Anti-Oomycete Activity and Pepper Root Colonization of Pseudomonas plecoglossicida YJR13 and Pseudomonas putida YJR92 against Phytophthora capsici
Article information
Abstract
Previously, Pseudomonas plecoglossicida YJR13 and Pseudomonas putida YJR92 from a sequential screening procedure were proven to effectively control Phytophthora blight caused by Phytophthora capsici. In this study, we further investigated the anti-oomycete activities of these strains against mycelial growth, zoospore germination, and germ tube elongation of P. capsici. We also investigated root colonization ability of the bacterial strains in square dishes, including cell motility (swimming and swarming motilities) and biofilm formation. Both strains significantly inhibited mycelial growth in liquid and solid V8 juice media and M9 minimal media, zoospore germination, and germ tube elongation compared with Bacillus vallismortis EXTN-1 (positive biocontrol strain), Sphingomonas aquatilis KU408 (negative biocontrol strain), and MgSO4 solution (untreated control). In diluted (nutrient-deficient) V8 juice broth, the tested strain populations were maintained at >108 cells/ml, simultaneously providing mycelial inhibitory activity. Additionally, these strains colonized pepper roots at a 106 cells/ml concentration for 7 days. The root colonization of the strains was supported by strong swimming and swarming activities, biofilm formation, and chemotactic activity towards exudate components (amino acids, organic acids, and sugars) of pepper roots. Collectively, these results suggest that strains YJR13 and YJR92 can effectively suppress Phytophthora blight of pepper through direct anti-oomycete activities against mycelial growth, zoospore germination and germ tube elongation. Bacterial colonization of pepper roots may be mediated by cell motility and biofilm formation together with chemotaxis to root exudates.
Pepper (Capsicum annuum L.) is an important cash crop in Korea, in which the yields are significantly reduced annually due to several soilborne and airborne plant diseases (Lim and Kim, 2010). Among these diseases, Phytophthora blight, caused by an oomycete soilborne Phytophthora capsici, is one of the most serious diseases affecting pepper production (Hausbeck and Lamour, 2004). Soilborne P. capsici can infect pepper plants at all developmental stages and invade plants via diverse infection courts (roots, stems, and leaves), making its management more complicated. Since soil is the most common inoculum source of soilborne pathogens, disease control measures usually rely on soil application of fungicides (Hausbeck and Lamour, 2004). However, fungicide usage has proven problematic due to the vast range of adverse side effects. These negative fungicidal effects may include threats to non-target organisms, soil fertility alteration, plant toxicity, the occurrence of resistant strains of the pathogen, and environmental pollution (Barnhoorn and van Dyk, 2020; Parra and Ristaino, 2001; Singh et al., 2003; Thind and Hollomon, 2018; Vogel et al., 2021). Thus, biological control has been recommended as an alternative control measure for the destructive soilborne disease of pepper.
In general, biological control implies the application of antagonistic microbes against plant pathogens, without impact on non-target organisms and the environment (Barratt et al., 2018; Sang and Kim, 2014; Sang et al., 2013; Volynchikova and Kim, 2022). Biocontrol by microbes comprises several mechanisms, including antibiosis, competition, and hyperparasitism (Chemeltorit et al., 2017; Köhl et al., 2019). Diverse types of bacterial secondary metabolites, including antibiotics, cell wall-degrading enzymes, and lipopeptides, can inhibit the development of plant pathogens, including P. capsici (Arora et al., 2007; Li et al., 2020). Because dispersal and infection of P. capsici in the field are usually initiated through zoospores (Hausbeck and Lamour, 2004), secondary metabolites of antagonistic microbes that inhibit its zoospore germination and subsequent mycelial growth might be of special interest.
Despite positive performance of biocontrol agents in laboratory conditions, several prominent agents fail to maintain the same performance under in vivo conditions (Alaux et al., 2018). One possible reason might be the inability of these organisms to establish sufficient cell densities in target plant rhizosphere or phylloplane (Guyer et al., 2015; Hunziker et al., 2015; Li et al., 2013). In the case of soilborne disease biocontrol, microbial colonization in rhizospheres predominantly depends on root exudates, which comprise carbohydrates, amino acids, lipids, and organic acids (Dietz et al., 2020). Some components of root exudates act as attractants for biocontrol agents, stimulate microbial root-colonizing activity, and promote biocontrol performance (Ma et al., 2018). Upon successful bacterial colonization, they can form biofilms on the root surfaces, creating an established micro-niche for their long-term survival and physically protecting roots from plant pathogen penetration (Li et al., 2013; Sang and Kim, 2014). Moreover, exudate consumption by beneficial rhizobacteria results in nutrient limitation and starvation of the target pathogen, making colonization ability of biocontrol agents highly beneficial. In this regard, the efficient performance of biocontrol Pseudomonas and Bacillus against plant pathogens was reported in pepper, tomato, and other solanaceous crops (Ngo et al., 2020; Oliver et al., 2019; Sheoran et al., 2015). Pseudomonas spp. can actively colonize the surfaces and interiors of roots of economically important plants including pepper, tomato, and ginger (Sheoran et al., 2015; Sun et al., 2017). For example, tomato bacterial wilt can be controlled by Pseudomonas putida A1, providing strong chemotaxis to plant extracts and subsequent successful colonization (Sun et al., 2017). Furthermore, P. putida can produce volatile compounds that inhibit the growth of pathogenic fungi, oomycetes (for example, P. capsici and Pythium myriotylum), and nematodes (Sheoran et al., 2015; Zhai et al., 2018). Thus, these traits could allow Pseudomonas species to be promising candidates as biocontrol agents for plant diseases.
