The Crucial Role of Chloroplast-Related Proteins in Viral Genome Replication and Host Defense against Positive-Sense Single-Stranded RNA Viruses
Article information
Abstract
Plant viruses are responsible for worldwide production losses of numerous economically important crops. The most common plant RNA viruses are positive-sense single-stranded RNA viruses [(+)ss RNA viruses]. These viruses have small genomes that encode a limited number of proteins. The viruses depend on their host’s machinery for the replication of their RNA genome, assembly, movement, and attraction to the vectors for dispersal. Recently researchers have reported that chloroplast proteins are crucial for replicating (+)ss plant RNA viruses. Some chloroplast proteins, including translation initiation factor [eIF(iso)4E] and 75 DEAD-box RNA helicase RH8, help viruses fulfill their infection cycle in plants. In contrast, other chloroplast proteins such as PAP2.1, PSaC, and ATPsyn-α play active roles in plant defense against viruses. This is also consistent with the idea that reactive oxygen species, salicylic acid, jasmonic acid, and abscisic acid are produced in chloroplast. However, knowledge of molecular mechanisms and functions underlying these chloroplast host factors during the virus infection is still scarce and remains largely unknown. Our review briefly summarizes the latest knowledge regarding the possible role of chloroplast in plant virus replication, emphasizing chloroplast-related proteins. We have highlighted current advances regarding chloroplast-related proteins’ role in replicating plant (+)ss RNA viruses.
Viruses cause major crop losses worldwide and thus are a threat to sustainable and productive agriculture. New plant viruses are being discovered and continue to pose a clear danger to our food systems globally (Hilaire et al., 2022; Jones and Naidu, 2019; Rubio et al., 2020; Whitfield et al., 2015). Once positive-sense single-stranded RNA viruses [(+)ss RNA viruses] are inside the cell, the viral genome serves as a template for the production of a large number of progeny viruses (den Boon et al., 2010; Nagy and Pogany, 2008b), and RNA replication is done by viral proteins and host plant proteins in chloroplast membranes of the infected cell. Many viruses invade specific host chloroplast membranes in the process of viral genome replication (Hyodo and Okuno, 2016; Nagy et al., 2012; Sanfaçon, 2005; Xu and Nagy, 2014). Therefore, chloroplasts play central roles in replicating several plant virus species and biosynthesis of most plant hormones, making chloroplast factors crucial for plant defense response.
The chloroplast is a vital organelle of plant cells carrying out photosynthesis. In addition to the outer and inner membranes, mature chloroplasts have an internal membrane network of thylakoids, where the light energy is converted into chemical energy stored in ATP (Fig. 1) (Li et al., 2016). Many researchers have published quality data using confocal microscopes, and they have captured stunning and excellent images (Bhat et al., 2013; Kaido et al., 2014; Thivierge et al., 2008) which show that majority of the plant (+)ss RNA viruses replicate, assemble, and mature in chloroplast components called viral replication complexes (VRCs), including plum pox virus (PPV) (Martin et al., 1995), tobacco etch virus (Gadh and Hari, 1986), turnip mosaic virus (TuMV) (Kitajima and Costa, 1973), maize draft mosaic virus (Mayhew and Ford, 1974), and tobacco mosaic virus (TMV) (Bhat et al., 2013). Moreover, chloroplast-related proteins are reportedly involved in defense against plant viruses because chloroplasts are sites where defense-related hormones are synthesized (Bhattacharyya and Chakraborty, 2018; Bobik and Burch-Smith, 2015; Kozuleva et al., 2011; Nambara and Marion-Poll, 2005; Padmanabhan and Dinesh-Kumar, 2010; Seyfferth and Tsuda, 2014; Stael et al., 2015; Torres et al., 2006; Wasternack and Hause, 2013; Wildermuth et al., 2001; Yang et al., 2021). Recently, the chloroplast membrane-associated protein pSaC in photosystem I (PSI) and the ATP-synthase α-subunit whose chloroplast DNA encoded protein is part of the ATP-synthase complex induced resistance in soybean mosaic virus (SMV) belonging to the Potyvirus genus (Bwalya et al., 2022).
