 |
 |
| Plant Pathol J > Volume 38(6); 2022 > Article |
|
This article has been corrected. See "Application of Rapid and Reliable Detection of Cymbidium Mosaic Virus by Reverse Transcription Recombinase Polymerase Amplification Combined with Lateral Flow Immunoassay" in Volume 39 on page 158.
Abstract
Cymbidium mosaic virus (CymMV) is one of economically important viruses that cause significant losses of orchids in the world. In the present study, a reverse transcription recombinase polymerase amplification (RT-RPA) assay combined with a lateral flow immunostrip (LFI) assay was developed for the detection of CymMV in orchid plants. A pair of primers containing fluorescent probes at each terminus that amplifies highly specifically a part of the coat protein gene of CymMV was determined for RT-RPA assay. The RT-RPA assay involved incubation at an isothermal temperature (39°C) and could be performed rapidly within 30 min. In addition, no cross-reactivity was observed to occur with odontoglossum ringspot virus and cymbidium chlorotic mosaic virus. The RT-RPA with LFI assay (RT-RPA-LFI) for CymMV showed 100 times more sensitivity than conventional reverse transcription polymerase chain reaction (RT-PCR). Furthermore, the RT-PCR-LFI assay demonstrated the simplicity and the rapidity of CymMV detection since the assay did not require any equipment, by comparing results with those of conventional RT-PCR. On-site application of the RT-RPA-LFI assay was validated for the detection of CymMV in field-collected orchids, indicating a simple, rapid, sensitive, and reliable method for detecting CymMV in orchids.
Orchid, which belong to the family Orchidaceae, is one of the most important ornamental crops in the world’s flower industry, because of their attractive flowers with diverse shapes and long vase life (Lawson et al., 1995; Li et al., 2019). In Korea, the annual production of orchids was 112 million pots, and the wholesale value was about 27.1 million US dollars (Ministry for Food, Agriculture, Forestry and Fisheries, Korea, 2021). Orchids have been reported to be infected with more than 50 viruses (Zettler et al., 1990). Among orchid viruses, cymbidium mosaic virus (CymMV) is the most prevalent, economically important in orchid production (Ajjikuttira et al., 2002; Ryu and Park, 1995).
CymMV is a member of the genus Potexvirus of the family Alphaflexiviridae and the virus has a positive-sense single-strand RNA genome of approximately 6.3 kb. The genome of CymMV consists of 5 open reading frames flanked by 5′ and 3′ non-coding regions plus 3′ poly(A) tail (Adams et al., 2004; Wong et al., 1994). Open reading frame (ORF) 1 encodes a 160-kDa replicase containing a methyltransferase domain in the N-terminal region, the RNA helicase domain and the RNA-dependent RNA polymerase domain in the C-terminal region. ORFs 2-4, referred to as the triple gene block, are involved in cell-to-cell movement and suppression of RNA silencing. ORF 5 encodes the 24-kDa viral coat protein (CP), which is indispensable for virus assembly and systemic spread in orchids (Verchot-Lubicz et al., 2007; Wong et al., 1994). CymMV frequently causes reductions in yield and quality of orchid flowers in Korea as well as in countries where orchids are cultivated (Sherpa et al., 2006; Wisler et al., 1987; Wong et al., 1994; Yoon et al., 2012; Zettler et al., 1990). Orchids infected with CymMV showed color-breaking or necrotic spots in flowers, and streak, yellowing or stripe mosaic in leaves (Wisler et al., 1987; Wong et al., 1994; Yoon et al., 2012; Zettler et al., 1990). Therefore, the detection of CymMV for the control of virus infection is important as it helps to reduce the economic losses in orchid production.
