 |
 |
| Plant Pathol J > Volume 39(1); 2023 > Article |
|
Abstract
In the present investigation, bacterial isolates from infected apple trees causing apple canker during winter were studied in the northern Gyeongbuk Province, Korea. The pathogen was identified as Pseudomonas syringae pv. syringae (Pss) through various physiological and biochemical characterization assays such as BIOLOG, gas chromatography of fatty acid methyl esters, and 16S rRNA. Bioassays for the production of phytotoxins were positive for syringopeptin and syringomycin against Bacillus megaterium and Geotrichum candidum, respectively. The polymerase chain reaction (PCR) method enabled the detection of toxin-producing genes, syrB1, and sypB in Pss. The differentiation of strains was performed using LOPAT and GATTa tests. Pss further exhibited ice nucleation activity (INA) at a temperature of −0.7°C, indicating an INA+ bacterium. The ice-nucleating temperature was −4.7°C for a non-treated control (sterilized distilled water), whereas it was −9.6°C for an INA− bacterium Escherichia coli TOP10. These methods detected pathogenic strains from apple orchards. Pss might exist in an apple tree during ice injury, and it secretes a toxin that makes leaves yellow and cause canker symptoms. Until now, Korea has not developed antibiotics targeting Pss. Therefore, it is necessary to develop effective disease control to combat Pss in apple orchards. Pathogenicity test on apple leaves and stems showed canker symptoms. The pathogenic bacterium was re-isolated from symptomatic plant tissue and confirmed as original isolates by 16S rRNA. Repetitive element sequence-based PCR and enterobacterial repetitive intergenic consensus PCR primers revealed different genetic profiles within P. syringae pathovars. High antibiotic susceptibility results showed the misreading of mRNA caused by streptomycin and oxytetracycline.
Apple is one of the major fruit crops in Korea and occupies first place with 29% of the total fruit production when compared with other fruit crops, and produces around 475,303 metric tons (Ritchie and Roser, 2020). It has been reported that in Korea, apples are grown in large amounts with a cultivation area of 33,600 ha, of which the major portion is in the northern Gyeongbuk Province, Korea (Choi and Hinkle, 2017) and thus plays an important role in Korean economic development. Currently, the need for apple production is increasing as they are being exported to various countries from Korea. Apple has been attacked by various diseases in the past several years. Subsequently, there was an occurrence of gummosis spreading on the stems, followed by cracks in the bark of matured apple trees in many apple orchards in winter, which has become a major concern for apple growers in Korea. Similar symptoms were also observed in crabapples in China (Liu et al., 2018). Chemical treatments could not solve this issue.
A research team led by Prof. Yongho Jeon, a senior plant pathologist from Andong National University, Korea, took up a challenge to investigate the same issue in various apple orchards (cv. “Fuji”) in the Gyeongbuk Province, Korea, and determined the reason for the above symptoms on apple trees based on various physiological tests. This team found that the disease incidence characterized by this issue is due to the involvement of the bacterial pathogen Pseudomonas syringae pv. syringae (Pss). These symptoms were found to be the same as the symptoms of apple canker, whose causal pathogen was Pss, described in a recent report on apples in Brazil (Araujo et al., 2020). Similar symptoms were observed in fruit crops, such as apricot, peach, nectarine, sweet cherry, and almonds (Popović et al., 2021). Apple trees showing these unusual symptoms end up collapsing. The symptoms of apple canker were characterized by stunted growth, cracks on the bark, shedding of bark around the graft union, and trees collapsing with a full load of fruits. P. syringae is a highly heterogeneous species comprising more than 60 pathovars (Young, 2010). The ability of pathovar to exhibit virulence and necrotic symptoms in a specific host plant is determined by the pathogenicity test (Gašić et al., 2012).
P. syringae is one of the phytopathogenic bacteria associated with several plant species (Gašić et al., 2018; Kennelly et al., 2007; Lee et al., 2015; Ruinelli et al., 2019), including fruits and ornamental plants (Scortichini et al., 2003; Vicente and Roberts, 2007). It is a prevalent bacterial pathogen with the capacity to incite stem and leaf diseases in various crop plants, particularly in temperate regions (Scholz-Schroeder et al., 2001). The diseases of fruit trees caused by the strains of P. syringae are of major concern in fruit-producing countries, including Korea, resulting in economic losses (Gomila et al., 2017; Lee et al., 2015; Young, 2010). P. syringae complex infects woody tissue, exhibits the symptoms of cankers, and eventually spreads to the entire wood and kills branches (Gomila et al., 2017; Perminow et al., 2018). However, previous reports have displayed that the two different pathovars of the same species, such as Pss (Lee et al., 2015) and P. syringae pv. papulans (Kerkoud et al., 2002), exhibit distinct disease symptoms in different tissues on the same apple tree.
In addition, adaptation mechanisms to its plant hosts and microbial evolution have been displayed by several research groups (Xin et al., 2018). In particular, the virulent strains of Pss produce large quantities of toxins, including syringopeptin and syringomycin, and are localized to plant tissues infected with Pss pathovars (Fogliano et al., 1999; Scholz-Schroeder et al., 2003; Wang et al., 2006a). Furthermore, the best bacterial ice nucleators are P. syringae, which enable ice nucleation at a lower temperature (−2°C). The ability of bacteria to facilitate ice formation is attributed to specialized ice-nucleating proteins anchored to the outer bacterial cell membrane (Lukas et al., 2022). Therefore, as a plant pathogen, P. syringae plays an important role in frost damage to plant tissue by increasing the nucleation temperature of water (Warren and Wolber, 1991). For example, freezing temperature causes an increase in the severity of Pss infection on the leaves of cherry (Süle and Seemüller, 1987) and bacterial apical necrosis in mango (Cazorla et al., 1998).
Freeze injury in most horticultural crops, including apple is a serious problem during winter, leading to a significant yield loss mainly due to ice nucleation by the presence of ice nucleation-active (INA) bacteria (Lindow, 1983; Rodrigo, 2000). The life cycle of Pss on an apple is explained in Fig. 1. During the year 2016, many apple growers in Korea reported cases of apple canker, where the infected trees showed the yellowing of leaves and stunted growth, followed by a lot of cracking on the stems and gummosis, eventually leading to the death of the apple trees. Recently, similar observations were reported in Science magazine (Stokstad, 2019). The decline seems to be more common in trees with a popular rootstock called M9, which goes into dormancy in the fall slower than the others (Foster et al., 2017).
Various conventional agrochemicals are being used in Korea for the control of apple diseases. Previously, a survey identified 28 apple diseases, including fungal and bacterial diseases in Korea (Lee et al., 2006), but apple canker disease caused by Pss was not yet identified. The cause of apple canker in Korea during winter has not been revealed in any reports to date in Korea. Therefore, the objective of this study was to identify and characterize the bacterial pathogen responsible for causing apple canker by the development of severe symptoms on apple trees in Korea.
In the early spring season of the years 2013 and 2014, a survey has been conducted to investigate the symptoms of apple canker. These are similar to the symptoms of bacterial blight, in the apple orchards of various places (Yeongju, Yeongyang, Cheongsong, Mungyeong, Andong, and Yecheon) in the northern Gyeongbuk Province, Korea (Fig. 2A). For the disease survey analysis, observations have been made randomly by selecting 20 trees from each orchard. Disease symptoms have been observed during the early stage and late winter season.
Samples were collected from leaves and stems of 5-year-old disease-infected apple trees from various regions (Supplementary Table 1). The symptomatic leaf tissues were cut into squares using a scalpel and the surface disinfected by immersing them in 70% ethyl alcohol for 30 s, followed by treatment with 2% sodium hypochlorite (NaOCl) solution for 1 min and rinsing in sterile distilled water (SDW) two or three times. Thus, the surface-sterilized tissues were placed onto the King’s B agar (KB; 20 g protease peptone, 1.6 g K2HPO4, 1.5 g MgSO4·7H2O, 15 g agar, 15 ml glycerol, and water to make 1,000 ml volume) plates and incubated at 28°C for 2-3 days. To isolate pathogens from infected apple stems, the cross-sectioned stem at infected region and graft region were placed onto the KB agar plates for a few seconds (Supplementary Fig. 1). Then, the cultured plates were observed under UV light 48 h after incubating at 28°C to differentiate fluorescence colonies from non-fluorescence colonies. Thus the fluorescent colonies on the KB medium were selected and stored in glycerol (20%) at −50°C for long-term storage.
The representative bacterial isolate WSPS007 and reference strains (P. syringae pv syringae, P. syringae pv. morsprunorum, and P. syringae pv. persicae) were tested for LOPAT (levan production, oxidase reaction, pectolytic activity, arginine hydrolysis activity, and tobacco hypersensitivity), following the method described by Lelliott and Stead (1987). LOPAT tests are used to discriminate P. syringae from other species of fluorescent pseudomonads. To determine the levan production from sucrose, the bacterial colonies were inoculated onto Petri dishes containing nutrient agar (NA) supplemented with 5% sucrose and observed for levan production after incubating the Petri dishes at 28°C for 24-48 h. For the presence of oxidase (O), bacteria were cultured onto the KB medium for 48 h at 25°C. The colonies were picked up with a sterile loop and rubbed on a filter paper impregnated with 1% (w/v) solution of N,N,N′,N′-tetramethyl-p-phenylenediamine dihydrochloride. The appearance of a violet color within 10 s indicates a positive test whereas the absence of color or delayed color development indicates a negative test. Pectinase activity was determined on the crystal violet pectate medium. When the bacterial colonies show a jelly-like substance 7 days after incubation at 25°C, it indicates a positive test. The presence of arginine dihydrolase was detected by introducing a 48 h-old grown bacterial culture into two test tubes containing a semi-liquid medium. Once inoculated, one tube is overlaid with sterile mineral oil. Color variations, from orange-red to magenta-amarant after 2-5 days in anaerobic conditions, indicate a positive reaction. Tobacco hypersensitivity (HR) reaction was detected in tobacco leaves. Bacterial cell suspensions (108 cfu/ml) were infiltrated into the mesophyll of a fully-grown tobacco leaf. Rapid glassy to white necrosis at the infiltrated area 24 h after inoculation indicates the ability to induce an HR.
For distinguishing pathovars within P. syringae, GATTa (gelatin and aesculine hydrolysis, tyrosinase activity, and tartrate acid metabolism) tests are commonly used (Lelliott and Stead, 1987). For gelatin hydrolysis, a 24 h-old bacterial culture was introduced into a tube containing a solidified medium consisting of 0.3% yeast extract, 0.5% peptone, and 12% gelatin. The characteristic liquefaction of gelatin after 7-14 days of incubation at 18°C is an indication of a positive result. For aesculin hydrolysis, bacteria grown for 24-48 h on NA were inoculated onto a semi-solid medium containing 1% peptone, 0.1% aesculine, 0.05% ferric citrate, and 2% agar. The brown color of the medium after 24-48 h incubation at 26-28°C indicates the presence of the aesculinase (Sneath, 1956). For tyrosinase activity, a 24-48-hour-old bacterial culture was inoculated onto a semi-solid medium containing 0.5% sucrose, 1% casamino acid, 0.1% L-tyrosine, 0.05% potassium phosphate, and 0.0125% magnesium sulfate. The pH of the medium was adjusted to 7.2 before adding 2% agar. Change in the color of the medium to red after 7-10 days of incubation at 26-28°C shows the presence of tyrosinase (Lelliott et al., 1966). For tartrate utilization, 24-48-hour-old-grown bacterial culture was inoculated in the broth (pH 7.0) containing 0.1% ammonium dihydrogen phosphate, 0.02% potassium chloride, 0.02% magnesium sulfate, 0.2% (w/v) sodium tartrate, and 1 ml of 4% alcohol bromothymol blue solution. After 21 days of incubation at 25°C, a color change in the medium from green to blue indicates a positive test result (Lelliott and Stead, 1987).
To determine the INA, bacterial colonies grown on NA plates for 48 h at 28°C were selected randomly. Escherichia coli TOP10 grown on the NA medium was used to measure INA, whereas SDW, deionized water (DI; ultra-pure grade water), and apple leaf juice were used as controls. Bacterial suspensions (106 cfu/ml) were prepared with SDW, or DI, or a mixture of SDW + apple leaf juice in glass test tubes (15 ml). All test tubes were placed in a styrofoam box (30 × 20 × 15 cm), and the box was placed into a freezer at −70°C (Kim and Kim, 1997). The freezing point was measured using a Pt100 thermocouple (Tanaka Metal Jewelry). Strains were scored positive for INA if immediate ice formation occurred in test tubes. The freezing point in a test tube was represented as a peak on the graph. The freezing point for Pss was compared with controls, such as a tube with distilled water and a tube with Escherichia coli suspensions. The experiment was performed two times with six replicates per treatment.
For microscopic observation, the bacterial colony was picked up with a sterile loop, placed in SDW on a Formvar-coated copper grid, and stained with 2% uranyl acetate for negative staining (Kim et al., 2016). The preparation was examined under a HITACHI transmission electron microscope (TEM; JEOL, Tokyo, Japan). Morphological and biochemical properties were identified, evaluated, and compared as described in Bergey’s Manual of Systematic Bacteriology (Bergey et al., 1939) and the laboratory guide for the identification of plant pathogenic bacteria (Schaad et al., 2001).
The putative Pseudomonas isolate was tested for the utilization of 95 carbon sources using the BIOLOG program (Jeon et al., 2003). Briefly, the bacterial cells cultured on bacterial universal growth agar supplemented with 0.25% maltose and 0.9% thioglycolate at 28°C for 48 h were suspended in an inoculating fluid (0.4% NaCl, 0.03% Pluronic F-68, and 0.01% gellan gum), inoculated onto microplates (Biolog GP MicroPlate), and incubated at 28°C. After 24-48 h of incubation, the plates were read using a MicroLog 3-Automated MicroStation system (BIOLOG, Hayward, CA, USA). The bacterium was identified based on the MicroLog Gram-negative database (version 4.0, BiOLOG). Gas chromatography of fatty acid methyl esters (GC-FAME) was conducted to confirm bacterial identification. Bacteria were cultured on tryptic soy agar plates at 28°C for 48 h. The colonies were harvested and placed in screw-cap culture tubes, and 1 ml of saponification reagent (NaOH aqueous methanol) was added. A methylation reagent (hydrochloric acid in aqueous methanol) was added after heat treatment and fatty acids were extracted with extraction solvent (hexane/MTBE), mild base (10.8 g NaOH in 900 ml), and a saturated NaOH solution. The Sherlock system analyzed the fatty acid composition, followed by the generation of a similarity index for the isolates that corresponded to a microorganism in the database Sherlock version 3.1 (New York, DE, USA) (Kim et al., 2016).
The selected WSPS007 isolate was subjected to molecular identification based on the sequence homology of its 16S rRNA gene (Weisburg et al., 1991). The genomic DNA of WSPS007 isolate was isolated using a kit (Genomic DNA Extraction Kit for bacteria, iNtRON Biotechnology, Seongnam, Korea) as per the manufacturer’s instructions. The 16S rRNA gene was amplified using a polymerase chain reaction (PCR) with Taq DNA polymerase and with primer set 27F (5′-AGA GTT TGA TCM TGG CTC AG-3′) and 1492R (5′-GGY TAC CTT GTT ACG ACT T-3′) (Weisburg et al., 1991). The conditions for thermal cycling were as follows: denaturation at 94°C for 5 min followed by 30 cycles at 94°C for 1 min, annealing at 56°C for 1 min, and extension at 72°C for 1 min. At the end of the cycle, the reaction mixture was held at 72°C for 5 min and then cooled to 4°C. The PCR product obtained was sequenced by an automated sequencer (Genetic Analyzer 3130, Applied Biosystems, Foster City, CA, USA). The above primers were used for this purpose. The sequence was compared with the reference species of bacteria contained in a genomic database using an NCBI-BLAST tool. The re-isolated pathogens, such as Re_1 WSPS007 and Re_2 WSPS007 from the pathogenicity test were also subjected to molecular identification using the same method as above. Sequence alignment and phylogenetic tree construction were performed by maximum parsimony using a MEGA 4.0 program (Biodesign Institute, Tempe, AZ, USA).
To assess the pathogenicity of Pss on the apple plant, disease-free apple twigs (cv. Fuji) from 2-year-old apple trees were artificially inoculated with bacterial suspensions (106 cfu/ml) by foliar spray and soil drench methods. For artificial inoculation, the pathogenic bacterial cell suspensions were prepared by spreading the suspensions on KB agar plates, incubating them for 48 h at 28°C, and adjusting to a final concentration (106 cfu/ml) in SDW. For the foliar spray method, disease-free apple twigs with leaves were washed with water and dried. All the leaves on a twig were wounded with a sterile needle and inoculated with bacterial suspensions (106 cfu/ml) of 50 ml by the foliar spray method, dried for some time, and incubated for 2 weeks at 28°C. Apple twigs sprayed with SDW served as a control. Disease symptoms were assessed 2 weeks after inoculation. For the soil drench method, a pathogenicity test was conducted on 2-year-old healthy apple plants grown in pots under greenhouse conditions at 24 ± 2°C. Apple plants were drenched with 500 ml bacterial suspensions (106 cfu/ml) near the root region in pots. Pots drenched with SDW served as a non-treated control. Disease severity (%) was recorded 1 week after incubating the plants under humid conditions (70-80%) in the greenhouse. Pathogenicity was evaluated based on a disease rating index scale from 0 to 4 (where 0, no lesions; 1, leaves showing yellowing symptoms appear from 0 to 25%; 2, leaves showing yellowing from 26 to 50%; 3, leaves show yellow and then become brown from 51 to 75%; and 4, leaves showing blight and branch becomes wilt from 76 to 100%). Each experiment was performed at least two times with six replicates per treatment. To fulfill Koch’s postulates, the pathogen Pss was re-isolated from all symptomatic tissues to determine whether they are morphologically identical to the original isolates.
The Pss strains were screened for their ability to produce syringomycin and syringopeptin. To test syringomycin production, a procedure described by Toben et al. (1989) was followed. Pss strains were grown overnight in 5 ml of nutrient broth yeast extract. Bacterial cells were harvested by centrifugation and washed once with SDW. The bacterial suspensions were adjusted to a final concentration of 1 × 106 cfu/ml and 10 μl aliquots of bacterial suspensions were inoculated onto syringomycin minimal agar plates and allowed to dry for 5 min. The plates were incubated for 5 days at 28°C and then lightly over-sprayed with the suspension of the arthrospores of the fungus Geotrichum candidum (KCTC6195), which is sensitive to syringomycin. As a result of syringomycin production, the fungal growth inhibition zone was measured 5 days after incubation at 28°C. The Pss strains were evaluated for syringopeptin production as described by Grgurina et al. (1996), but were modified to use peptone-glucose-NaCl agar (PGNA) instead of potato dextrose agar. Pss strains were cultured as described above, except the bacterial suspensions were transferred to PGNA plates and incubated for 4 days at 25°C. The PGNA plates were then over-sprayed with the cell suspensions of Bacillus megaterium Km (KACC13749), which is highly sensitive to syringopeptin. To avoid confusion in distinguishing between the syringomycin and syringopeptin inhibition zones, syringopeptin mutations were introduced into the strain BR334 (Zhang et al., 1995). The strain BR344 is a syrC mutant of B301D−R, which provides a clear bioassay for syringopeptin but does not produce syringomycin (Grgurina et al., 1996).
The toxin-producing genes, such as syrB1, recA, hrpZ, and sypB, were detected by PCR amplification method using a BIONEER AccuPower PCR premix kit (Bioneer Co., Daejeon, Korea). Primers used for PCR amplification were designed using the Laser Gene Expert Sequence Analysis Package (DNASTAR, Madison, WI, USA) and are listed in Table 1. PCR was completed within 35 cycles with annealing temperatures of 55°C, 52°C, 58°C, and 63°C for syrB1, sypB, recA, and hrpZ, respectively, with an initial denaturation step at 94°C for 5 min, extension at 94°C and 72°C for 30 and 1 min, respectively, and the final extension step at 72°C for 10 min. The amplified DNA samples were loaded on 1% agarose gel and electrophoresed using loading STAR for confirmation. The 100 bp DNA ladder (Dyne Bio, Seongnam, Korea) was used as a marker.
All the isolated strains were subjected to ERIC-PCR (enterobacterial repetitive intergenic consensus PCR) and REP-PCR (repetitive element sequence-based PCR) genomic fingerprinting using primer sets corresponding to REP (REP1R-I: 5′-IIIICGICGICATCIGGC-3′ and REP2-I: 5′-ICGICTTA TCIGGCCTAC-3′) and ERIC (ERIC1R: 5′-ATG TAA GCT CCT GGG GAT TCA C-3′ and ERIC2: 5′-AAG TAA GTG ACT GGG GTG AGC G3′) elements (Schaad et al., 2001). The reaction mixture (25 μl) consisted of a Taq reaction buffer containing 1.0 μl of template DNA, 100 pmol of each primer, 200 μM of each deoxynucleotide triphosphate, 2.0 mM MgCl2, and 1.0 U of Taq DNA polymerase (Bioneer Co.). The PCR amplifications were performed in a MyGenie96/384 Thermal Block (Bioneer Co.) with the following parameters: initial denaturation at 95°C for 7 min, template denaturation at 94°C for 1 min, primer annealing at 52°C (ERIC) or 41°C (REP) for 1 min, and DNA extension for 8 min at 65°C. The PCR was repeated for 30 cycles, with a final extension step of 15 min at 65°C. Amplicons were detected by electrophoresis on a 1.5% agarose gel with 0.5× TAE buffer and visualized by a UV transilluminator after staining with ethidium bromide (1 μg/ml) solution.