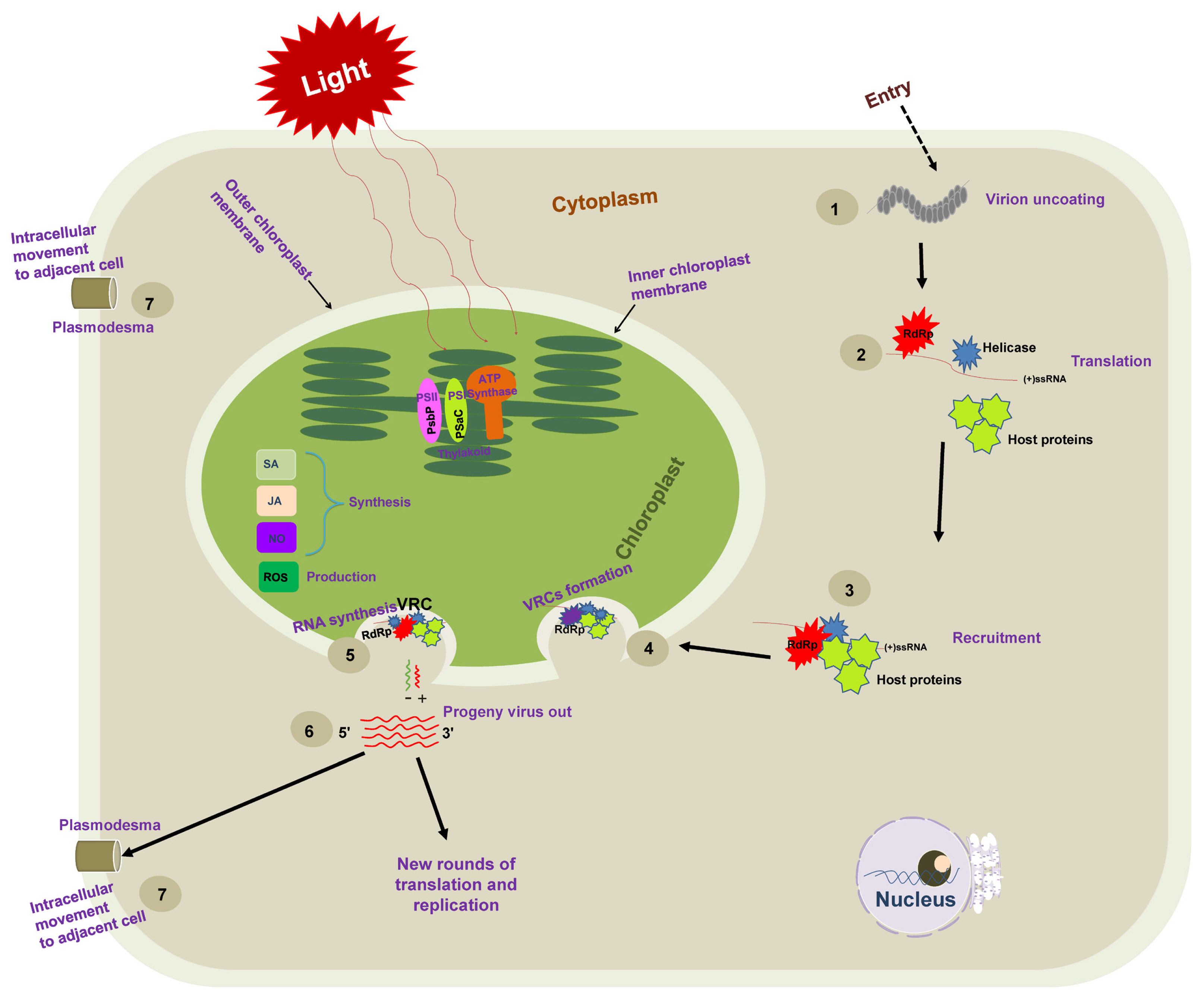
Schematic representation of necessary steps for positive-sense single-stranded RNA [(+)ss RNA] virus genome replication. Following the entry into host cells, (1) viral genomic RNAs are released from virions into the cytoplasm, and (2) viral RNA [(+)ss RNA] translates RNA-dependent RNA polymerase (RdRp) at the early stage of infection and recruits additional factors (such as methyltransferase and chaperone). (3) The resulting viral replication proteins target themselves to recruit the host translation machinery for the successful production of viral replication proteins. (4) The recruited viral genomic (+) RNAs and host proteins are then trafficked to chloroplast membranes, where they assemble viral replication complexes (VRCs) on the host chloroplast membrane. VRCs are shown by an invagination of the plant chloroplast membrane containing the viral protein (blue shape), the viral RdRP (red shape), host proteins (green shape), and viral RNA (red line). The VRC synthesizes a complementary negative-strand RNA (green line) using the original (+)RNA as a template. The (−) RNA is then used as a template to synthesize many new (+) RNAs (red lines). Progeny viruses are released from VRCs, undergo additional translation and replication, or move to adjacent cells.