To date, some diagnostic methods have been developed to detect CymMV in orchid plants. These techniques include immunological assays (Eun and Wong, 1999; Park et al., 1998; Tanaka et al., 1997; Yoon et al., 2014), nucleic acid hybridization (Eun et al., 2002; Hu and Wong, 1998), reverse transcription (RT)-polymerase chain reaction (PCR) (Kim and Choi, 2015; Seoh et al., 1998; Yoon et al., 2011), and one-step RT loop-mediated isothermal amplification assay (LAMP) (Lee at al., 2011). However, all of these techniques have a few disadvantages, such as time-consuming, low-sensitivity, cross reactivity, laborious or requirement of expensive equipment. Most of these technologies are not applied directly to detect CymMV in the orchid farms, but have to be applied in the laboratory. For example, RT-LAMP is a reliable detection method. However, this technology requires four to six primers, and the primer design process is relatively complex. RT-LAMP is also limited to inapplicability for cloning, multiplex application and low specificity (Wong et al., 2018).
Recently, isothermal recombinase polymerase amplification (RPA) and RT-RPA assay have been shown to be a rapid, simple, sensitive and cost-effective approach to DNA and RNA detection in plant viruses and viroids (Babu et al., 2018; Ivanov et al., 2020; Kim et al., 2019, 2020; Kovalskaya and Hammond, 2022; Lodoño et al., 2016; Lu et al., 2018; Mekuria et al., 2014; Silva et al., 2014; Zhang et al., 2014). The RPA or RT-RPA can be generally carried out at low temperature (37-42°C) and the reaction is completed within 20 min depending on the starting copy number of the targeted template and the size of the amplicon (Li et al., 2019). In general, RPA products are analyzed using agarose gel electrophoresis, probe-based fluorescence or lateral flow immunoassay (LFI). Of the analyzing methods, RPA products combined with LFI (RPA-LFI) is able to avoid the time and equipment required for electrophoresis (Milenia Biotec, Giessen, Germany). To achieve the RPA-LFI technology, primers should be end-labeled with chemically modified reporter molecules, such as 5′-FAM or 5′-biotin. Subsequently, the resulting amplicon containing biotin and FAM can be shown using a nitrocellulose membrane coated with anti-biotin antibody and anti-FAM nanogold conjugates (Fig. 1). This study is aimed to develop a RT-PRA method for CymMV using specifically modified primers and LFI, and to validate for on-site CymMV detection in orchid samples from farms without any equipment.
Cymbidium sp. and Phalaenopsis sp. which showed necrotic stripe or chlorotic patch symptoms were kindly obtained from National Institute of Horticultural and Herbal Science, Rural Development Administration, Korea in 2020. The plants were maintained in a growth chamber with a 16 h light/8 h dark cycle at 24°C.
Total RNA was extracted from symptomatic leaves of 8 Cymbidium sp. and 7 Phalaenopsis sp. using easy-BLUE total RNA extraction kit, according to manufacturer’s instructions (iNtRON Biotechnology Inc., Seongnam, Korea). Briefly, 100 mg of fresh leaf tissue was ground to a fine powder using liquid nitrogen, transferred to a sterile 1.5 ml tube followed by the addition of 1.0 ml of extraction buffer (100 mM Tris [pH 8.0], 50 mM EDTA, 500 mM NaCl, and 10 mM 2-mercaptoethanol). The tube was vortexed vigorously, and then incubated at room temperature for 5 min. Two hundred microliter of chloroform was added and vortexed vigorously, followed by incubation at room temperature for 3 min. The sample was centrifuged at 13,000 rpm for 15 min at 4°C. The upper aqueous phase was collected into a new tube and 0.5 ml of 100% isopropanol was added to the tube. The mixture then was incubated at room temperature for 10 min and centrifuged at 13,000 rpm for 10 min at 4°C. Total RNA was washed using 1.0 ml of 75% ethanol, and centrifuging the tube at 10,000 rpm for 5 min at 4°C. The RNA pellet was air-dried for 10 min. The RNA pellet was then resuspended in 50 μl of RNase-free water. RNA quality and concentration were determined using a NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA), and total RNA was stored at −70°C for use later.