To assess the antibiotic sensitivity, a total of nine antibiotics (oxolinic acid [WDG], validamycin-A [WSP], dithianon [WSP], validamycin-A [WDG], copper hydroxide [WP], streptomycin [WP], iminoctadine triacetate [SC], oxytetracycline [WSP], and kasugamycin [WDG]) were used against Pss. Approximately 100 μl bacterial suspension (107 cfu/ml) cultured for 48 h was spread onto the freshly prepared NA medium. The sterile paper disks (6 mm in diameter) were impregnated with antibiotics in different ratios of concentrations (1:500, 1:1,000, and 1:2,000) and placed on agar plates. The discs loaded with SDW served as a negative control. A clear zone of inhibition was observed 48 h after incubating at 28°C and sensitivity to antibiotics was recorded.
In Korea, from 2013 onwards, apple trees at various locations showed disease symptoms causing apple cankers. The cause of this was identified and confirmed in this study. The symptoms that appeared were identical to the symptoms of bacterial infections, such as apple cankers caused by Pss (Araujo et al., 2020), which mainly affect leaves and stems. The percentage (%) of disease incidence of apple canker (from a total of 20 apple trees) varied from place to place in the Gyeongbuk Province, Korea (Fig. 2B). Disease incidence (%) in Cheongsong was recorded at a greater level than in other locations. The symptoms of apple canker at the initial stage appeared as yellowing of the leaves on branches of the plant (Supplementary Figs. 1B and 2A), especially during fruit formation (Supplementary Figs. 1D and 2C). Under severe conditions, the leaves completely wither and shed from the tree and start showing cracking symptoms (Supplementary Fig. 2E). The symptoms of apple canker were associated with gummosis spreading on the stem (Fig. 3A) which induced the formation of cracks on the bark of the stem (Fig. 3B) from which the sap oozes out (Fig. 3C), resulting in the spread of the bacteria in the tissue causing its browning (Fig. 3D). Under severe conditions, sap oozes out from the trunk of the tree with symptoms (Fig. 3E). Furthermore, apple canker symptoms extended to the infected roots, graft region, and stem, which could be visually observed in their cross-sections (Fig. 3F-H), followed by the collapse of the entire tree.
A total of 82 isolates were collected from different locations in the northern Gyeongbuk Province, Korea (Supplementary Table 1). Of these, only 50 isolates that exhibit fluorescence under UV light were considered pseudomonads (Supplementary Table 2); while the isolates which do not exhibit fluorescence are considered non-pseudomonads. Based on pathogenicity levels of various isolates from the orchards of Andong region of the Gyeongbuk Province, Korea (Supplementary Fig. 3), only one representative isolate (WSPS007) Pss was used for further studies. The isolate WSPS007 has been found to exhibit higher virulence causing in terms of disease severity (%) on detached apple leaves in comparison with other Pss isolates (Supplementary Table 3).
All the studied Pseudomonas syringae pathovars were gram-negative and the results of LOPAT and GATTa tests were reported in Table 2. In LOPAT tests, it was noticed that all the isolates were positive for levan production; the colonies were large white dome-shaped on the NSA medium. They showed negative results in oxidase, pectolytic capability, and arginine dihydrolase test. In the tobacco HR test, the tobacco leaves became hypersensitive 24 h after the introduction of the pathogen into the leaves (Table 2, Supplementary Fig. 4A). In GATTa tests, the gelatin showed liquefaction, and aesculin was hydrolyzed, which indicates that the reaction was positive, but no tyrosinase activity was observed (Table 2, Supplementary Fig. 4B). The strains were confirmed to be Pss.
We studied the impact of INA by adding Pss cell suspensions to SDW or deionized water, and a mixture of SDW and apple leaf juice. The result of the latter was compared with two controls. For all three samples, INA and the freezing point were found to be higher than that of E. coli and the control. When Pss cell suspensions were added to SDW, INA was found at −0.7°C; while the INA for E. coli and SDW was found at −9.6°C and −4.7°C, respectively (Supplementary Fig. 5A). When Pss cell suspensions were added to deionized water, INA was found at −2.8°C; while the INA for E. coli and sterilized deionized water was found at −5.9°C and −6.3°C, respectively (Supplementary Fig. 5B); and finally when Pss cell suspensions were added to a mixture of SDW and apple leaf juice, the ice nucleation temperature was found to be −0.3°C; while INA for E. coli and SDW + apple leaf juice was found at −2.3°C and −2.2°C, respectively (Supplementary Fig. 5C). The ice crystallization event is well known to be an uncertain phenomenon when carried out without nucleating agents. When added to aqueous solutions, the cells of Pss can exert accurate control of nucleation, which is a key step of the crystallization phenomenon. The freezing point of the three samples was 0°C, but the supercooling break was different for treated and control samples. This means that the degree of supercooling was reduced by 2.5°C upon the addition of Pss cell suspensions.
Bacterial colonies cultured on the KB solid medium appeared as translucent, closely white, circular, convex shape, and fluorescent under UV light. When viewed using a TEM, the bacterial population was found to be rod-shaped (Fig. 4A), approximately 2.0 μm (width) × 3.0 μm (length). An individual bacterium was observed with lophotrichous flagella at one end (Fig. 4B and C); whereas the dividing bacterium was observed with flagella (Fig. 4D). The BIOLOG system determined the classification of the strain as Pseudomonas syringae, because it closest matched from the reference results based on the utilization of various carbohydrates examined (Table 3). In particular, carbon sources, such as sorbitol, L-lactate, and erythritol, resulted in clear classification. When bacterial strains were subjected to FAMEs analysis, the strains were found to be close to P. syringae, and further, they showed a distinct difference from Erwinia sp. (Table 4). Comparing these traits to those mentioned in the BIOLOG database revealed that this strain had a match probability of 100% to P. syringae.
The WSPS007 isolate was further characterized by 16S rRNA gene sequencing. The 16S rRNA gene sequences suggested that the isolate WSPS007 (accession no. KF500097.1) belonged to the Pseudomonas genus and had the highest homology to Pss (99%). Thus, both morphological and molecular characteristics confirmed this species as Pss. In the phylogenetic tree, the isolate was clustered with other Pseudomonas sp., which are closely related to P. syringae (Fig. 5). Almost all isolates were identified as Pss with 99% homology (Supplementary Table 2).
Pathogenicity of the isolated pathovar Pss (WSPS007) was tested on detached stems of 2-year-old apple plants containing leaves (Fig. 6). When the leaves on the branch were inoculated with 50 ml of bacterial suspensions (106 cfu/ml) by foliar spray, Pss started to induce apple canker symptoms on leaves of the branch, appearing as brown to black spots, which later turned to yellow and fell off 2 weeks after inoculation (Fig. 6A). The infected leaves showed the symptoms of diseased spots with yellow halos around them, whereas no symptoms were developed in non-inoculated (control) leaves. In the case of the inoculation by the soil drench method, the leaves at the top appeared healthy, whereas the bottom leaves showed disease symptoms (Fig. 6B). Initially, the leaves close to the soil became yellow, and the symptoms progressed from the bottom to the top. The yellowing symptoms gradually turned to blight symptoms. The symptoms appeared slowly in the upper part of the plant. The bark of the stem showed canker and longitudinal symptoms after two months (Fig. 6C). These symptoms mainly appeared near the grafting region. Then, the xylem of the bark canker showed browning symptoms. No disease symptoms were observed in the non-treated control. One week after inoculation, the pathogen was re-isolated from the symptomatic tissues and cultured onto the KB medium at 28°C for 3 days to fulfill Koch's postulates.
Pathogenic Pss strains were screened for the secretion of toxic substances, such as syringomycin and syringopeptin against the growth of G. candidum and B. megaterium, respectively, as indicator microorganisms (Supplementary Fig. 6). Out of 50 Pss strains screened for syringomycin production under in vitro conditions, 47 strains exhibited the formation of an inhibition zone against G. candidum, indicating that they secrete syringomycin, which might be due to the presence of syrP, whereas three strains (SHPS021, SHPS025, and SHPS057) did not show any inhibition zone (Supplementary Fig. 6A). Among all the Pss strains, the strain WSPS007 exhibited a greater inhibition zone (6.6 mm) than other strains. In the case of syringopeptin production, four strains (SHPS021, SHPS025, SHPS042, and SHPS057) did not show any inhibition zone, while other strains exhibited inhibition zones against the bacterium B. megaterium, 48 h after incubation on PGN agar plates, and 12 strains showed a minor level of inhibition zone (0.6-1.0 mm) (Supplementary Fig. 6B). On the other hand, the strain WSPS007 has been found to produce both syringopeptin and syringomycin at a greater level than other strains.
Expressions of syringomycin synthetase (syrB1) and syringopeptin synthetase (sypB) genes were observed in Pss (Fig. 7A). Similarly, the harpin-encoding hrpZ, which secretes “harpin” protein, enables Pss to elicit the HR in tobacco leaves. The principal sigma factor and housekeeping gene, recA, used for the determination of the ability of Pss to induce pathogenicity, was also expressed at an equal level to that of syrB1. The gene recA is required for DNA repair and homologous recombination.
Rep-PCR using REP and ERIC primers revealed differences in the genetic profiles of the tested Pseudomonas syringae pathovars (Fig. 7B). ERIC- and REP-PCR showed significant diversity in the banding patterns of all Pseudomonas strains from different infected apple orchards in the Gyeongbuk Province, Korea. The corresponding PCR products ranged approximately from 100 to 2,000 bp in size, and the banding patterns differed from strain to strain in both ERIC- and REP-PCR. Based on the secretion of syringopeptin, inhibition zones were formed, and the strains were grouped as follows: T1, the group that did not produce syringopeptin; T2, the group producing about 1-4 mm inhibition zone; and T3, the group producing >4 mm inhibition zone. The cumulative dendrogram of REP-PCR was constructed based on the REP and ERIC of P. syringae from different apple orchards.
In the antibiotic sensitivity test, the pathogenic Pss strain used in our study showed the highest sensitivity to oxytetracycline with an inhibition zone of 22.6 mm, the next highest sensitivity was to streptomycin with an inhibition zone of 15.8 at 1:2,000 dilutions, and the inhibition zone was 12.4 mm against oxolinic acid (Table 5). The pathogen Pss was strongly resistant to the remaining six antibiotics, for which no inhibition zone was observed.
In this study, we describe apple canker caused by members of the Pss on apple orchards is responsible for relevant yield losses in Korea. The symptoms appeared as yellowing of leaves, leaf blight, blister bark, and longitudinal cracks on stems of infected apple trees. We isolated and characterized the pathogen Pss from the leaves and stems of apple trees that apple canker, resulting in severe damage to the apple orchard, leading to a reduction in product quality. Under severe conditions, apple trees get collapsed in the entire orchard (Singh et al., 2019). In recent years, P. syringae has been found to infect apple orchards in various countries causing various diseases, such as shoot blight in Korea (Lee et al., 2015), blister bark in Southern Norway (Perminow et al., 2018), and blossom blast on apple buds in United States (Gašić et al., 2018). On the other hand, previous reports have displayed that the symptoms caused by P. syringae can occur due to extended periods of cold or wet weather conditions (Perminow et al., 2018; Rommel et al., 2010). Sometimes, P. syringae pathovars have been found to survive in plant tissue without showing any symptoms (Gallelli et al., 2011).
Generally, the appearance of necrotic lesions and wood decay was found to show signs of the declining number of apple trees in the orchard, which follows a specific dispersion pattern from bark or vascular cambium towards the heartwood (Singh et al., 2019). Recent reports have displayed that the incompatibility of graft union between rootstock and scion led to canker on apple trees (Araujo et al., 2020), which might be due to poor vascular connections and phloem degeneration (Dolgun et al., 2009). However, the exact cause of necrotic lesions causing apple canker in apple orchards is not yet known. The probe for apple canker has been detected and confirmed in our study by the presence of bacterial pathogen Pss based on various parameters.
Various methods have been used to characterize Pss isolates from leaves and stems of apple trees. Biochemical and physiological characteristics of Pss have been studied by culturing and observing colonies on the KB medium. The molecular analysis was effective in the identification of Pss isolates, which is supported by previous studies (Gormez et al., 2013; Ivanović et al., 2017). Pss is one of the most dangerous disease-causing pathovars within the P. syringae complex that causes severe economic damage to various stone fruits around the world, including apples (Kennelly et al., 2007). In a previous study, Gašić et al. (2012) investigated the differentiation of Pseudomonas species and compared the assessment of the difference in virulence towards lemon fruits and nectarine fruits to show a clear intra-pathovar variation.
Gram’s reaction, fluorescence on the KB medium, LOPAT, and GATTa tests were performed to differentiate Pss pathovars from other pseudomonads. In a previous study by Gašić et al. (2012), it has been stated that the pseudomonads, such as P. syringae pv. persicae, grew slower on the KB medium than the other two pathovars, such as Pss and P. syringae pv. morsprunorum, without producing green fluorescent pigment. On the other hand, another study by Bultreys et al. (2001) reported that a highly virulent non-fluorescent P. syringae pv. aptata strain was isolated and grown on a GASN medium, while Pss also produced “levan type” colonies on the NSA medium, which is indicative of the strain.
The GATTa tests have been performed to clearly distinguish between P. syringae pv. morsprunorum and Pss. The strain WSPS007 used in this study, which hydrolyzed gelatin and aesculin, produced tyrosinase, and did not utilize tartaric acid belongs to Pss (Table 2). However, these results are contradictory to a previous report by Bultreys and Kaluzna (2010), where GATTa tests for P. syringae pv. morsprunorum strains are homogeneous as they do not hydrolyze gelatin and aesculin, but produce tyrosinase and utilize tartaric acid. Therefore, only gelatin hydrolysis can be used for the differentiation of P. syringae pv. morsprunorum from other strains. In addition to GATTa test, the INA test, which is rapid, reliable, inexpensive, and simple to perform, can be utilized for assaying differentiation. The relationship between frost damage and diseases caused by Pss in woody plants has been demonstrated previously in the bacterial canker of peach and prune (Cao et al., 1999). The INA and freezing point were found to be higher in Pss cell suspensions than E. coli cell suspensions and SDW. This showed that the pathovar morsprunorum can be differentiated from the other two pathovars by its inability to initiate ice nucleation. On the other hand, not all strains within these species are ice nucleation active (Hirano and Upper, 2000), but ice nucleation activity has been used as one of the traits to distinguish strains among the P. syringae pathovars (Jones et al., 1983). For example, strains within pv. syringae frequently exhibit the ice phenotype, while some other strains within pv. tomato or morsprunorum, do not exhibit ice nucleation activity (Hirano and Upper, 2000). Yellow and white growth of strains in the SNB medium can be used for pathovars syringae and morsprunorum (Bultreys and Kaluzna, 2010). It has been suggested that winter damage is a contributing factor to outbreaks in many host plants (Cambours et al., 2005; Kennelly et al., 2007), where frost damage incites a greater degree of the blast.
Previously, Jones et al. (1993) tested many strains belonging to various genera, including Pseudomonas using a BIOLOG system; for example, Pss (Gormez et al., 2013), P. syringae pv. tomato strains (Gormez et al., 2013), and P. syringae pv. actinidae (Flores et al., 2018) were identified by BIOLOG program. Pss, isolated from infected apple leaves, was confirmed based on its morphology and the results obtained from the BIOLOG program and GC-FAME. The BIOLOG database system identified accurately all the Pss strains to species level, hence, this system is thought to be more suitable for characterization studies. Previously, Slabbinck et al. (2009) identified species in three genera, including Pseudomonas, using a GC-FAME analysis, which was superior to the commercially available FAME analysis system. Furthermore, Pss is a known bacterium that produces different phytotoxins depending on the type of host. For example, isolates from stone fruits and peaches produce syringomycin (Popović et al., 2021), isolates from citrus trees produce syringotoxin (Gonzalez et al., 1981), and isolates from lilac blight produce syringostatin (Sorensen et al., 1998). In this study, Pss recovered from infected apple trees was investigated for their genetic diversity using REP-PCR. The virulent strains of Pss produce large quantities of syringopeptin and syringomycin in vitro, and the toxins are localized to plant tissues infected with Pss or related pathovars (Fogliano et al., 1999; Scholz-Schroeder et al., 2001). This study is supportive of our study, where the toxins, such as syringopeptin and syringomycin from Pss are involved in plant-pathogen interaction to contribute to the virulence of Pss strain WSPS007 causing disease symptoms in apple trees.
Based on the ERIC- and REP-PCR amplifications, strains belonging to Pss appeared as the most heterogeneous to the host plant and their origin (Kaluzna et al., 2010; Scortichini et al., 2003). The discriminatory potential of REP-PCR for different P. syringae pathovars have been shown in previous reports (Kaluzna et al., 2010; Vicente and Roberts, 2007), and another study by Çepni and Gürel (2012) examined a limited number of strains that use modified REP-PCR primers to detect intra-pathovar diversity. Tobes and Pareja (2006) reported that the P. syringae genome contains high copy numbers of REP elements that are related to insertion sequence elements. This feature of REP elements may explain the abundance and diversity of fingerprints observed upon amplification of the intervening regions.
Our pathogenicity test results showed that the detached leaves and stems from an apple tree are susceptible to Pss. Hence, the pathogen is considered highly virulent for an apple tree, and we successfully reproduced the symptoms from inoculated host plant tissues. To the best of our knowledge, this is the first report on the identification and characterization of the Pss strain that causes apple canker in many orchards in the northern Gyeongbuk Province of Korea, which encompasses representative strains of all pathovars classified in the species. Furthermore, an antibiotic sensitivity test revealed that Pss was sensitive to a few antibiotics, such as oxolinic acid, streptomycin, and oxytetracycline, while resistant to other antibiotics, such as validamycin-A, dithianon, copper hydroxide, iminoctadine triacetate, and kasugamycin (Table 5). Copper resistance is a widespread phenomenon in P. syringae pathogens of many hosts in which copper is utilized in reducing bacterial populations (Kennelly et al., 2007; Scheck and Pscheidt, 1998). Similarly, a previous report displayed to show P. syringae strains resistance to antimicrobial compounds (Neu, 1992). This finding could provide a basis for further research and understanding of the variation among Pss populations and their epidemiology and a deeper insight into the occurrence of apple canker infections in Korea.
The results of this study determined the widespread Pss in apple orchards at various locations in Korea, causing apple cankers. A wide host range and favorable weather conditions contributed to the survival and spread of this Pss. The strain Pss was considered the best virulence-contributing strain causing necrotic symptoms in plant tissues. During winter, the pathogen survives in the plant in a dormant stage without exhibiting any necrotic symptoms. LOPAT and GATTa tests indicated that the bacterial isolates were Pss, while the BIOLOG system determined the classification of the strain. The increased temperature led to better growth of Pss in the apple tree, and it caused frost injury in severe conditions during winter. The frost injury appeared due to the low temperatures of the bark. The presence of Pss caused cracks in the bark of the apple tree. Pss is highly sensitive to oxytetracycline, an antibiotic that can reduce bacterial population growth. The characterization of the isolates indicates that most of the isolates analyzed belong to pv. syringae. In a pathogenicity test, the disease symptoms were developed from the inoculated stems and leaves, and the same pathogen was re-isolated to fulfill Koch’s postulates. The production of syringopeptin and syringomycin is considered a major virulence determinant of Pss, causing plant cell death.
Acknowledgments
This work was supported by a grant from research fund of Andong National University.
Electronic Supplementary Material
Supplementary materials are available at The Plant Pathology Journal website (http://www.ppjonline.org/).
Fig. 1
The life cycle of Pseudomonas syringae pv. syringae (Pss) on the apple tree. Bacteria grow on the surface of stems and leaves (epiphytically) during the early spring season. The primary inoculum originates from bacteria located in dormant buds. The Pss population is mainly present on the buds and leaf surfaces. Canker formation occurs on the stem following pathogen colonization. In summer, Pss causes lesions on leaves leading to yellowing and dryness symptoms, and sap oozes out from the stem canker. When the temperature falls in autumn, the bark bursts or gets peeled off as the water level of the apple tree decreases, and bacteria with ice nucleation activity, which is believed to cause winter damage, survive in a dormant stage in the apple tree.