In our previous study, bacterial strains YJR13 and YJR92 (identified as P. putida), isolated from the root surfaces of pepper plants, exhibited biocontrol activity against Phytophthora blight of pepper (Sang et al., 2013). In our 3 year-field tests, strain YJR92 was proven to have significant biocontrol efficacy against Phytophthora blight of pepper, with no impact on indigenous microbial communities (Sang et al., 2013); however, the biocontrol traits of this strain have not yet been studied. Therefore, the objectives of this study were (1) to evaluate the anti-oomycete activity of the selected strains YJR13 and YJR92 against mycelial growth of P. capsici and its zoospore germination and germ tube elongation, (2) to determine bacterial colonization of pepper roots in a square dish system, and (3) to examine bacterial cell motility (swimming and swarming activities) and biofilm formation with regard to the enhancement of bacterial colonization ability on plant roots.
Materials and Methods
Bacterial strains and pathogen inoculum
Bacterial strains YJR13 and YJR92 antagonistic to P. capsici on pepper were isolated from the root surfaces of pepper plants in Youngjongdo, Korea in 2004 (Sang et al., 2013). Strain YJR92 was previously identified as P. putida (Sang et al., 2013), whereas strain YJR13 was identified in this study as Pseudomonas plecoglossicida, based on 16S rRNA gene analysis using universal primers 27f (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1492r (5′-GGTTACCTTGTTACGACTTC-3′) (Supplementary Fig. 1). The obtained sequence (1,421 nucleotides) of strain YJR13 was analyzed as described by Jeong et al. (2016) and deposited in NCBI GenBank (accession no. OM443073).
Bacillus vallismortis EXTN-1 (EXTN, Dongbu HiTech Co., Seoul, Korea), a commercial biocontrol agent, was used as a positive bacterial control; Sphingomonas aquatilis KU408 isolated from rice grains (Mannaa et al., 2017) was used as a negative bacterial control; and a 10-mM MgSO4 solution was utilized as an untreated control in all experiments. Bacterial suspensions used in this study were prepared according to the procedures described by Kim et al. (2008). Briefly, single colonies of bacterial strains on nutrient agar (NA) grown for 48 h at 28°C were inoculated into nutrient broth (NB) and incubated in a shaking incubator (160 rpm) for 24 h. Pre-cultured bacterial strains were transferred to NB and incubated for 48 h under the same conditions. Bacterial cultures were centrifuged (5,000 ×g) for 15 min at 4°C, followed by washing twice with 10-mM MgSO4. The bacterial suspensions were then adjusted to 108 cells/ml (OD600 = 0.5) using a spectrophotometer (Optizen 2120UV, Mecasys Co., Daejeon, Korea).
The P. capsici S197 inoculum was prepared following the procedures described by Kim et al. (2008). Briefly, P. capsici S197 cultured on oatmeal agar at 28°C for 7 days was flooded with 20 ml of sterile distilled water (SDW) and further incubated for 7 days at 25°C under continuous fluorescent light to induce zoosporangium formation. To induce zoospore release, the plates were treated with 5 ml chilled SDW and placed at 4°C for 30 min, followed by incubation at 25°C for an additional 30 min. Mycelial debris was removed by filtration through two layers of cheesecloth. To determine the zoospore concentration, the filtered zoospore suspension (1 ml) was vortexed for 30 s to allow zoospores to encyst, and zoospore numbers were then determined with a hemocytometer under a microscope.
Biocontrol activity of bacterial strains against P. capsici on pepper plants
To evaluate biocontrol activity of the tested bacterial strains, plant tests were conducted in a growth room (25°C and 16-h photoperiod fluorescent light). Briefly, germinated pepper seeds (cv. Nockwang) were sown in 128-cell trays containing potting mix (Baroker, Seoul Bio, Seoul, Korea). Three-week-old seedlings grown in the room were transplanted into 10-cm-diameter pots containing 50 g of the mix. A week prior to inoculation, the plants were treated with 25 ml bacterial suspension (108 cells/g dry soil wt.) or 10-mM MgSO4 solution (untreated control). Five-week-old plants were inoculated with the prepared zoospore suspension (25 zoospores/g dry soil wt.) of P. capsici. Disease severity was assessed on a scale of 0 (symptomless) to 5 (plant dead), as described by Kim et al. (1989) for 14 days after inoculation (DAI).
Anti-oomycete activity of bacterial strains on solid media and in liquid media
To determine in vitro inhibition of mycelial growth by diffusible metabolites produced by the tested bacterial strains, a dual-culture assay was conducted. A colony of each bacterial strain or SDW (untreated control) was streaked in the center of a Petri dish containing V8 juice agar or M9 minimal medium (Marley et al., 2001). At 48 h after bacterial streaking on the media, two mycelial plugs (5 mm in diameter) from the margin of the 5-day-old culture of P. capsici S197 were inoculated on opposite sides of the media. The Petri dishes were then incubated in the dark at 28°C until mycelia of untreated controls nearly reached the center of the dishes. On the other hand, to determine minimal inhibitory bacterial cell density, bacterial suspensions were prepared as described previously and serial dilutions were prepared to obtain appropriate cell densities (100–10 cells/ml) of the strains. Five microliters of each diluted cell suspension were applied to sterile filter paper disks (8 mm in diameter) placed on V8 juice agar. A mycelial plug of the 5-day-old culture of P. capsici was placed at the center of the plates. Mycelial growth was photographed when the mycelia reached 100 cell-treated disks (MgSO4-treated control).
Similarly, the antagonistic activity of the tested strains against P. capsici was conducted in V8 juice broth. Bacterial suspensions (200 μl of 108 cells/ml) were inoculated into Erlenmeyer flasks containing V8 juice broth (19.8 ml). A mycelial plug (5 mm in diameter) from the 5-day-old culture of P. capsici was inoculated in each flask. The flasks were incubated in a shaker (75 rpm) in the dark at 28°C for 5 days. Mycelia from flasks were harvested after removing agar plugs and mycelial weights were determined 3 days after drying at 50°C.