A single infection of CymMV in Cymbidium sp. and Phalaenopsis sp. was confirmed by RT-PCR with primers specific to CymMV, odontoglossum ringspot virus (ORSV), or cymbidium chlorotic mosaic virus (CyCMV), as describe previously (Chung et al., 2010; Yoon et al., 2018) (Supplementary Table 1). A healthy Cymbidium sp. was used as a negative control. Orchid mitochondrial nad5 mRNA was used as the internal control for RT-PCR assay (Lee and Chang, 2006) (Supplementary Table 1). RT-PCR was carried out using 1 μl of total RNA, 1 μl of each of 10 pmoles of forward and reverse primers, 13 μl of DEPC-treated water, and 5 μl of Topscript one-step RT-PCR mix (Enzynomics, Daejeon, Korea), with a total reaction volume of 20 μl. The thermo-cycling conditions were as follows: 30 min at 50°C for RT, 10 min at 95°C (1 cycle), 94°C for 30 s, 50°C for 30 s, and 72°C, for 40 s (40 cycles), and a final extension at 72°C for 5 min.
To design primers specific for RT-RPA-LFI assay, CP sequences and TGBp1 sequences of 13 CymMV isolates (AF016914, LC125633, AB198937, EF125180, AM055720, HQ681906, AY571289, U62963, AM055640, JQ860108, EU314803, MK816927, and KR185347) originating from some countries were downloaded from the GenBank database. Then, conserved regions of CP genes or TGBp1 genes were analyzed using multiple sequence alignments of MEGA10 (Kumar et al., 2018) (Supplementary Table 2). After designing a total of 10 primer sets targeting CP and TGB1, primer set showing the optimal RT-RPA result was selected among these primer sets, and biotin or FAM was attached to the end of the primers (CymMVT1-3sFAM-F and CymMVT1-3sBiotin-R) (Table 1, Supplementary Table 3). They were used for optimizing for RT-RPA-LFI. The RT-RPA-LFI carried out with those primer set according to the manufacturer’s instructions of TwistAmp Basic kit (TwistDX, Ltd., Cambridge, UK). The RT-RPA-LFI assay for CymMV detection in orchids was performed in a 50 μl reaction volume containing 1 μl of total RNA isolated from each CymMV-infected orchid tissue, 29.5 μl rehydration buffer, 2.4 μl of 2 pmoles forward primer, 2.4 μl of 2 pmoles reverse primer, 1.0 μl of RevertAid reverse transcriptase (Thermo Fisher Scientific), 2.5 μl of 280 mM magnesium acetate, and 11.2 μl DEPC-treated water. After a brief spin, the reaction was incubated for 30 min at 38°C in heat block with a closed lid. For lateral flow assay, with reference to the Milenia GenLine HybriDetect 1 (Milenia Biotec), 10 μl of RT-RPA products were mixed with 100 μl of assay buffer (HybriDetect assay buffer, Milenia Biotec) in a new reaction tube. Then the LFI strip was immersed into the mixture and incubated for 5 min at room temperature. A band with one visible line in the control line was considered negative, and a band with two visible lines in both the control line and the test line was determined as positive (Fig. 1B, Supplementary Fig. 1).
The detection specificity of the primers was verified in a single CymMV, ORSV, or CyCMV-infected orchid sample by RT-RPA assay (Fig. 2A). The RT-PCR was performed to verify the viral presence on the orchids (Fig. 2A). The RPA-LFI assay successfully detected CymMV in each orchid using primers specific to the partial TGBp1 gene of CymMV, and did not detect CymMV in ORSV-infected, CyCMV-infected and a healthy orchid plant, similar to the results of RT-RPA on agarose gel electrophoresis (Fig. 2B). These results suggest that the specificity of RT-RPA for CymMV detection is equivalent to that of conventional RT-PCR analysis and has no cross-reactivity in ORSV and CyCMV at the test band of the RT-RPA-LFI assay.