Fig. 2
Disease survey of Pseudomonas syringae pv. syringae (Pss) from various orchards in Korea. (A) The disease survey has been conducted from various places (Mungyeong, Yecheon, Yeongju, Bonghwa, Andong, and Yeongyang) in the northern Gyeongbuk Province, Korea. Infected samples were collected from the above places (marked in red color) to isolate the pathogens. (B) Percentage (%) of disease incidence of apple canker caused by Pss at various apple orchards in Gyeongbuk Province, Korea. Observations were made from 20 apple trees. Bars with the same letters indicate no statistical difference between the two orchards, according to the least significant difference test (P < 0.05).
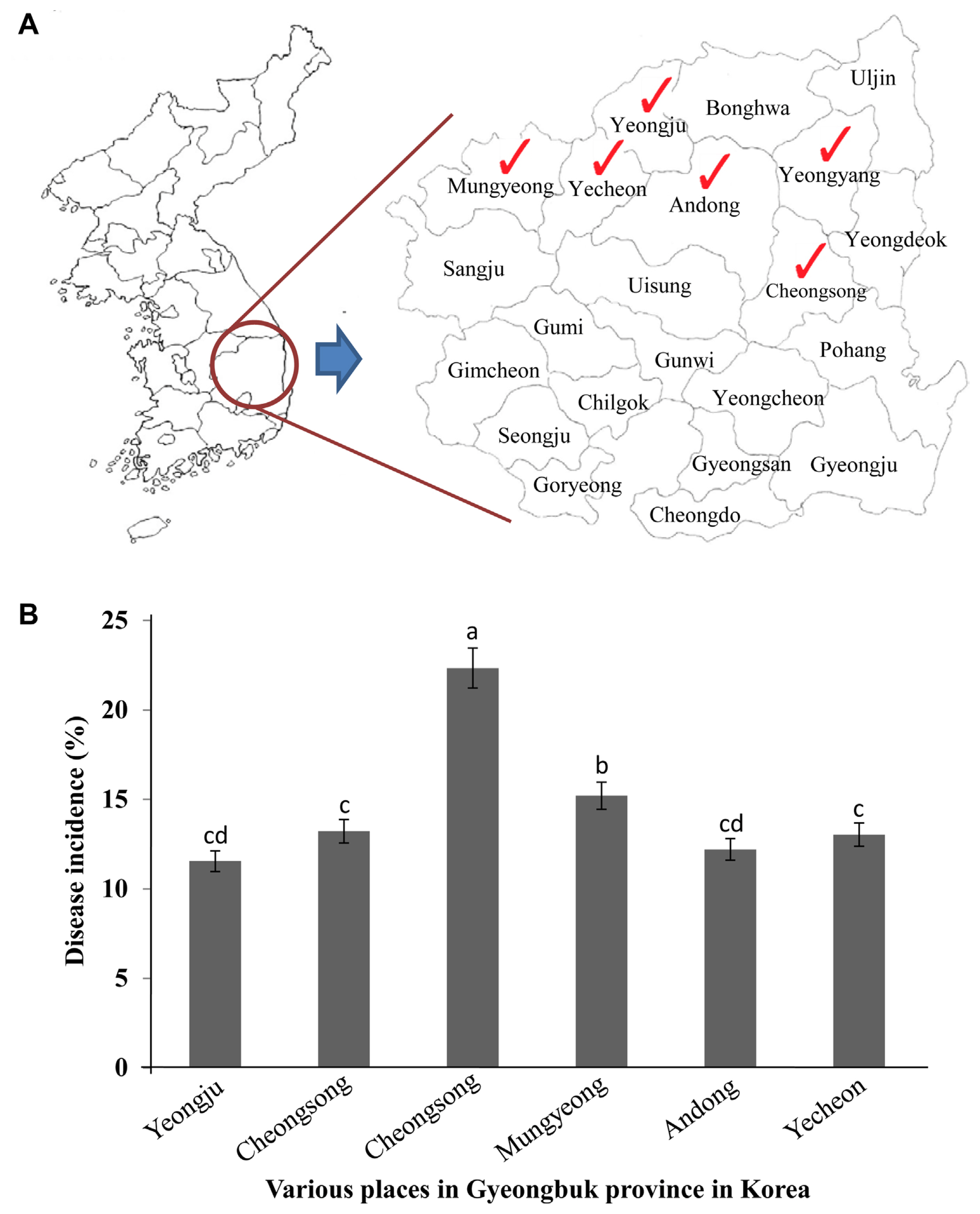
Fig. 3
Typical symptoms of apple canker disease on apple trees at different stages. (A) Bacterial canker symptoms on the stem are associated with gummosis spreading. (B) Formation of cracks on the bark of the stem where the infection moves from the epidermis and begins to induce a canker on the stem. (C) Oozing out the sap from the cracked portion on the stem. (D) Bacteria have moved internally into the wood tissues, causing browning of the tissues after the bark has peeled off. (E) Oozing out the sap from the trunk of the tree in severe cases (indicated by arrows). Cross-section of the infected root (F), graft region (G), and stem (H) of applewood showing visual symptoms (browning) of apple canker.