Diluted media on mycelial growth of P. capsici and bacterial populations
To evaluate inhibitory activity of the tested bacterial strains under nutrient-deficient conditions, they were co-cultivated in variously diluted V8 juice broth with P. capsici. Two hundred microliters of bacterial suspension (108 cells/ml) or 10-mM MgSO4 solution were inoculated in diluted (0 [undiluted], 10, 50, 100, and 200 times) V8 juice broth, which contained a plug (5 mm in diameter) of the 5-day-old culture of P. capsici. The flasks were then incubated under stationary conditions in the dark at 28°C for 5 days. Mycelia were harvested without agar plugs and mycelial weights were determined as described previously. Simultaneously, V8 juice broth was serially diluted and smeared on NA. The number of colony-forming units (CFU) of bacterial populations was determined 2 days after incubation at 28°C.
Bacterial cell suspensions and cell-free culture filtrates against zoospore development of P. capsici
To evaluate the effect of bacterial strains or cell-free culture filtrates on zoospore germination and germ tube elongation of P. capsici, bacterial suspensions (108 cells/ml) and zoospores of P. capsici were prepared as described previously. Alternatively, cell-free culture filtrates (108 cells/ml) were prepared by aseptically filtering bacterial cultures through 0.2 μm pore-sized filters (Sartorius Stedim Biotech GmbH, Goettingen, Germany). A drop of the mixture (1:1, v/v) of bacterial cells or cell-free culture filtrates and P. capsici zoospores was incubated on a glass slide at 28°C for 2 h. The SDW was utilized as an untreated control. Zoospore germination (%), germ tube length (μm), and zoospore lysis were assessed using a microscope (BX50, Olympus, Tokyo, Japan). Three hundred zoospores per sample (replication) were counted for germination rate and 30 germinated zoospores per sample (replication) were determined for germ tube length.
Bacterial colonization on pepper roots in a square dish system
Pepper seeds were surface-sterilized with a 0.5% NaOCl solution for 1 min and uniformly germinated seeds were soaked in bacterial suspensions (108 cells/ml) for 3 h. These seeds were then blotted on sterile filter papers to remove excess moisture and placed on 2% water agar in 125 × 125 × 20 mm dishes. The plates were incubated in the dark at 28°C for 2 days and then placed at 28°C under fluorescent light with a 16 h/day photoperiod. To evaluate the bacterial colonization of roots developed from the seeds, root samples were collected 1 and 7 days after bacterial treatment. Roots were aseptically detached from the seedlings, placed into tubes containing 10-mM MgSO4 solution, and shaken at 160 rpm for 30 min. The number of CFU in the resulting suspensions was determined as described previously and expressed as CFU/cm root.
Bacterial motility and biofilm formation assays
The swimming and swarming activities of the bacterial strains were evaluated to estimate the potential of these strains to target pepper roots after their application to soil. After bacterial strains were pre-incubated in Luria-Bertani (LB) medium at 28°C for 48 h, cells were harvested by centrifugation at 5,000 ×g for 10 min and resuspended in LB medium to obtain 108 cells/ml density. Two microliters of the resulting bacterial suspensions were drop-inoculated onto 1/10 tryptic soy broth surface containing 0.3 or 0.5% agar to assess swimming or swarming activity. For the swimming activity, 150 × 25 mm Petri dishes were used and 90 × 15 mm Petri dishes were used for the swarming activity. The plates were sealed with parafilm and incubated for 24 (swimming activity) or 48 h (swarming activity) at 28°C. The swimming or swarming activity of the tested strains was then determined as follows: swimming or swarming activity = halo diameter (mm) at 24 or 48 h after incubation – halo diameter (mm) at 0 h after incubation.
Biofilm formation was evaluated in 96-well microtiter plates (Becton Dickinson Labware, Franklin Lakes, NJ, USA), as described by O’Toole et al. (1999). Bacterial strains were pre-incubated in LB medium at 28°C for 24 h and harvested as described previously. The cells were then resuspended in biofilm formation medium (Hamon and Lazazzera, 2001), and 100 μl of the resulting suspension (104 cells/ml) was inoculated into each well of the microtiter plates. After stationary incubation of the plates for 48 h, the inoculum was removed and all wells were thoroughly rinsed with SDW. The remnant adherent cells were stained with 0.1% (w/v) crystal violet in a buffer solution (0.15 M ammonium sulfate, 100 mM potassium phosphate [pH 7], 34 mM sodium citrate, and 1 mM MgSO4). To solubilize adherent cells, 200 μl of ethanol:acetone (80%:20%, v/v) mixture was added into each well. Biofilm formation was assessed by measuring the absorbance (OD595) level with a microplate reader (HIDEX Oy, Turku, Finland).
Bacterial motility elicited by amino acids, organic acids, and sugars of pepper root exudates
Amino acids (aspartic acid, glutamic acid, glycine, and threonine), organic acids (citric acid, fumaric acid, oxalic acid, and succinic acid), and sugars (arabinose, fructose, glucose, and maltose) in pepper root exudates (Kamilova et al., 2006; Vančura and Hovadik, 1965) were tested to determine the chemotactic motility of bacterial strains. Swimming and swarming activities were evaluated on M9 minimal medium containing 0.3 and 0.5% agar, respectively, supplemented with amino acids, organic acids, or sugars. These supplements were incorporated at a final concentration of 10 or 1,000 μM after filtering through 0.2-μm microfilters (Sartorius Stedim Biotech GmbH). The plates were inoculated with 2 μl bacterial suspension (108 cells/ml) prepared in LB medium. After incubation of the plates at 28°C for 48 h, swimming and swarming activities were determined as described previously.