The conditions of the RPA reaction were optimized based on the reaction temperature and time. The assay was used with a selected primer set (CymMVT1-3s-F and CymMVT1-3s-R) and visualized on the 2% agarose gel after 15 min of RT-RPA incubation time at a temperature of 35-42°C (Supplementary Fig. 2). The RT-RPA-LFI assay was performed on CymMV-infected Cymbidium leaves at 1, 5, 10, 20, and 30 min, respectively. The assay was visualized with a red band at the test line of the immunostrip from 10 min onwards and remained until 30 min at a temperature of 39°C. The strips with no-template control (NTC) were not visualized any line in the same range of time (Fig. 3). Therefore, it was determined that the optimal reaction conditions for the detection of CymMV RT-RPA-LFI assay were 39°C for 15 min.
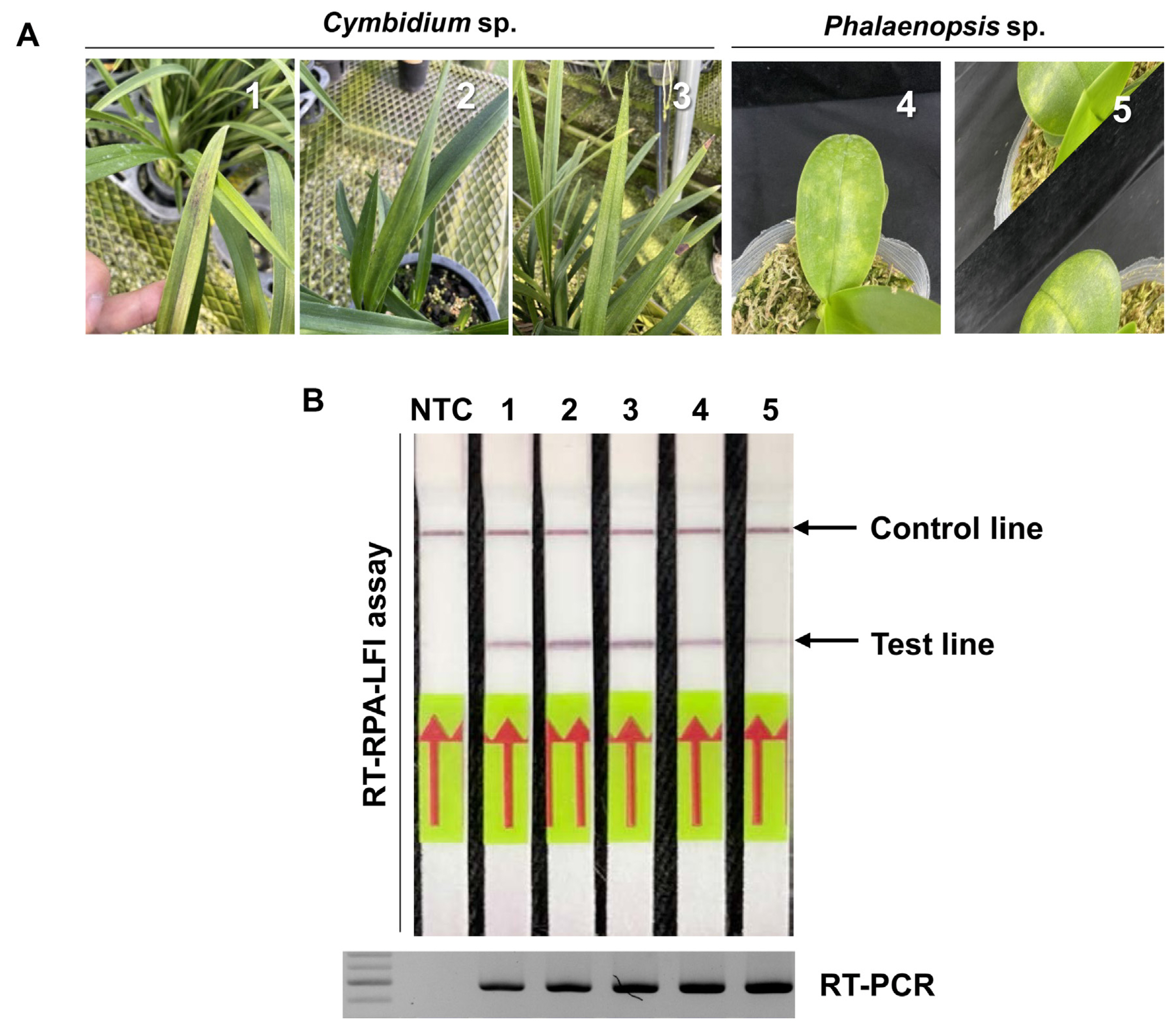
To survey the CymMV infection, a total of five samples were randomly selected among 8 Cymbidium sp. and 7 Phalaenopsis sp. plants and subjected to RT-RPA-LFI assay (Fig. 4A). All leaves have confirmed the infection of CymMV by RT-RPA-LFI assay without a false-positive band in the test line and consistent with the RT-PCR results (Fig. 4B). These results indicated that the RT-RPA-LFI assay could be used reliably for CymMV detection in field-collected samples.
In the present study, we successfully developed a RT-RPA-LFI assay to detect CymMV from orchid plants, showing a simple, rapid, sensitive, and accurate method. The RT-RPA-LFI assay for CymMV detection was not showed cross-reactivity with three orchid-infecting viruses (Fig. 2). On-site detection of CymMV from orchid leaves in farms proved applicability of the RT-RPA-LFI assay and the determination for the presence of CymMV in farms can be verified from 10 minutes after the reaction, compared with 150 min for the RT-PCR assay. These features allow wide use both in the laboratory and in the field.
To date, there are a number of reports on the development of RPA-based assays for plant viruses (Babu et al., 2018; Babujee et al., 2019; Kim et al., 2019, 2020, 2022, 2016; Lu et al., 2018; Mekuria et al., 2014; Silva et al., 2014; Zhang et al., 2014) and viroids (Hammond and Zhang, 2016; Ivanov et al., 2020; Kovalskaya and Hammond, 2022; Lee et al., 2020). The RT-RPA-LFI assays for detection of plant viruses and viroids were performed in accordance with the TwistDx kit and lateral flow strips were utilized as an end-point readout method (Kim et al., 2022; Kovalskaya and Hammond, 2022). Most RPA assays were developed with a TwistAmp nfo kit which is required nfo probe as well as a specific primer set (Babu et al., 2018; Ivanov et al., 2020; Kim et al., 2022). Here, we validated RPA reaction for the detection of CymMV with a TwistAmp basic kit without a probe. To decrease the excessive amplification in RPA reaction, half the concentration (4.8 pmole) of primer was used and RPA generates a limited amount of the target. The low quantity of amplicons was not problematic as a non-specific reaction in the NTC reaction. To our knowledge, this is the first report of the development of RT-RPA-LFI assay to detect CymMV RNA in orchid plants using TwitAmp basic kit, combined with the LFI method.