Fig. 4
Morphological characteristics of Pseudomonas syringae pv. syringae isolates observed using transmission electron microscopy. (A) Rod-shaped bacterial cells. (B) A single bacterium with two flagella. (C) An enlarged image of the bacterium with two flagella. (D) Dividing bacterium. The experiment was carried out at least twice, with three replicates per treatment. Scale bars = 2.0 μm (A), 0.5 μm (B, D), 0.2 μm (C).
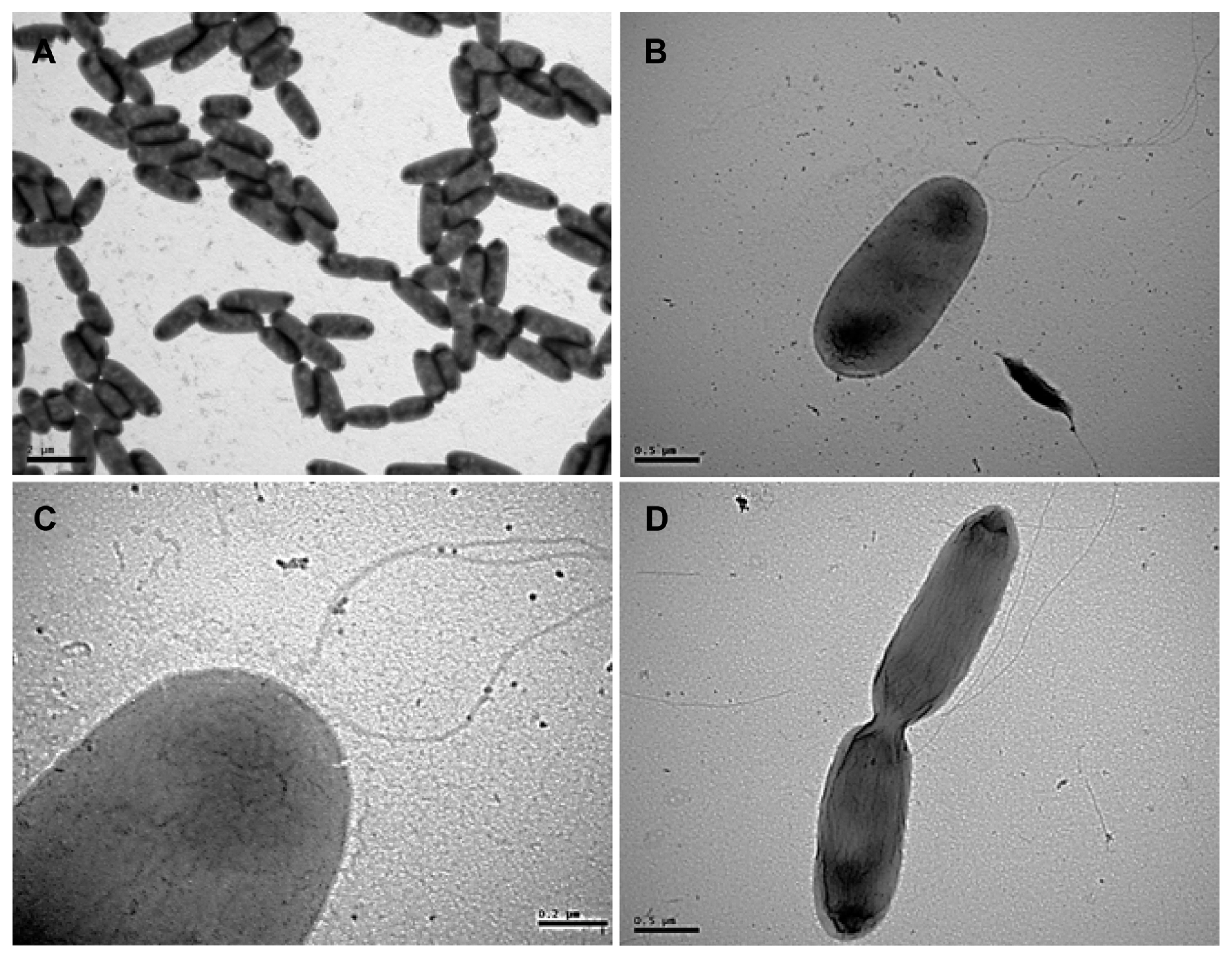
Fig. 5
Phylogenetic analysis of Pseudomonas syringae pv. syringae (Pss) based on 16S rRNA sequences. Phylogenetic relationships of Pss were determined based on similarity using the MEGA-X program. WSPS007 is the observed isolate; Re_1 and Re_2 WSPS007 were re-isolated from the pathogenicity test.