Statistical analysis
All experiments were conducted using a completely randomized design twice with three replicates per treatment, except for the biocontrol plant test (10 replicates). Data analysis was performed using the Statistical Analysis System software (SAS Institute, Cary, NC, USA). The homogeneity of variances for data from repeated experiments was verified using Levene’s test (Levene, 1960); then, the data were combined and further analyzed. For analysis of the bacterial population data, data were analyzed after log10 transformation. The zoospore germination rate (%) was analyzed after arcsine square-root transformation. Analysis of variance was performed using the general linear model procedure and the means were separated using the least significant difference test at P < 0.05.
Results
Biocontrol activity of P. plecoglossicida YJR13 and P. putida YJR92 against P. capsici on pepper
Strains YJR13 and YJR92 significantly (P < 0.05) reduced disease severity on 5-week-old pepper plants at 14 DAI with P. capsici compared with MgSO4-treated plants (untreated control) or KU408 (negative bacterial control)-treated plants (Fig. 1). No significant difference was found in the control efficacy between YJR13 and YJR92. Strain EXTN-1 (positive bacterial control) showed a similar level of control efficacy to strains YJR13 and YJR92 (Fig. 1).
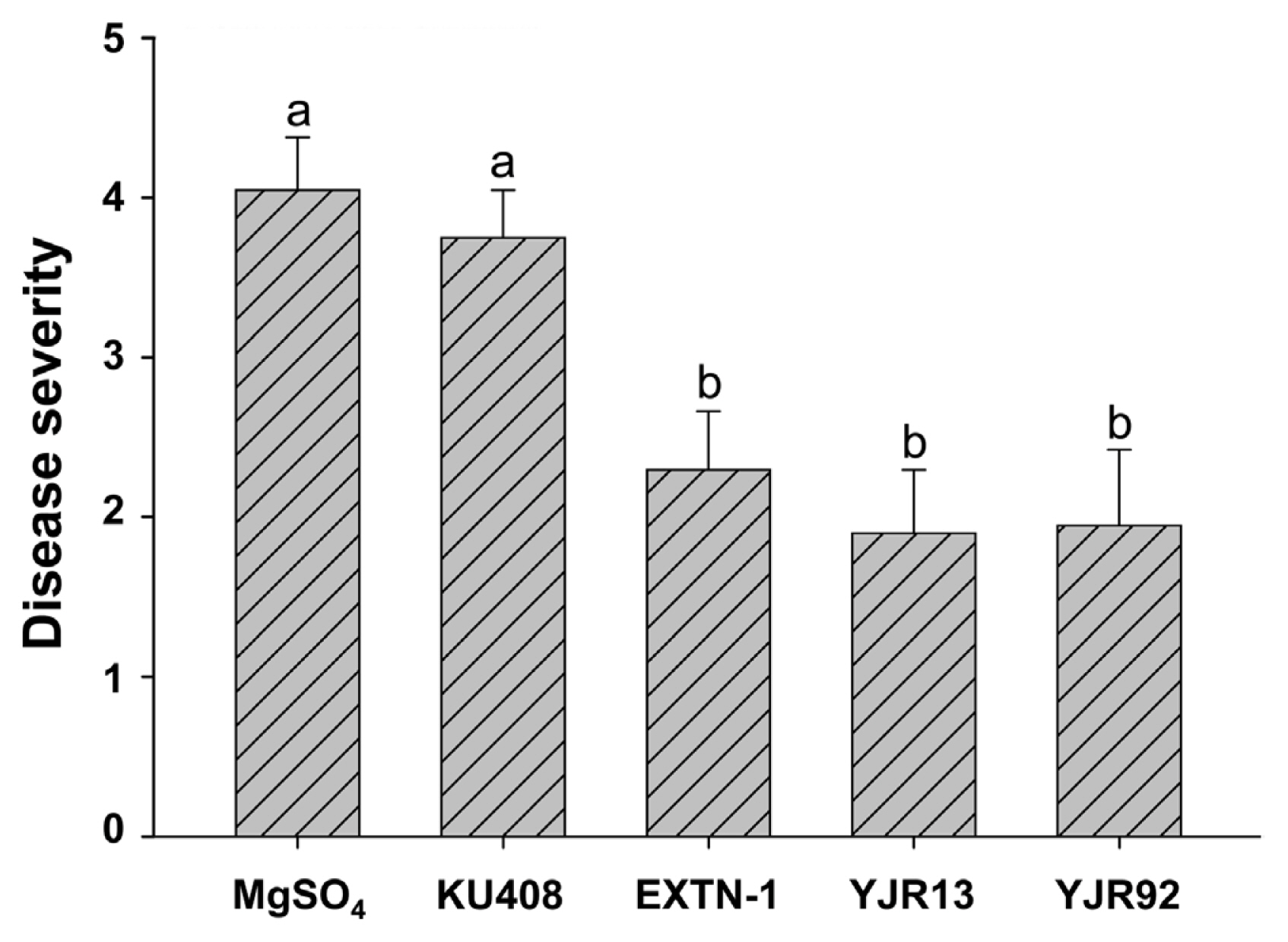
Biocontrol activity of Pseudomonas plecoglossicida YJR13 and Pseudomonas putida YJR92 compared with Sphingomonas aquatilis KU408 (negative bacterial control) and Bacillus vallismortis EXTN-1 (positive bacterial control) 14 days after inoculation with Phytophthora capsici on 5-week-old pepper (cv. Nockwang) plants. Ten-mM MgSO4 solution was used as an untreated control. Different letters above error bars (mean + standard error, n = 20) indicate significant (P < 0.05) differences between treatments according to the least significant difference test.