Detection methods for CymMV include generally double-sandwich enzyme-linked immunosorbent assay (DAS-ELISA) (Park et al., 1998; Sherpa et al., 2006), nucleic acid hybridization (Eun et al., 2002; Hu and Wong, 1998), RT-PCR (Chung et al., 2010; Park et al., 1998; Seoh et al., 1998; Yoon et al., 2012), multiplex RT-PCR (Ali et al., 2014; Kim and Choi, 2015; Lee and Chang, 2006), RT-LAMP (Lee et al., 2011), and real-time RT-PCR (Eun et al., 2000). Most of these assays require expensive equipment, purification of nucleic acids, trained persons and analytic tools of amplified products by gel electrophoresis or specialized equipment. Furthermore, commercial DAS-ELISA kits and lateral flow immunoassay kits for CymMV and ORSV are available from a few companies. The latter kits have been known for on-site diagnosis of CymMV and ORSV, similar to the RT-RPA-LFI assay for CymMV detection. Nonetheless, one of disadvantages of the RT-RPA-LFI assay is the cost. The price of RT-RPA reagents per sample is higher than that of RT-PCR analysis. There have been some reports for cost cut-off per sample in lateral flow strip detection, such as in the use of inexpensive paper and glass fiber (Lobato and O’Sullivan, 2018). Development of the simple sensitive, rapid and accurate RT-RPA-LFA assay helps early diagnosis and subsequent prevention of CymMV in orchid production in Korea.
Acknowledgments
This study was supported by a grant of Cooperative Research Program for Agriculture Science and Technology Development (Project no. PJ01499503), Rural Development Administration, Republic of Korea and supported by “Research Base Construction Fund Support Program” funded by Jeonbuk National University in 2022.
Electronic Supplementary Material
Supplementary materials are available at the website of the Plant Pathology Journal (hhtp://www.ppjonline.org).
Fig. 1
Schematic representation of the reverse transcription recombinase polymerase amplification (RT-RPA) assay for the detection of cymbidium mosaic virus. (A) Schematic representation of the working principle of RT-RPA which driven FAM-/biotin-labeled primers without the nfo probe. (B) End point detection of the biotin linked RT-RPA product by lateral flow immunoassay (LFI). The migration direction of RT-RPA products was indicated by an arrow from the sample pad of the gray color at the bottom to up.
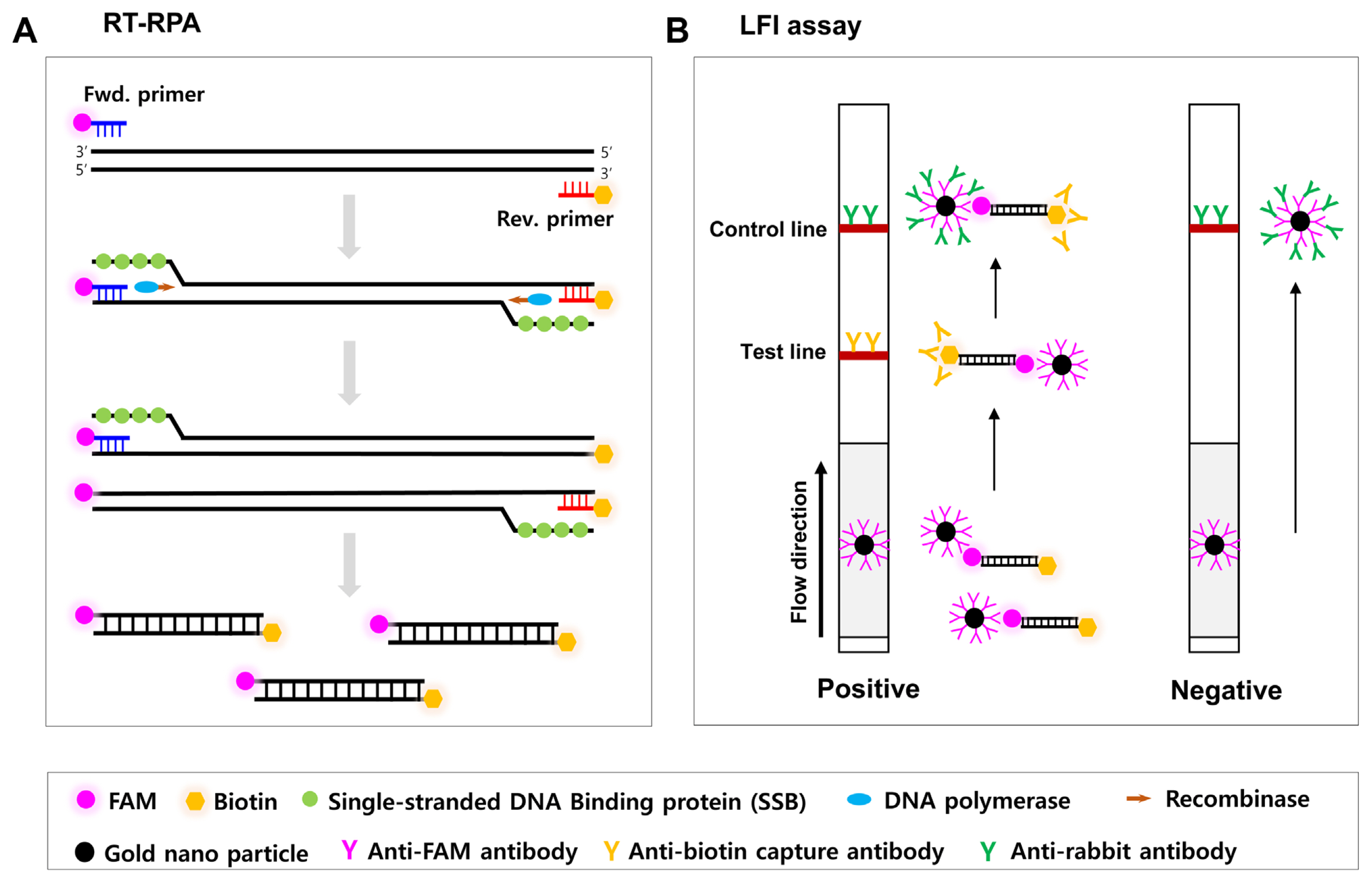
Fig. 2
Detection specificity of the reverse transcription recombinase polymerase amplification (RT-RPA) and RT-RPA assay combined with a lateral flow immunostrip (RT-RPA-LFI) assay for the detection of cymbidium mosaic virus (CymMV). A single CymMV, odontoglossum ringspot virus (ORSV), or cymbidium chlorotic mosaic virus (CyCMV)-infected orchid sample was used as a template for the CymMV detection of RT-RPA and RT-RPA-LFI assay. (A) RT-RPA and reverse transcription polymerase chain reaction (RT-PCR) were visualized on 2% agarose gels. The primers used in RT-RPA and RT-PCR were described in Supplementary Tables 1 and 2. Lane 1, CymMV-infected sample; lane 2, ORSV-infected sample; lane 3, CyCMV-infected sample; lane 4, healthy Cymbidium plant. The mitochondrial NADH dehydrogenase gene (nad5) of orchid was used as an internal control (Lee and Chang, 2006). DNA marker (M) was used 100 bp DNA ladder marker (Enzynomics, Daejeon, Korea). (B) RT-RPA-LFI assay. Fifty-fold dilutions of RT-RPA product in 1× phosphate buffered saline containing 0.1% Tween 20. The NTC is the no-template control used as sterilized water.
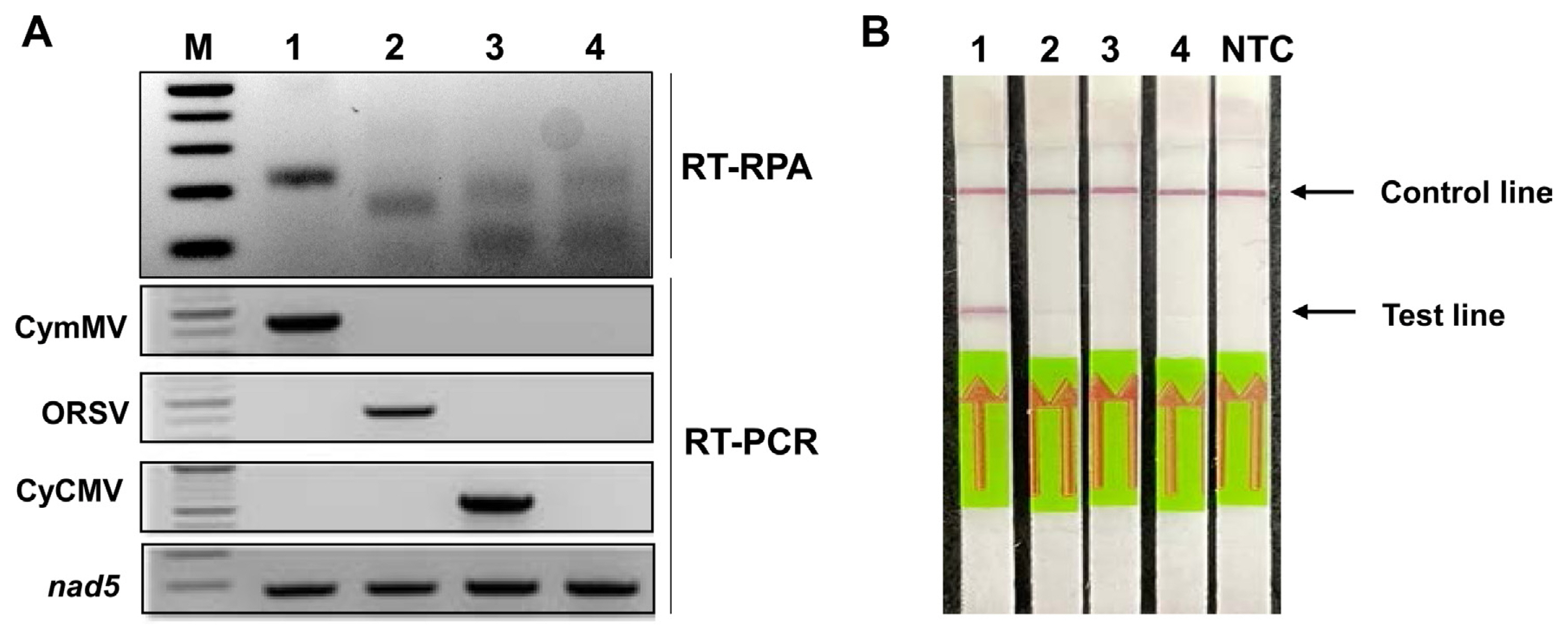
Fig. 3
Optimization of reaction time of reverse transcription recombinase polymerase amplification (RT-RPA) assay combined with a lateral flow immunostrip assay. The RT-RPA was performed on cymbidium mosaic virus (CymMV)-infected Cymbidium leaves at 5, 10, 15, 20, and 30 min, respectively. The assay was visualized with a distinct line at the test line of the strip from 10 min onwards and remained the same thick band from 15 min to 30 min at a temperature of 39°C. The strips with no-template control (NTC) were not visualized any line in the same range of time.