Fig. 6
Pathogenicity test of Pseudomonas syringae pv. syringae (Pss) WSPS007 cell suspensions by foliar spray and soil drench methods on detached leafy branches of an apple tree. (A) Development of disease symptoms on leaves of detached leaves 7 days after inoculation (DAI) by foliar spray method. Inoculated leaves showed yellow halos around the black spots, whereas no symptoms appeared on non-inoculated (control) leaves and stems. (B) Symptoms on bottom leaves (shown in red square) appear as yellowing, 14 days after inoculation by using the soil drench method. In enlarged images, the bottom (indicated by an arrow) and upper leaves of the infected plant appeared healthy. No symptoms were observed in non-treated plants (control). (C) This disease symptoms (indicated by arrows) appeared on the stem two months after inoculation. The experiment was performed two times with six replicates per treatment.
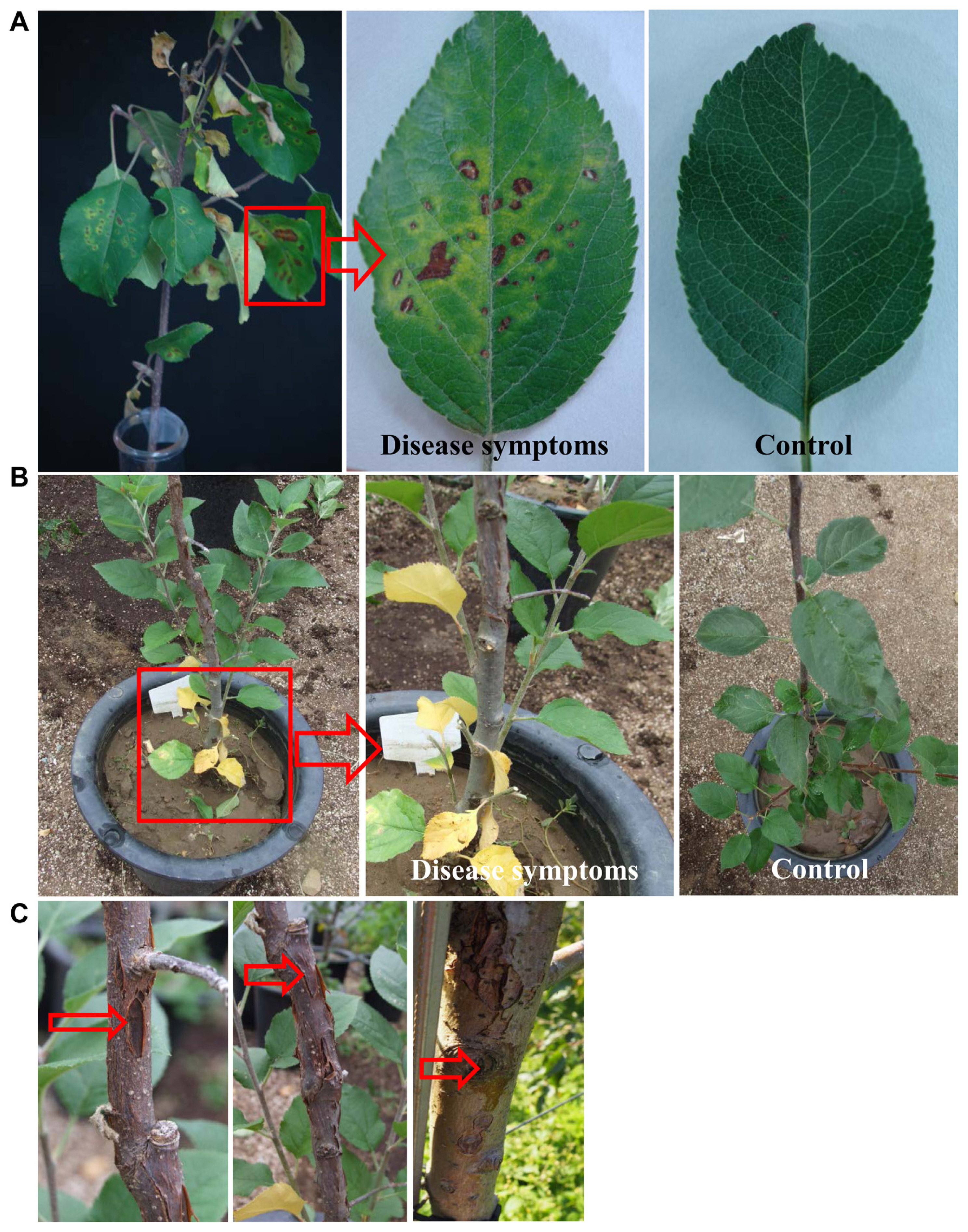
Fig. 7
Expression of toxin-producing genes and repetitive element sequence-based polymerase chain reaction (PCR) and enterobacterial repetitive intergenic consensus PCR in Pseudomonas syringae pv. syringae (Pss) isolates from apple canker infected tissues from various apple orchards in the Gyeongbuk Province, Korea. (A) Expression of toxin-producing genes of Pss induced by plant signal molecules, syrB1 (syringomycin biosynthesis enzyme), recA (principal sigma factor), hrpZ (harpin), and sypB (syringopeptin synthesis). (B) Three types of banding patterns were formed as a result of the random PCR, and these patterns were grouped using the NTSYS program. The index of the pattern was determined according to the presence or absence of a band, and the pattern was grouped using unweighted pair group method with arithmetic mean. ERIC, enterobacterial repetitive intergenic consensus; REP, repetitive element sequence.
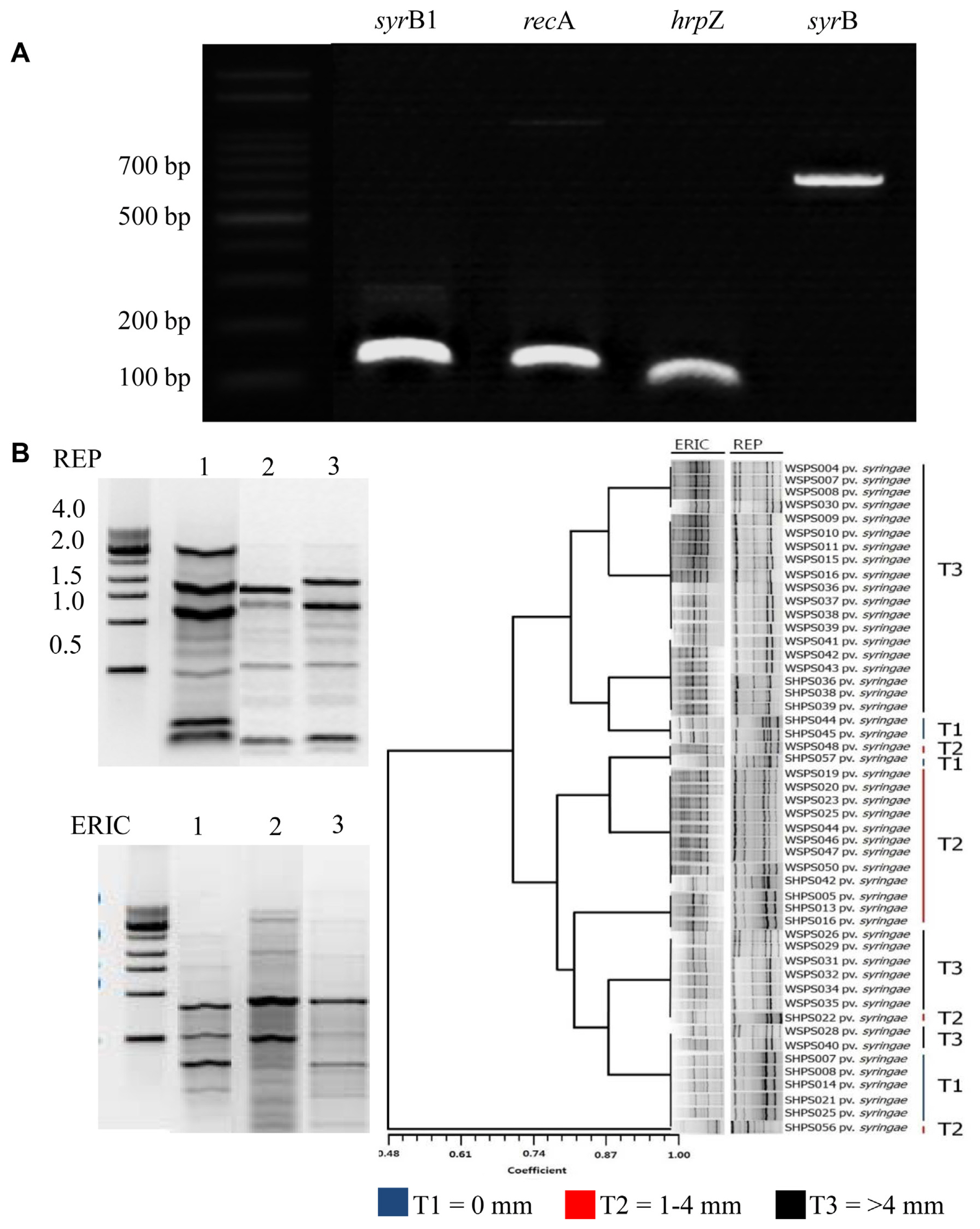
Table 1
Primer sequences used to detect toxin-producing genes from Pss
| Gene | Primera | Sequence (5' to 3') | Reference |
|---|---|---|---|
| syrB1 | syrB1F/syrB1R |
TTAGCGCCGCGTCAGCCCCTCTCAAG GCTCAACGTCCGGGCTGCATCGCTCAC |
Kang and Gross (2005) |
| recA | recAF/recAR |
CTTCGGTACGCCTGGACA AACTCGGCCTGACGGAAC |
Lu et al. (2005) |
| hrpZ | hrpZF/hrpZR |
TCCTGAAACCGAGACGACTGG GACCGTTGCGCATCAGTTCCTC |
Wang et al. (2006b) |
| sypB | syrBF/syrBR |
CTTTCCGTGGTCTTGATGAGG TCGATTTTGCCGTGATGAGTC |
Schaad et al. (2001) |
Table 2
Identification of bacterial isolates at pathovar level using LOPAT and GATTa tests
Table 3
BIOLOG test analysis
Table 4
Hydroxyl fatty acid (FAMEs) composition in bacteria of different genera
| FAMEsa | Responsesb | ||
|---|---|---|---|
|
|
|||
| Isolate | Pseudomonasc | Erwiniac | |
| 8:0 3OH | − | − | − |
| 10:0 3OH | + | + | − |
| 12:0 2OH | + | + | − |
| 12:0 3OH | + | + | − |
| 14:0 2OH | − | − | (+) |
| 14:0 3OH | − | (+) | + |
| 16:0 2OH | − | − | − |
| 16:0 3OH | − | (+) | − |
Table 5
Antibiotic sensitivity test of Pss
References
Araujo, L., Cardoza, Y. F., Duarte, V. and de Moraes, M. G. 2020.
Pseudomonas syringae causing bacterial canker on apple trees in Brazil. Bragantia 79:592-598.