Anti-oomycete activity of P. plecoglossicida YJR13 and P. putida YJR92 against P. capsici on solid media and in liquid media
Dual-culture assays showed that strains YJR13 and YJR92 significantly (P < 0.05) inhibited mycelial growth of P. capsici on V8 juice agar and M9 minimal agar, compared with SDW (untreated control) and strain KU408 (negative bacterial control) (Table 1, Fig. 2). Likewise, strain EXTN-1 (positive bacterial control) significantly (P < 0.05) inhibited P. capsici growth on both media. Moreover, strains YJR13 and YJR92 inhibited mycelial growth more on M9 minimal agar (Fig. 2B) than on V8 juice agar (Table 1, Fig. 2A). Similar results were observed for the mycelial dry weights of P. capsici grown in liquid V8 juice media. Strains YJR13 and YJR92, and the positive bacterial control EXTN-1 greatly inhibited mycelial dry weight of P. capsici, compared with MgSO4 solution (untreated control) and strain KU408 (negative bacterial control) (Table 1).
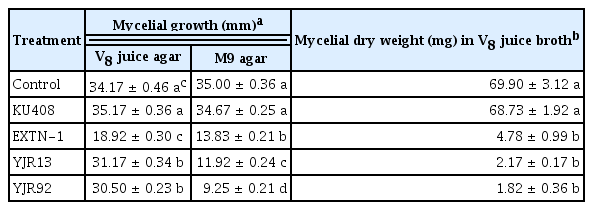
Anti-oomycete activity of Pseudomonas plecoglossicida YJR13, Pseudomonas putida YJR92, Sphingomonas aquatilis KU408 (negative bacterial control), and Bacillus vallismortis EXTN-1 (positive bacterial control) against Phytophthora capsici mycelial growth on V8 juice agar and M9 agar and in V8 juice broth
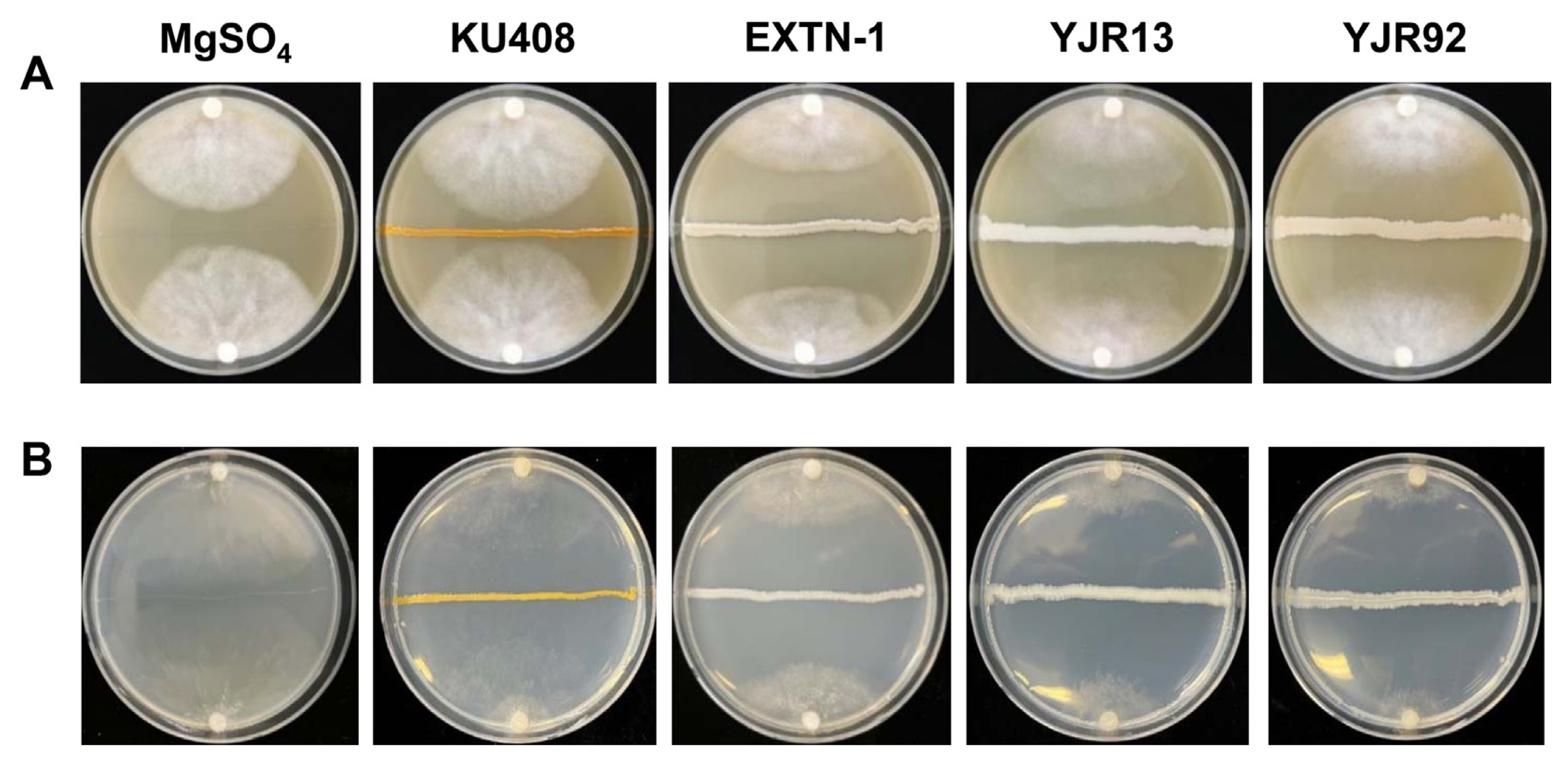
Anti-oomycete activity of Pseudomonas plecoglossicida YJR13 and Pseudomonas putida YJR92 compared with Sphingomonas aquatilis KU408 (negative bacterial control) and Bacillus vallismortis EXTN-1 (positive bacterial control) against Phytophthora capsici mycelial growth on (A) V8 juice agar and (B) M9 agar. Photographs were captured when mycelia in 10-mM MgSO4 solution plates (untreated control) reached the center of the plates. Bacterial strains or MgSO4 solution were streaked on the centers of the media 48 h before inoculation with P. capsici.