Fig. 4
Application of the reverse transcription recombinase polymerase amplification assay combined with a lateral flow immunostrip (RT-RPA-LFI) assay for the detection of cymbidium mosaic virus (CymMV) on the leaves of Cymbidium sp. and Phalaenopsis sp. (A) Symptomatic leaves of Cymbidium sp. and Phalaenopsis sp. showed the necrotic or chlorotic patch symptoms. (B) CymMV was detected on the leaf samples of Cymbidium sp. and Phalaenopsis sp. by RT-RPA-LFI assay. All leaves confirmed the infection of CymMV by reverse transcription polymerase chain reaction (RT-PCR). Immunostrips did not have a false-positive band in the test line. Lanes 1-3, symptomatic leaves from Cymbidium sp.; lanes 4-5, leaves from Phalaenopsis sp.; lane NTC, no-template control.
References
Adams, M.J., Antoniw, J.F., Bar-Joseph, M., Brunt, A.A., Candresse, T., Foster, G.D., Martelli, G.P., Milne, R.G., Zavriev, S.K. and Fauquet, C.M. 2004. The new plant virus family Flexiviridae and assessment of molecular criteria for species demarcation. Arch. Virol 149:1045-1060.



Ajjikuttira, P.A., Lim-Ho, C.L., Woon, M.H., Ryu, K.H., Chang, C.A., Loh, C.S. and Wong, S.M. 2002. Genetic variability in the coat protein genes of two orchid viruses: Cymbidium mosaic virus and Odontoglossum ringspot virus
. Arch. Virol 147:1943-1954.