In another example, infection with TMV reduced the expression levels of the ATPsyn-γ gene and silencing ATPsyn-γ in Nicotiana benthamiana increased TMV accumulation and pathogenicity (Bhat et al., 2013). Jiménez et al. (2006) have indicated a positive role for PSI-K protein (a product of the gene psaK) in resistance against the PPV. For PPV infection, transient expression of PSI-K decreased accumulation level of PPV RNA in N. benthamiana. But PPV RNA accumulation was enhanced when psaK was knocked down (Jiménez et al., 2006). Importantly, it should be noted that many viruses intrude photosynthetic machinery functionality through protein-protein interactions and participate in peddling these molecules to the chloroplast, where replication occurs (Fig. 2). This review summarizes the active roles of chloroplast proteins that play in the replication of (+)ss RNA viruses and plant defense against viruses.
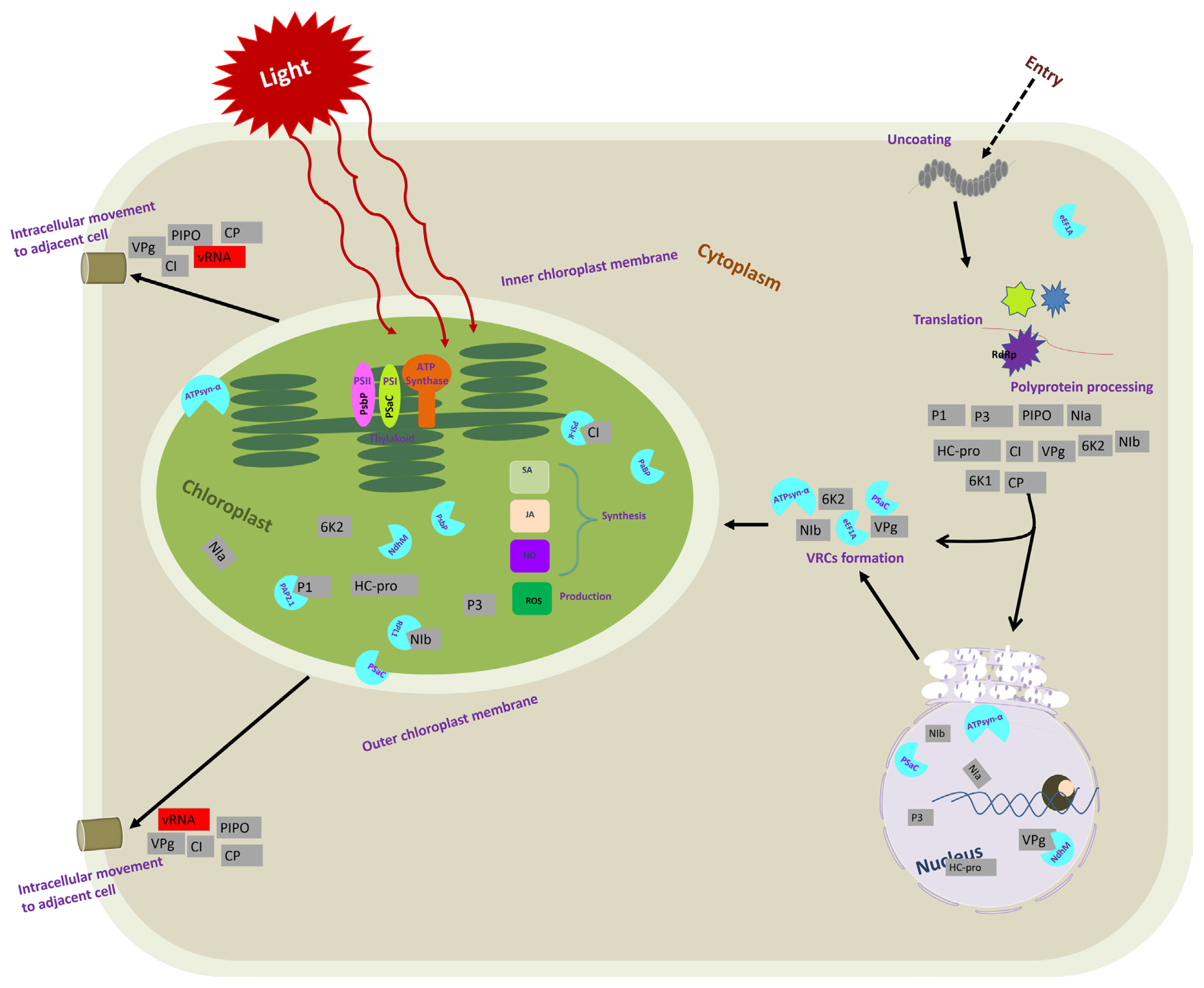
Schematic representation of the important events during chloroplast host factors and potyvirus interactions in a plant cell. After entry into host plant cells, a potyvirus virion undergoes the disassembly of viral particles and releases the viral genome. Viral genomic RNA is then used as the template for translation to produce viral polyproteins (11 viral proteins indicated by grey squares). The 6K2 remodels the subcellular membranes to form the viral replication complexe (VRC)-containing vesicles for potyvirus genome replication. The 6K2-induced vesicles may subsequently target chloroplasts for robust viral replication. The NIb is recruited to the VRC, likely via its interaction with the VPg domain of 6K2–VPg-Pro. Then, NIb recruits many host factors, such as eEF1A, PSaC, and ATP-synthase α-subunit. In the figure, each viral protein is represented by a grey color-coded square in a location where they play a crucial role. A light blue color-coded semi-circle indicates chloroplast-related host proteins identified for virus infection. They are depicted in sites where they interact with viral protein or play a crucial role.
Chloroplasts and Chloroplast-Related Proteins Facilitate the Viral Replication Cycle of Plant (+)ss RNA Viruses
The viral replication process of (+)ss RNA viruses starts from the switch from translation to replication, including a selection of template RNA (Nagy and Pogany, 2006; Nishikiori et al., 2006; Panavas et al., 2005). The viral and host components required for replication are targeted to chloroplast (subcellular) membranes, where the VRCs are assembled. VRCs synthesize the negative-sense (−) and (+) RNAs and release (+) RNA progenies (Fig. 1).