When the minimal inhibitory cell density of the bacterial strains was examined, a cell density of 106–10 cells/ml of strains YJR13 and YJR92, including the positive bacterial control strain EXTN-1 demonstrated distinct inhibition of P. capsici mycelial growth compared with that of 100–4 cells/ml of the strains (Fig. 3). Similar inhibitory zones to the mycelial growth were observed from 106 to 1010 cells/ml of strains YJR13, YJR92, and EXTN-1. On the other hand, negative bacterial control strain KU408 failed to inhibit mycelial growth of P. capsici at all tested cell densities (Fig. 2).

Cell density-dependent inhibitory activity of Pseudomonas plecoglossicida YJR13 and Pseudomonas putida YJR92 compared with Sphingomonas aquatilis KU408 (negative bacterial control) and Bacillus vallismortis EXTN-1 (positive bacterial control) on V8 juice agar. Five microliters of various concentrations of cell suspensions (100 [10-mM MgSO4 solution] to 1010 cells/ml) were applied on sterile filter paper disks. Mycelial plugs of the 5-day-old cultures of Phytophthora capsici were placed in the center of V8 juice agar and incubated at 28°C in the dark until mycelia reached until mycelia covered the 100 cell-treated paper disks.
Effects of diluted media on mycelial growth of P. capsici and populations of P. plecoglossicida YJR13 and P. putida YJR92
When the inhibitory activity of strains YJR13 and YJR92 was tested in diluted (nutrient-deficient) V8 liquid medium, these strains significantly (P < 0.05) inhibited mycelial growth of P. capsici in the medium regardless of the medium dilution compared with MgSO4 solution (untreated control) and strain KU408 (negative bacterial control) (Fig. 4A). However, strain EXTN-1 (positive bacterial control) had similar mycelial inhibition as observed in strains YJR13 and YJR92 regardless of the medium dilution. Additionally, strains YJR13, YJR92, and EXTN-1 significantly (P < 0.05) reduced the mycelial dry weights in 1/10 to 1/200 diluted V8 media, compared with that of 0 (undiluted) media (Fig. 4A). Similar results were observed in the MgSO4 control and the negative bacterial strain KU408; however, the inhibition rate was not similar to that of strains YJR13, YJR92, and EXTN-1 (Fig. 4A).

(A) Inhibitory activity of Pseudomonas plecoglossicida YJR13 and Pseudomonas putida YJR92 compared with Sphingomonas aquatilis KU408 (negative bacterial control) and Bacillus vallismortis EXTN-1 (positive bacterial control) against Phytophthora capsici mycelial growth 5 days after inoculation in diluted (0, 1/10, 1/50, 1/100, and 1/200) V8 juice broth. (B) Bacterial populations (log colony-forming units [CFU]/ml) of P. plecoglossicida YJR13, P. putida YJR92, S. aquatilis KU408, and B. vallismortis EXTN-1 in the diluted V8 broth 5 days after inoculation. Ten-mM MgSO4 solution was used as an untreated control. Different lowercase and uppercase letters indicate significant (P < 0.05) differences between the dilutions in each treatment and between treatments according to the least significant difference test, respectively.
In the bacterial population analysis, strains YJR13, YJR92, and EXTN-1 significantly (P < 0.05) showed higher populations than the negative bacterial strain KU408, regardless of the dilution (Fig. 4B). Populations of the negative control KU408 declined to 106 cells/ml when the strains were cultured in diluted V8 liquid media. However, strains YJR13, YJR92, and EXTN-1 demonstrated similar population levels (1012 cells/ml) in 1/10 diluted V8 juice broth compared with the 0 (undiluted) media, and exceeded 108 cells/ml in 1/50 to 1/200 diluted media (Fig. 4B).
Inhibitory activity by cell suspensions and cell-free culture filtrates of P. plecoglossicida YJR13 and P. putida YJR92 against zoospore development of P. capsici
Cell suspensions of strains YJR13 and YJR92 significantly (P < 0.05) inhibited zoospore germination of P. capsici compared with the SDW control (Table 2). However, the negative and positive control strains KU408 and EXTN-1, respectively, failed to inhibit zoospore germination. Similarly, strains YJR13 and YJR92 and the positive bacterial control EXTN-1 significantly (P < 0.05) inhibited germ tube length compared with the SDW control and negative bacterial control KU408 (Table 2). Moreover, zoospore lysis was observed 10 min after treatment with cell suspensions of strains YJR13 and YJR92 but not with SDW and strains KU408 and EXTN-1 (Supplementary Fig. 2). Likewise, when cell-free culture filtrates of the tested strains were applied to P. capsici zoospores, similar results except KU408 for zoospore germination were obtained in zoospore germination and germ tube length of P. capsici as observed in bacterial cell suspensions (Table 2).

Inhibitory activity of cell suspensions and cell-free culture filtrates of Pseudomonas plecoglossicida YJR13, Pseudomonas putida YJR92, Sphingomonas aquatilis KU408 (negative bacterial control), and Bacillus vallismortis EXTN-1 (positive bacterial control) against zoospore germination and germ tube elongation of Phytophthora capsici
Pepper root colonization by P. plecoglossicida YJR13 and P. putida YJR92 in a square dish system
When bacterial colonization was evaluated on pepper roots grown in a square dish system, strains YJR13 and YJR92 colonized pepper roots significantly (P < 0.05) more than MgSO4 solution (untreated control) and positive bacterial control EXTN-1 at 1 and 7 DAT (Table 3). The colonization ability of the negative bacterial strain KU408 was significantly (P < 0.05) lower than that of both YJR13 and YJR92 at 7 DAT. However, strain KU408 had similar bacterial numbers to strains YJR13 and YJR92 at 1 DAT but declined at 7 DAT. Pepper roots treated with MgSO4 solution (untreated control) had the lowest bacterial populations both 1 and 7 DAT. In addition, bacterial root colonization in all treatments between 1 and 7 DAT was not significantly (P > 0.05) different (Table 3).