Ali, R.N., Dann, A.L., Cross, P.A. and Wilson, C.R. 2014. Multiplex RT-PCR detection of three common viruses infecting orchids. Arch. Virol 159:3095-3099.



Babu, B., Ochoa-Corona, F.M. and Paret, M.L. 2018. Recombinase polymerase amplification applied to plant virus detection and potential implications. Anal. Biochem 546:72-77.


Babujee, L., Witherell, R.A., Mikami, K., Aiuchi, D., Charkowski, A.O. and Rakotondrafara, A.M. 2019. Optimization of an isothermal recombinase polymerase amplification method for real-time detection of Potato virus Y O and N types in potato. J. Virol. Methods 267:16-21.


Chung, B.N., Yoon, J.-Y. and Kim, M.S. 2010. Viral infection of tissue cultured orchids and evaluation of damages. Plant Pathol. J 16:194-197.

Eun, A.J.-C., Huang, L., Chew, F.T., Li, S.F.-Y. and Wong, S.M. 2002. Detection of two orchid viruses using quartz crystal microbalance (QCM) immunosensors. J. Virol. Methods 99:71-79.


Eun, A.J.-C., Seoh, M.-L. and Wong, S.-M. 2000. Simultaneous quantitation of two orchid viruses by TaqMan real-time RT-PCR. J. Virol. Methods 87:151-160.


Eun, A.J. and Wong, S.M. 1999. Detection of cymbidium mosaic potexvirus and odontoglossum ringspot tobamovirus using immuno-capillary zone electrophoresis. Phytopathology 89:522-528.


Hammond, R.W. and Zhang, S. 2016. Development of a rapid diagnostic assay for the detection of tomato chlorotic dwarf viroid based on isothermal reverse-transcription-recombinase polymerase amplification. J. Virol. Methods 236:62-67.


Hu, W.W. and Wong, S.M. 1998. The use of DIG-labelled cRNA probes for the detection of cymbidium mossaic potexvirus (CymMV) and odontoglossum ringspot tobamovirus (ORSV) in orchids. J. Virol. Methods 70:193-199.

Ivanov, A.V., Shmyglya, I.V., Zherdev, A.V., Dzantiev, B.B. and Safenkova, I.V. 2020. The challenge for rapid detection of high-structured circular RNA: assay of potato spindle tuber viroid based on recombinase polymerase amplification and lateral flow tests. Plants 15:1369.



Kim, N.-K., Kim, S.-M. and Jeong, R.-D. 2020. Reverse transcription recombinase polymerase amplification assay for rapid and sensitive detection of barley yellow dwarf virus in oat. Plant Pathol. J 36:497-502.




Kim, N.-K., Lee, H.-J., Kim, S.-M. and Jeong, R.-D. 2022. Rapid and visual detection of barley yellow dwarf virus by reverse transcription recombinase polymerase amplification with lateral flow strips. Plant Pathol. J 38:159-166.




Kim, N.-Y., Lee, H.-J. and Jeong, R.-D. 2019. A portable detection assay for apple stem pitting virus using reverse transcription-recombinase polymerase amplification. J. Virol. Methods 274:113747.


Kim, S.M. and Choi, S.H. 2015. Simultaneous detection of cymbidium mosaic virus and odontoglossum ringspot virus in orchids using multiplex RT-PCR. Virus Genes 51:417-422.



Kovalskaya, N. and Hammond, R.W. 2022. Rapid diagnostic detection of tomato apical stunt viroid based on isothermal reverse transcription-recombinase polymerase amplification. J. Virol. Methods 300:114353.


Kumar, S., Stecher, G., Li, M., Knyaz, C. and Tamura, K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol 35:1547-1549.



Lawson, R.H. and Hsu, H.T. 1995. Orchid. In: Virus and virus-like diseases of bulb and flower crops, eds. by G. Loebenstein, R.H. Lawwon and A.A. Brunt, pp. 409-420. John Wiley & Sons, New York, USA.
Lee, H.-J., Kim, H.-J., Lee, K. and Jeong, R.-D. 2020. Rapid detection of peach latent mosaic viroid by reverse transcription recombinase polymerase amplification. Mol. Cell. Probes 53:101627.


Lee, M.-S., Yang, M.-J., Hseu, Y.-C., Lai, G.-H., Chang, W.-T., Hsu, Y.-H. and Lin, M.-K. 2011. One-step reverse transcription loop-mediated isothermal amplification assay for rapid detection of Cymbidium mosaic virus. J. Viol. Methods 173:43-48.