Bergey, D. H., Breed, R. S., Murray, E. G. D. and Hitchens, A. P. 1939.
Bacterium methylicum (Loew) Migula. (Bacillus methylicus Loew). In: Bergey’s manual of determinative bacteriology: a key for the identification of organisms of the class Schizomycetes, eds. by D. H. Bergey, R. S. Breed, E. G. D. Murray and A. P. Hitchens, 5th ed. pp. 597. Baillière, Tindall & Cox, London, UK.
Bultreys, A., Gheysen, I., Maraite, H. and de Hoffmann, E. 2001. Characterization of fluorescent and nonfluorescent peptide siderophores produced by Pseudomonas syringae strains and their potential use in strain identification. Appl. Environ. Microbiol 67:1718-1727.




Bultreys, A. and Kaluzna, M. 2010. Bacterial cankers caused by Pseudomonas syringae on stone fruit species with special emphasis on the pathovars syringae and morsprunorum race 1 and race 2. J. Plant Pathol 92:S21-S33.
Cambours, M. A., Nejad, P., Granhall, U. and Ramstedt, M. 2005. Frost-related dieback of willows. Comparison of epiphytically and endophytically isolated bacteria from different Salix clones, with emphasis on ice nucleation activity, pathogenic properties and seasonal variation. Biomass Bioenerg 28:15-27.