Bacterial motility and biofilm formation by P. plecoglossicida YJR13 and P. putida YJR92
Strains YJR13 and YJR92 exhibited significantly (P < 0.05) greater swimming and swarming activities on tryptic soy agar containing 0.3 and 0.5% agar, respectively, than strain EXTN-1 (positive bacterial control) (Table 4, Supplementary Fig. 3). Swimming or swarming activity of strain KU408 (negative bacterial control) was almost undetectable. In the microtiter plate test, strains YJR13 and YJR92 exhibited significantly (P < 0.05) greater biofilm formation than the negative control strain KU408 and positive control strain EXTN-1 (Table 4).
Effects of amino acids, organic acids, and sugars on bacterial motility of P. plecoglossicida YJR13 and P. putida YJR92
All tested amino acids (aspartic acid, glutamic acid, glycine, and threonine), organic acids (citric acid, fumaric acid, oxalic acid, and succinic acid), and sugars (arabinose, fructose, glucose, and maltose), which are components of pepper root exudates, greatly increased the swimming and swarming activities of strains YJR13 and YJR92 compared with the positive bacterial control strain EXTN-1 (Fig. 5). However, these compounds, except for maltose, did not affect the swimming and swarming activities of the negative bacterial control KU408 (Fig. 5). In general, these compounds affected the swimming activity of the tested strains to a greater extent than their swarming activity (Fig. 5). These compounds stimulated the swimming and swarming activities of strains YJR13 and YJR92, regardless of the low (10 μM) and high (1,000 μM) concentrations (Fig. 5). In general, the high (1,000 μM) concentration of the tested compounds affected bacterial motility more than the low (10 μM) concentration. However, among the tested compounds, the low concentrations of glutamic acid, fumaric acid, oxalic acid, fructose, and maltose enhanced the swimming activity of YJR92 more than the high concentration (Fig. 5A). Threonine and fructose concentrations increased the swarming activity of the strain (Fig. 5B). Similarly, low fumaric acid, oxalic acid, succinic acid, and maltose concentrations enhanced the swimming activity of strain YJR13 more than high concentrations (Fig. 5A) and threonine, arabinose, and fructose increased the swarming activity of the strain (Fig. 5B).
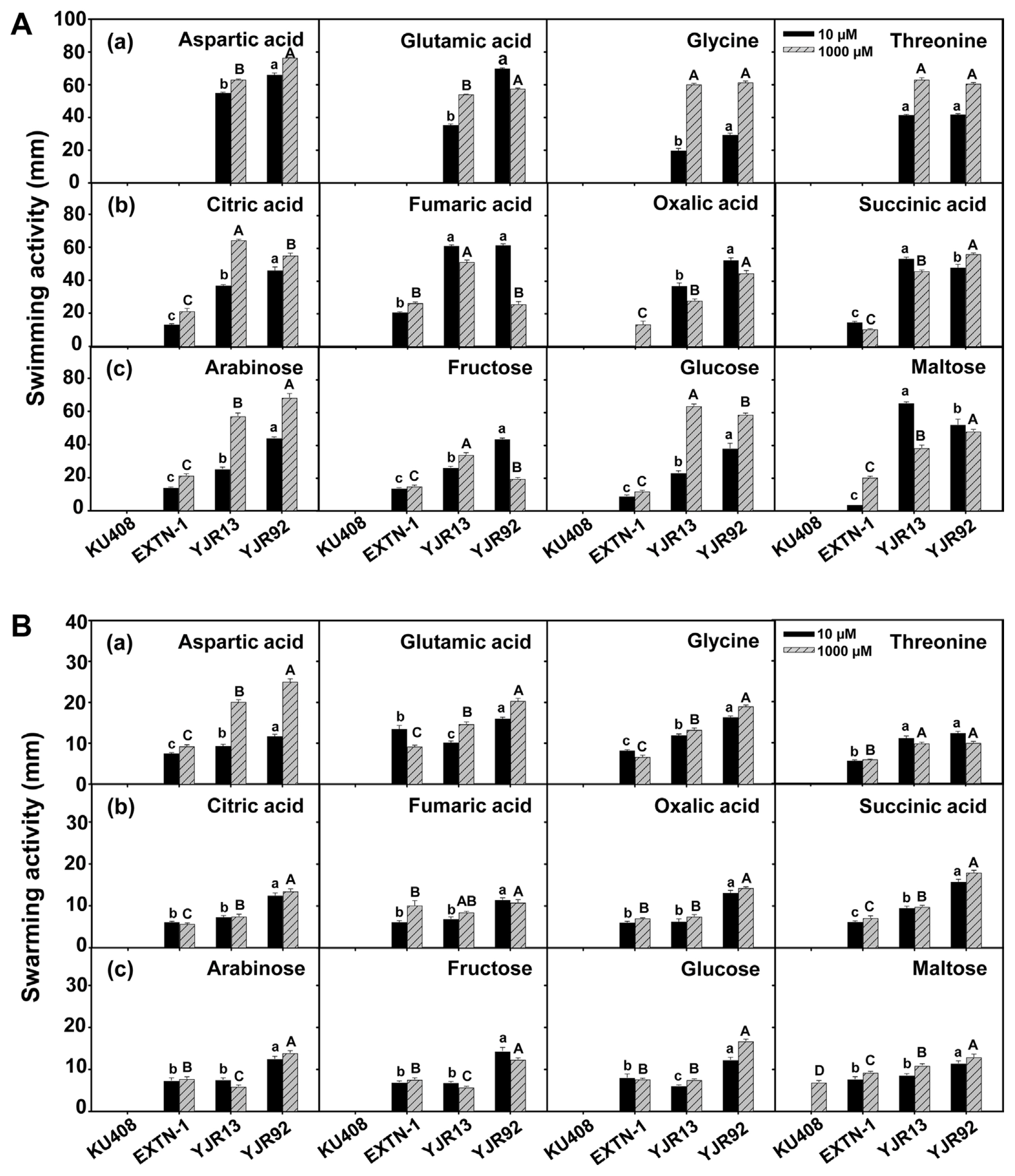
(A) Swimming and (B) swarming activities of Pseudomonas plecoglossicida YJR13 and Pseudomonas putida YJR92 compared with Sphingomonas aquatilis KU408 (negative bacterial control) and Bacillus vallismortis EXTN-1 (positive bacterial control) 48 h after incubation on M9 minimal medium containing 0.3 and 0.5% (w/v) agar, respectively. These media were amended with 10 and 1,000 μM of (a) amino acids (aspartic acid, glutamic acid, glycine, and threonine), (b) organic acids (citric acid, fumaric acid, oxalic acid, and succinic acid), and (c) sugars (arabinose, fructose, glucose, and maltose), respectively. Different lowercase and uppercase letters on the columns (mean + standard error, n = 6) indicate significant differences between bacterial treatments at 10 and 1,000 μM concentrations at P < 0.05, respectively.