Lee, S.-C. and Chang, Y.-C. 2006. Multiplex RT-PCR detection of two orchid viruses with an internal control of plant nad5 mRNA. Plant Pathol. Bull 15:187-196.
Li, T.-T., Wang, J.-L., Zhang, N.-Z., Li, W.-H., Yan, H.-B., Li, L., Jia, W.-Z. and Fu, B.-Q. 2019. Rapid and visual detection of Trichinella spp. using a lateral flow strip-based recombinase polymerase amplification (LF-RPA) assay. Front. Cell. Infect. Microbiol 9:1.



Lobato, I.M. and O’Sullivan, C.K. 2018. Recombinase polymerase amplification: basics, applications and recent advances. Trends Analyt. Chem 98:19-35.



Londoño, M.A., Harmon, C.L. and Polston, J.E. 2016. Evaluation of recombinase polymerase amplification for detection of begomoviruses by plant diagnostic clinics. Virol. J 13:48.



Lu, Y., Yao, B., Wang, G. and Hong, N. 2018. The detection of ACLSV and ASPV in pear plants by RT-LAMP assays. J. Virol. Methods 252:80-85.


Mekuria, T.A., Zhang, S. and Eastwell, K.C. 2014. Rapid and sensitive detection of Little cherry virus 2 using isothermal reverse transcription-recombinase polymerase amplification. J. Virol. Methods 205:24-30.


Ministry for Food, Agriculture, Forestry and Fisheries, Korea 2021 Production of ornamental plants in Korea, 2021 URL https://data.mafra.go.kr/opendata/data/indexOpenDataDetail.do?data_id=20210819000000001556&filter_ty=#
. [30 October 2022].
Park, W.M., Shim, K.B., Kim, S.J. and Ryu, K.H. 1998. Detection of cymbidium virus and odontoglossum ringspot virus by ELISA and RT-PCR from cultivated orchids in Korea. Korean J. Plant Pathol 14:130-135.
Ryu, K.H. and Park, W.M. 1995. The complete nucleotide sequence and genome organization of Odontoglossum ringspot tobamovirus RNA. Arch. Virol 140:1577-1587.



Seoh, M.L., Wong, S.M. and Zhang, L. 1998. Simultaneous TD/RT-PCR detection of cymbidium mosaic potexvirus and odontoglossum ringspot tobamovirus with a single pair of primers. J. Virol. Methods 72:197-204.


Sherpa, A.R., Bag, T.K., Hallan, V. and Zaidi, A.A. 2006. Detection of Odontoglossum ringspot virus in orchids from Sikkim, India. Aust. Plant Pathol 35:69-71.

Silva, G., Bömer, M., Nkere, C., Kumar, P.L. and Seal, S.E. 2014. Rapid and specific detection of yam mosaic virus by reverse-transcription recombinase polymerase amplification. J. Virol. Methods 222:138-144.

Tanaka, S., Nishii, H., Ito, S., Kameya-Iwaki, M. and Sommartya, P. 1997. Detection of Cymbidium mosaic potexvirus and Odontoglossum ringspot tobamovirus from Thai orchids by rapid immunofilter paper assay. Plant Dis 81:167-170.


Verchot-Lubicz, J., Ye, C.-M. and Bamunusinghe, D. 2007. Molecular biology of potexviruses: recent advances. J. Gen. Virol 88:1643-1655.


Wisler, G.C., Zettler, F.W. and Mu, L. 1987. Virus infections of Vanilla and other orchids in French Polynesia. Plant Dis 71:1125-1129.

Wong, S.M., Chng, C.G., Lee, Y.H., Tan, K. and Zettler, F.W. 1994. Incidence of cymbidium mosaic and odontoglossum ringspot viruses and their significance in orchid cultivation in Singapore. Crop Prot 13:235-239.

Wong, Y.-P., Othman, S., Lau, Y.-L., Radu, S. and Chee, H.-Y. 2018. Loop-mediated isothermal amplification (LAMP): a versatile technique for detection of micro-organisms. J. Appl. Microbiol 124:626-643.




Yoon, J.-Y., Choi, G.-S., Cho, I.-S. and Choi, S.-K. 2014. Development of rapid immune-gold strip kit for on-site diagnosis of tomato spotted wilt virus. Res. Plant Dis 20:15-20.

Yoon, J.-Y., Chung, B.-N., Choi, G.-S. and Choi, S.-K. 2012. Genetic variability in the coat protein genes of Cymbidium mosaic virus isolates from orchids. Virus Genes 44:323-328.



Yoon, J.-Y., Chung, B.-N. and Choi, S.-K. 2011. High sequence conservation among Odotoglossum ringspot virus isolates from orchids. Virus Genes 42:261-267.



Yoon, J.Y., Kwon, S.J., Cho, I.S. and Choi, G.S. 2018. First report of cymbidium chlorotic mosaic virus infection in Cymbidium goeringii in South Korea. Plant Dis 102:2665.

- TOOLS
-
METRICS

- ORCID iDs
-
Ju-Yeon Yoon

https://orcid.org/0000-0003-1646-7310 - Related articles



 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Supplement
Supplement Print
Print