Cao, T., Sayler, R. J., Dejong, T. M., Kirkpatrick, B. C., Bostock, R. M. and Shackel, K. A. 1999. Influence of stem diameter, water content, and freezing-thawing on bacterial canker development in excised stems of dormant stone fruit. Phytopathology 89:962-966.


Cazorla, F. M., Torés, J. A., Olalla, L., Pérez-García, A., Farré, J. M. and de Vicente, A. 1998. Bacterial apical necrosis of mango in Southern Spain: a disease caused by Pseudomonas syringae pv. syringae
. Phytopathology 88:614-620.


Cepni, E. and Gürel, F. 2012. Variation in extragenic repetitive DNA sequences in Pseudomonas syringae and potential use of modified REP primers in the identification of closely related isolates. Genet. Mol. Biol 35:650-656.



Choi, S. and Hinkle, A. F. 2017 South Korea: 2017 apple report-revised. Gain report No. KS1733 URL https://www.fas.usda.gov/data/south-korea-2017-apple-report-revised
. 23 November 2018.
Dolgun, O., Yildirim, A., Polat, M., Yildirim, F. and Aşkın, A. 2009. Apple graft formation in relation to growth rate features of rootstocks. Afr. J. Agric. Res 4:530-534.
Flores, O., Prince, C., Nuñez, M., Vallejos, A., Mardones, C., Yañez, C., Besoain, X. and Bastías, R. 2018. Genetic and phenotypic characterization of indole-producing isolates of Pseudomonas syringae pv. actinidiae obtained from chilean kiwifruit orchards. Front. Microbiol 9:1907.



Fogliano, V., Gallo, M., Vinale, F., Ritieni, A., Randazzo, G., Greco, M., Lops, R. and Graniti, A. 1999. Immunological detection of syringopeptins produced by Pseudomonas syringae pv. lachrymans
. Physiol. Mol. Plant Pathol 55:255-261.

Foster, T. M., McAtee, P. A., Waite, C. N., Boldingh, H. L. and McGhie, T. K. 2017. Apple dwarfing rootstocks exhibit an imbalance in carbohydrate allocation and reduced cell growth and metabolism. Hortic. Res 4:17009.




Gallelli, A., Talocci, S., L’Aurora, A. and Loreti, S. 2011. Detection of Pseudomonas syringae pv. actinidiae, causal agent of bacterial canker of kiwifruit, from symptomless fruits and twigs, and from pollen. Phytopathol. Mediterr 50:462-472.
Gašić, K., Pavlović, Ž., Santander, R. D., Meredith, C. and Aćimović, S. G. 2018. First report of Pseudomonas syringae pv. syringae associated with bacterial blossom blast on apple (Malus pumila) in the United States. Plant Dis 102:1848.

Gašić, K., Prokić, A., Ivanović, M., Kuzmanović, N. and Obradović, A. 2012. Differentiation of Pseudomonas syringae pathovars originating from stone fruits. Pestic Phytomed. (Belgrade) 27:219-229.

Gomila, M., Busquets, A., Mulet, M., García-Valdés, E. and Lalucat, J. 2017. Clarification of taxonomic status within the Pseudomonas syringae species group based on a phylogenomic analysis. Front. Microbiol 8:2422.



Gonzalez, C. F., DeVay, J. E. and Wakeman, R. J. 1981. Syringotoxin: a phytotoxin unique to citrus isolates of Pseudomonas syringae
. Physiol. Plant Pathol 18:41-50.

Gormez, A., Sahin, F., Gulluce, M. and Aslan, I. 2013. Identification and characterization of Pseudomonas syringae isolated from apricot trees in the Erzurum province of Turkey and evaluation of cultivar reaction. J. Plant Pathol 95:525-532.
Grgurina, I., Gross, D. C., Iacobellis, N. S., Lavermicocca, P., Takemoto, J. Y. and Benincasa, M. 1996. Phytotoxin production by Pseudomonas syringae pv. syringae: syringopeptin production by syr mutants defective in biosynthesis or secretion of syringomycin. FEMS Microbiol. Lett 138:35-39.

