Kim, Y.-S., Min, J.Y., Kang, B.K., Van Bach, N., Choi, W.B., Park, E.W. and Kim, H.T. 2007. Analysis of the less benzimidazole-sensitivity of the isolates of Colletotrichum spp. causing the anthracnose in pepper and strawberry. Plant Pathol. J 23:187-192.
Moreira, R.R., Peres, N.A. and May De Mio, L.L. 2019.
Colletotrichum acutatum and C. gloeosporioides species complexes associated with apple in Brazil. Plant Dis 103:268-275.


Park, K.S. and Kim, C.H. 1992. Identification, distribution, and etiological characteristics of anthracnose fungi of red pepper in Korea. Korean J. Plant Pathol 8:61-69.
Peres, N.A.R., Sauza, N.L., Peever, T.L. and Timmer, L.W. 2004. Benomyl sensitivity of isolates of Colletotrichum acutatum and C. gloeosporioides from citrus. Plant Dis 88:125-130.


Ross, R.G. and Newberry, R.J. 1985. Tolerance to benomyl of Venturia inaequalis in Nova Scotia. Can. J. Plant Pathol 7:435-437.

Sedláková, B. and Lebeda, A. 2008. Fungicide resistance in Czech populations of cucurbit powdery mildews. Phytoparasitica 36:272-289.


Staub, T. 1991. Fungicide resistance: practical experience with antiresistance strategies and the role of integrated use. Annu. Rev. Phytopathol 29:421-442.


- TOOLS
-
METRICS

-
- 0 Crossref
- 0 Scopus
- 1,710 View
- 98 Download
- ORCID iDs
-
Heung Tae Kim

https://orcid.org/0000-0001-7132-0587 - Related articles
-
Detection of Potato Virus S Using ELISA and Rt-PCR Technique1997 October;13(5)



 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Print
Print