For plant (+)ss RNA viruses, host translation machinery uses the viral genome as an mRNA to produce replicating proteins and other viral proteins. Host and viral proteins have been well-documented to regulate the switch from translation to replication (Budziszewska and Obrępalska-Stęplowska, 2018; Nagy and Pogany, 2008a). However, most host proteins are more likely to participate in the recruitment step. For example, the recruitment of brome mosaic virus RNAs for replication was affected by Lsm1p, Pat1p, and Dhh1p (Díez et al., 2000; Noueiry and Ahlquist, 2003). When the (+)ss RNA viruses come into the cell, the RNAs come out of the virions and are released into the cytoplasm. The genomes of (+)ss RNA viruses act as a template for translation to produce viral replication proteins, including RNA-dependent RNA polymerase (RdRp), using host machinery leading to a series of interactions between host translation factors and RNA replication (Dreher and Miller, 2006; Simon and Miller, 2013). The viral and host proteins are bound in a discriminatory manner to the (+)ss RNA template and target subcellular membranes. Translation and selection of the viral (+)ss RNA for replication takes place in the cytoplasm, whereas replication of (+)ss RNA viruses occurs on the surface of various intracellular membranes, including chloroplast membranes (Ahlquist, 2002; Ahlquist et al., 2003; Burgyan et al., 1996; den Boon et al., 2010; Rubino and Russo, 1998; Salonen et al., 2005; Widyasari et al., 2020). Intracellular targeting of the RdRp and viral (+)ss RNA to the replication site occurs in a favorable microenvironment to assemble VRCs. VRC formation at the replication site occurs through viral replicase, virus-encoded accessory proteins, and host factors recruited (Jin et al., 2017; Panavas et al., 2005; Salonen et al., 2005). The process of VRC formation is intimately associated with viral translation, intracellular movement, and intercellular movement. In early models of cell-to-cell movement, the viral MP alone was thought to be responsible for intracellular and intercellular movement. Proper coordination of these processes is required for efficient viral infection. Therefore, viruses must properly build VRCs to avoid or minimize disruption of a coordinated balance, in which host factors play a role in the assembly of viral replicase and regulate its function (Hafrén et al., 2010; Jungfleisch et al., 2015; Kaido et al., 2014).
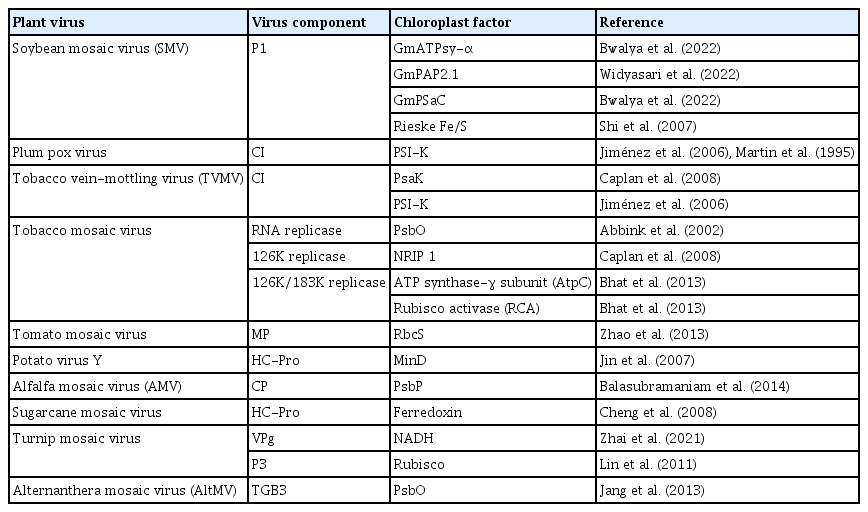
The critical step is the synthesis of viral RNA (De Graaff et al., 1993). The VRC synthesizes complementary (−) RNA using the original genomic (+) RNA as a template. The (−) RNA synthesizes more (+) RNAs. Newly synthesized (+) RNAs are then released from VRC to undergo rounds of translation and replication or move to adjacent cells or package into virions (Fig. 1). Nowadays, many studies have shown the association of the VRC with the outer chloroplast membrane (De Graaff et al., 1993; Moriceau et al., 2017; Nishikiori et al., 2006; Thivierge et al., 2008). Table 1 summarizes chloroplast-related proteins in the viral replication process. It was reported a long time ago that VRCs of alfalfa mosaic virus (AMV), the genus Alfamovirus, are associated with the chloroplast outer membrane (De Graaff et al., 1993). Turnip yellow mosaic virus (TYMV; a (+)ss RNA virus that shares replication features with other alphavirus-like supergroups) replication also occurs in close association with the chloroplast outer envelope membranes (Hatta et al., 1973; Koonin and Dolja, 1993; Prod’homme et al., 2001). The TYMV 140K protein was previously shown to be responsible for the recruitment of the polymerase to VRCs and targeting the VRCs to the outer chloroplast membrane where viral replication occurs (Jakubiec et al., 2004; Prod’homme et al., 2003). Moriceau et al. (2017) investigated determinants for the in vivo chloroplast targeting of the TYMV 140K replication protein. They identified the two amphipathic helices αA and αB within 140K/98K that constitute the determinants for chloroplast targeting of the TYMV VRCs. However, detailed delivery mechanisms remain to be determined.