Hirano, S. S. and Upper, C. D. 2000. Bacteria in the leaf ecosystem with emphasis on Pseudomonas syringae: a pathogen, ice nucleus, and epiphyte. Microbiol. Mol. Biol. Rev 64:624-653.




Ivanović, Ž., Perović, T., Popović, T., Blagojević, J., Trkulja, N. and Hrnčić, S. 2017. Characterization of Pseudomonas syringae pv. syringae, causal agent of citrus blast of mandarin in Montenegro. Plant Pathol. J 33:21-33.



Jeon, Y. H., Chang, S. P., Hwang, I. and Kim, Y. H. 2003. Involvement of growth-promoting rhizobacterium Paenibacillus polymyxa in root rot of stored Korean ginseng. J. Microbiol. Biotechnol 13:881-891.
Jones, J. B., Chase, A. R. and Harris, G. K. 1993. Evaluation of the Biolog GN microplate system for identification of some plant-pathogenic bacteria. Plant Dis 77:553-558.

Jones, J. B., Gitaitis, R. D. and McCarter, S. M. 1983. Evaluation of indirect immunofluorescence and ice nucleation activity as rapid tests for identifying foliar diseases of tomato transplants incited by fluorescent pseudomonads. Plant Dis 67:684-687.

Kaluzna, M., Ferrante, P., Sobiczewski, P. and Scortichini, M. 2010. Characterization and genetic diversity of Pseudomonas syringae from stone fruits and hazelnut using repetitive-PCR and MLST. J. Plant Pathol 92:781-787.
Kang, H. and Gross, D. C. 2005. Characterization of a resistance-nodulation-cell division transporter system associated with the syr-syp genomic island of Pseudomonas syringae pv. syringae
. Appl Environ. Microbiol 71:5056-5065.




Kennelly, M. M., Cazorla, F. M., de Vicente, A., Ramos, C. and Sundin, G. W. 2007.
Pseudomonas syringae diseases of fruit trees: progress toward understanding and control. Plant Dis 91:4-17.


Kerkoud, M., Manceau, C. and Paulin, J. P. 2002. Rapid diagnosis of Pseudomonas syringae pv. papulans, the causal agent of blister spot of apple, by polymerase chain reaction using specifically designed hrpL gene primers. Phytopathology 92:1077-1083.


Kim, Y. S., Kotnala, B., Kim, Y. H. and Jeon, Y. 2016. Biological characteristics of Paenibacillus polymyxa GBR-1 involved in root rot of stored Korean ginseng. J. Ginseng Res 40:453-461.

Kim, Y. and Kim, N. 1997. Cold hardiness in Spodoptera exigua (Lepidoptera: Noctuidae). Environ. Entomol 26:1117-1123.

Lee, D. H., Lee, S. W., Choi, K. H., Kim, D. A. and Uhm, J. Y. 2006. Survey on the occurrence of apple diseases in Korea from 1992 to 2000. Plant Pathol. J 22:375-380.

Lee, S., Cheon, W. and Jeon, Y. 2015. First report of bacterial shoot blight caused by Pseudomonas syringae pv. syringae of apple (Malus pumila) in Korea. Plant Dis 99:1641.

Lelliott, R. A. and Stead, D. E. 1987. Methods for the diagnosis of bacterial diseases of plants. Blackwell Scientific Publications, Oxford, UK. pp. 216.
Lelliott, R. A., Billing, E. and Hayward, A. C. 1966. A determinative scheme for the fluorescent plant pathogenic pseudomonads. J. Appl. Bacteriol 29:470-489.


Lindow, S. E. 1983. The role of bacterial ice nucleation in frost injury to plants. Ann. Rev. Phytopathol 21:363-384.

Liu, M., Zhang, W., Manawasinghe, I. S., Zhou, Y., Xing, Q. K., Li, X .H., Yan, J. Y. and Wang, S. 2018. First report of Nothophoma quercina causing trunk canker on crabapple (Malus micromalus) in China. Plant Dis 102:1462.

Lu, S.-E., Wang, N., Wang, J., Chen, Z. J. and Gross, D. C. 2005. Oligonucleotide microarray analysis of the salA regulon controlling phytotoxin production by Pseudomonas syringae pv. syringae
. Mol. Plant-Microbe Interact 18:324-333.


Lukas, M., Schwidetzky, R., Eufemio, R. J., Bonn, M. and Meister, K. 2022. Toward understanding bacterial ice nucleation. J. Phys. Chem. B 126:1861-1867.




Perminow, J. I. S., Børve, J., Brurberg, M. B. and Stensvand, A. 2018. First report of Pseudomonas syringae pv. syringae causing bacterial blister bark on apple in Norway. Plant Dis 102:1653.

Popović, T., Menković, J., Prokić, A., Zlatković, N. and Obradović, A. 2021. Isolation and characterization of Pseudomonas syringae isolates affecting stone fruits and almond in Montenegro. J. Plant Dis. Prot 128:391-405.


Ritchie, H. and Roser, M. 2020 Agriculture production Published online at OurWorldindata.org URL https://ourworldindata.org/agricultural-production
. 28 December 2022.
Rodrigo, J. 2000. Spring frosts in deciduous fruit trees - morphological damage and flower hardiness. Sci. Hortic 85:155-173.

Rommel, C. C., Valdebenito-Sanhueza, R. M. and Duarte, V. 2010. Detection of bacteria associated with European pear buds in Rio Grande do Sul State, Brazil. Trop. Plant Pathol 35:360-367 (in Portuguese).

Ruinelli, M., Blom, J., Smits, T. H. M. and Pothier, J. F. 2019. Comparative genomics and pathogenicity potential of members of the Pseudomonas syringae species complex on Prunus spp. BMC Genomics 20:172.




Schaad, N. W., Jones, J. B. and Chun, W. 2001. Laboratory guide for identification of plant pathogenic bacteria. 3rd ed. APS Press, St. Paul, MN, USA. pp. 373.
Scheck, H. J. and Pscheidt, J. W. 1998. Effect of copper bactericides on copper-resistant and -sensitive strains of Pseudomonas syringae pv. syringae
. Plant Dis 82:397-406.


Scholz-Schroeder, B. K., Hutchison, M. L., Grgurina, I. and Gross, D. C. 2001. The contribution of syringopeptin and syringomycin to virulence of Pseudomonas syringae pv. syringae strain B301D on the basis of sypA and syrB1 biosynthesis mutant analysis. Mol. Plant-Microbe Interact 14:336-348.


Scholz-Schroeder, B. K., Soule, J. D. and Gross, D. C. 2003. The sypA, sypS, and sypC synthetase genes encode twenty-two modules involved in the nonribosomal peptide synthesis of syringopeptin by Pseudomonas syringae pv. syringae B301D. Mol. Plant-Microbe Interact 16:271-280.

Scortichini, M., Marchesi, U., Dettori, M. T. and Rossi, M. P. 2003. Genetic diversity, presence of the syrB gene, host preference and virulence of Pseudomonas syringae pv. syringae strains from woody and herbaceous host plants. Plant Pathol 52:277-286.

Singh, J., Silva, K. J. P., Fuchs, M. and Khan, A. 2019. Potential role of weather, soil and plant microbial communities in rapid decline of apple trees. PLoS ONE 14:e0213293.



Slabbinck, B., De Baets, B., Dawyndt, P. and De Vos, P. 2009. Towards large-scale FAME-based bacterial species identification using machine learning techniques. Syst. Appl. Microbiol 32:163-176.


Sneath, PHA. 1956. Cultural and biochemical characteristics of the genus Chromobacterium. J. Gen. Microbiol 15:70-98.


Sorensen, K. N., Kim, K.-H. and Takemoto, J. Y. 1998. PCR detection of cyclic lipodepsinonapeptide-producing Pseudomonas syringae pv. syringae and similarity of strains. Appl. Environ. Microbiol 64:226-230.




Süle, S. and Seemüller, E. 1987. The role of ice formation in the infection of sour cherry leaves by Pseudomonas syringae pv. syringa
. Phytopathology 77:173-177.

Toben, H., Mavridis, A. and Rudolph, K. W. E. 1989. Basal glume rot (Pseudomonas syringae pv. atrofaciens) on wheat and barley in FRG and resistance screening of wheat. Bull. OEPP/EPPO Bull 19:119-125.

Tobes, R. and Pareja, E. 2006. Bacterial repetitive extragenic palindromic sequences are DNA targets for Insertion Sequence elements. BMC Genomics 7:62.




Vicente, J. and Roberts, S. J. 2007. Discrimination of Pseudomonas syringae isolates from sweet and wild cherry using rep-PCR. Eur. J. Plant Pathol 117:383-392.


Wang, N., Lu, S.-E., Records, A. R. and Gross, D. C. 2006a. Characterization of the transcriptional activators SalA and SyrF, which are required for syringomycin and syringopeptin production by Pseudomonas syringae pv. syringae
. J. Bacteriol 188:3290-3298.



Wang, N., Lu, S.-E., Wang, J., Chen, Z. J. and Gross, D. C. 2006b. The expression of genes encoding lipodepsipeptide phytotoxins by Pseudomonas syringae pv. syringae is coordinated in response to plant signal molecules. Mol. Plant-Microbe Interact 19:257-269.


Warren, G. and Wolber, P. 1991. Molecular aspects of microbial ice nucleation. Mol. Microbiol 5:239-243.


Weisburg, W. G., Barns, S. M., Pelletier, D. A. and Lane, D. J. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol 173:697-703.




Xin, X .F., Kvitko, B. and He, S. Y. 2018.
Pseudomonas syringae: what it takes to be a pathogen. Nat. Rev. Microbiol 16:316-328.




Young, J. M. 2010. Taxonomy of Pseudomonas syringae
. J. Plant Pathol 92:S5-S14.



 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Supplement1
Supplement1 Print
Print