Furthermore, published data for 3D electron tomography of barley stripe mosaic virus revealed replication factories and remodeling of the chloroplast outer membranes, characterized by clustering outer membrane-invaginated spherules in inner membrane-derived packets (Budziszewska and Obrępalska-Stęplowska, 2018). Our recent study also showed that chloroplast-related proteins pSaC and ATPsyn-α were localized in the chloroplast envelope (Bwalya et al., 2022). Nonetheless, helicases and chloroplast factors participate in each step of RNA synthesis. One such example of (+)ss RNA viruses infecting plant species is the eukaryotic translation elongation factor 1A (eEF1A), which was found to be a permanent resident of the tombusvirus replicase complex and eEF1A promotes the synthesis of (−) RNA by replicase complex (Li et al., 2009). The elevated abundance of eEF1A in the cells and eEF1A interaction with viral (+)RNAs, including the 3′ untranslated region of TYMV (Dreher, 1999), TMV, and TuMV, might facilitate the recruitment of eEF1A into VRC (Nishikiori et al., 2006; Thivierge et al., 2008). In TuMV infection, 6K2 vesicles were transported to the chloroplast and accumulated at the chloroplast membrane, where they induce the formation of chloroplast-bound elongated tubular structures and ultimately form chloroplast aggregation (Wei et al., 2013). Plant viruses co-opt host proteins such as methyltransferase and chaperone for viral replication (Budziszewska and Obrępalska-Stęplowska, 2018), which may perform similar tasks in viral replication. During viral infection, an RNA helicase protein, ISE2 (increased size exclusion limit 2), was highly expressed and accumulated in the chloroplast affecting viral replication and cell-to-cell communication (Ganusova et al., 2017).
Chloroplast-Related Proteins Interact with Viral Components during Virus Replication
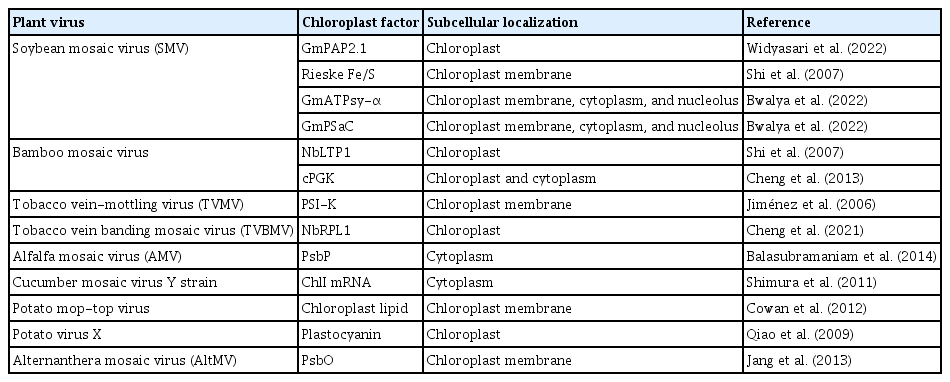
The accumulating evidence highlights that chloroplasts and chloroplast-related proteins can interact with viral components to favor the replication and movement of (+)ss RNA viruses (Bobik and Burch-Smith, 2015; Zhao et al., 2016, 2019) (Table 2, Fig. 2). In soybean (Glycine max), purple acid phosphatase (GmPAP2.1) conferred salicylic acid (SA)-dependent resistance to a susceptible cultivar by interacting with the SMV P1 protein in the chloroplasts, transient knockdown of endogenous SA-related genes resulted in systemic infection by SMV strain G5H (Widyasari et al., 2022). In N. benthamiana, overexpression of chloroplast nicotinamide adenine dinucleotide dehydrogenase-like complex M subunit gene (NdhM) inhibited TuMV accumulation, and the localization of NbNdhM is altered by its interaction with TuMV VPg in a way that promoted virus infection (Zhai et al., 2021). Cylindrical inclusion (CI) protein from PPV interacted with the PSI PSI-K protein (a product of the gene psaK) of N. benthamiana. Transient coexpression of PPV CI in N. benthamiana leaves decreased the level of PSI-K while silencing of psaK enhanced PPV accumulation (Jiménez et al., 2006). A chloroplast ribosomal protein large subunit 1 (NbRPL1), localized to the chloroplasts via its transit peptide, interacted with tobacco vein banding mosaic virus (TVBMV) nuclear inclusion protein b (NIb) and enhanced TVBMV infection. Silencing of NbRPL1 expression reduced TVBMV replication in N. benthamiana (Cheng et al., 2021). The photosystem II (PSII) oxygen-evolving complex protein (PsbP) interacted with the AMV capsid protein, and its overexpression reduced virus replication (Balasubramaniam et al., 2014). The helicase domain of the TMV replicase interacted with the psbO-encoded 33-kDa protein, a component of the oxygen-evolving complex. TMV infection depleted the psbO gene and the thylakoid’s entire PSII core complex. Silencing of the psbO gene increased TMV replication (Abbink et al., 2002; Balasubramaniam et al., 2014; Lehto et al., 2003). In maize (Zea mays), the multifunctional helper component-proteinase protein of sugar cane mosaic virus (SCMV) interacted with chloroplast precursor of ferredoxin-5 (FdV), and SCMV infection significantly downregulated the expression level of FdV mRNA in maize plants (Cheng et al., 2008). SMV P1 protein interacted with Rieske Fe/S protein in tobacco, retarded the Fe/S transport to the chloroplast (Shi et al., 2007). Many more interactions between chloroplast and viral proteins are listed in Table 2.
Chloroplast Plays a Vital Role in Plant Antiviral Defense
The chloroplast is one of the crucial organelles that provides energy and carbon through photosynthesis. However, the plant defense mechanism tends to decrease the photosynthetic rate and other anabolic processes to reduce the organic carbon supply to pathogens (Serrano et al., 2016).
Other than photosynthesis, chloroplast plays significant roles in plant defense. It provides plant defense against viruses because of the production of secondary metabolites, including calcium, reactive oxygen species (ROS), and biosynthesis of several defense-related hormones like SA, jasmonic acid (JA), and abscisic acid (ABA) that have an important connection with plant immunity (Bobik and Burch-Smith, 2015; Kozuleva et al., 2011; Nambara and Marion-Poll, 2005; Padmanabhan and Dinesh-Kumar, 2010; Seyfferth and Tsuda, 2014; Stael et al., 2015; Torres et al., 2006; Wasternack and Hause, 2013; Wildermuth et al., 2001).
Most plant virus-induced SA is synthesized through the isochorismate pathway in chloroplast and plays crucial roles in plant defense against viruses, and is essential for local and systemic acquired resistance (Alazem and Lin, 2015; Boatwright and Pajerowska-Mukhtar, 2013; Seyfferth and Tsuda, 2014; Zhao et al., 2016). For example, overexpression of GmPAP2.1, a chloroplast-localized protein, resulted in the upregulation of the SA pathway. Overexpression of GmPAP2.1 showed resistance to SMV infection, while transient knockdown of endogenous SA-related genes caused severe systemic symptoms by SMV (Widyasari et al., 2022). Another chloroplast-localized protein, named calcium-sensing receptor, acts upstream of SA accumulation and connects chloroplasts to cytoplasmic-nuclear immune responses in Arabidopsis thaliana (Nomura et al., 2012).
JA is synthesized from linolenic acid by the octadecanoid pathway, and biosynthesis starts with the conversion of linolenic acid to 12-oxo-phytodienoic acid in the chloroplast membranes (Turner et al., 2002). JA also plays a crucial role in plant-virus interaction. For example, in silencing the JA perception gene, COI1 (coronatine insensitive 1) accelerates the development of symptoms caused by the co-infection of potato virus X (PVX) and potato virus Y. It increases the level of viral titers at the early stages of infection (García-Marcos et al., 2013).
Pathways of ABA are also involved in plant resistance to viruses (Alazem and Lin, 2015; Alazem et al., 2017, 2018, 2019; Nomura et al., 2012; Zhang et al., 2012; Zhao et al., 2016). Previously published reports showed that ABA enhanced the expression of the antiviral RNA silencing genes in soybean and A. thaliana and that the enhanced expression confers partial resistance against SMV, bamboo mosaic virus, and PVX (Alazem et al., 2017, 2019).
The reaction centers of PSI and PSII in chloroplast thylakoids are the primary generation site of ROS. ROS is a sign of activated antiviral defense (Bwalya et al., 2022; Calil and Fontes, 2017; Wu et al., 2017). The reaction centers of PSI and PSII in chloroplast thylakoids produce ROS and the photosynthetic electron transport chain. Superoxide anion (O2−) is the primary reduced product of O2 photoreduction, and its disproportionation produces H2O2 in chloroplast thylakoids membranes (Asada, 2006; Mühlenbock et al., 2008; Zhao et al., 2016). ROS and calcium bursts in chloroplasts activate signaling cascades that regulate the expression of defense-related genes; therefore, ROS and calcium bursts act as chloroplast-to-nucleus retrograde signals when plants recognize the early step of virus infection (Bhattacharyya and Chakraborty, 2018; Medina-Puche et al., 2020; Nomura et al., 2012; Zhao et al., 2016).
The burst of intracellular ROS can be detected during virus infection in incompatible and compatible interactions (Allan et al., 2001; Hakmaoui et al., 2012). Chloroplast-sourced ROS are essential for hypersensitive response induced by the incompatible defensive response (Torres et al., 2006; Zurbriggen et al., 2010).
Conclusions and Future Perspectives
Chloroplasts have been recognized as a common target by many plant viruses. Hence chloroplast-virus interaction is an epicenter of plant-virus interplays. Viruses may directly modify chloroplast membranes to assemble their replication complex for viral genome replication. Based on previously published reports (Balasubramaniam et al., 2014; Bhat et al., 2013; Bhattacharyya and Chakraborty, 2018; Budziszewska and Obrępalska-Stęplowska, 2018; Bwalya et al., 2022; Cheng et al., 2021; Jiménez et al., 2006; Nagy et al., 2012; Serrano et al., 2016; Wei et al., 2013; Widyasari et al., 2022; Yang et al., 2021), we can conclude that some chloroplast-related proteins could function in virus replication, while some are involved in inhibiting viruses. Our review has summarized a few chloroplast-related proteins identified by researchers and their possible roles in virus infection. However, more chloroplast-related proteins must be determined to understand the proteins involved in host defense accurately. If the desired chloroplast-related gene functions are observed from host plants, they might be used to genetically engineer other plants to express these gene products after isolation and cloning. For example, scientists have boosted a carbon-craving enzyme called RuBisCO to turbocharge photosynthesis in corn (Salesse-Smith et al., 2018). The discovery promises to be a critical step in improving agricultural efficiency and yield. Moreover, more reports showed that overexpression of photosynthesis genes could enhance virus resistance in maize and soybean plants (Bwalya et al., 2022; Wang et al., 2018; Widyasari et al., 2022).
Acknowledgments
This research was supported by grants from the Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, and Forestry (120080-05-1-HD030), funded by the Ministry of Agriculture, Food and Rural Affairs, Republic of Korea. JB was supported by a research fellowship from the Brain Korea 21 Four Program.
Notes
Conflicts of Interest
No potential conflict of interest relevant to this article was reported.