Discussion
In our previous study, P. plecoglossicida YJR13 and P. putida YJR92 were demonstrated to have biocontrol potential against Phytophthora blight of pepper, which were selected from a sequential screening procedure (Sang et al., 2013). In this study, we demonstrated that these strains YJR13 and YJR92 suppressed disease severity caused by P. capsici through inhibition against mycelial growth, zoospore germination, and germ tube elongation of the pathogen, including bacterial colonization of pepper roots. Moreover, root colonization with biofilm formation by the tested strains was stimulated by enhanced motility and chemotaxis to the components of pepper root exudates.
Pseudomonads are common biocontrol agents with efficient performance against Phytophthora species, including Phytophthora cactorum, P. capsici, and Phytophthora infestans (Barahona et al., 2011; De Vrieze et al., 2018; Sang and Kim, 2014). In particular, P. putida is a well-known biocontrol agent that is active against bacterial (Sun et al., 2017), fungal (Oliver et al., 2019), oomycete (Hyder et al., 2020) pathogens, and even nematodes (Zhai et al., 2018). In addition, P. plecoglossicida biocontrol activity tested in this study has been rarely studied with regard to plant and pathogen systems. The P. plecoglossicida was first isolated as a causal agent of bacterial hemorrhagic ascites from ayu fish (Plecoglossus altivelis) (Nishimori et al., 2000). This species is known to be pathogenic to freshwater fishes, including the large yellow croaker (Pseudosciaena crocea), pejerrey (Odontesthes bonariensis), and rainbow trout (Oncorhynchus mykiss) (Huang et al., 2018). Subsequently, it was discovered in tannery soil and inside the roots of the sand dune plant Elymus mollis (Chowdhury et al., 2004; Park et al., 2005). The P. plecoglossicida is effective in soil bioremediation due to the strong gallic acid degradation (Chowdhury et al., 2004). Additionally, P. plecoglossicida can produce siderophores under iron-limited conditions and HCN, implying their biocontrol potential (Faramarzi and Brandl, 2006; Meyer et al., 2002). In this study, both strains YJR13 and YJR92 demonstrated effective biocontrol activity in planta and in vitro tests with comparable or even better efficiency than the commercial biocontrol agent B. vallismortis EXTN-1.
In our current study, P. plecoglossicida YJR13 and P. putida YJR92 could produce appropriate amounts of diffusible compounds antagonistic to mycelial growth of P. capsici on M9 minimal agar and in liquid medium, V8 juice broth, but not on solid medium, V8 juice agar (sparse mycelial growth detected). Previously, Aravind et al. (2009) reported that P. putida showed in vitro inhibitory activity in a dual-plate assay and performed more efficiently in plant tests. In our study, the tested strains YJR13 and YJR92 also demonstrated pronounced anti-oomycete activity in P. capsici mycelial growth on M9 minimal medium and in V8 juice broth. Low-nutrient conditions may be present in the plant rhizospheres, resulting in low cell densities of biocontrol agents and cause undesirable biocontrol activity. Therefore, the biocontrol activity of the bacterial strains introduced in poor nutrient conditions of the rhizospheres should be maintained. In this regard, our tested strains YJR13 and YJR92, which produce inhibitory compounds on the nutrient minimal medium, might exhibit more sustainable anti-oomycete activity on pepper root rhizospheres. Likewise, when the effect of low-nutrient conditions on the anti-oomycete activity of strains YJR13 and YJR92 was further tested using a diluted medium, the dilution factor had no significant effect on the growth and inhibitory activities of the tested strains. Guyer et al. (2015) demonstrated that Pseudomonas spp. suppressed potato late blight in vivo at 2 × 108 cells/ml, but not at lower concentrations. In this study, a decrease in cell populations of strains YJR13 and YJR92 was observed in 1/50 to 1/200 diluted media; simultaneously, the bacterial populations were maintained at more than 108 cells/ml and could significantly inhibit P. capsici mycelial growth. Obviously, the anti-oomycete activity of the tested strains YJR13 and YJR92 was distinct when either cell suspensions or cell-free culture filtrates of the strains were applied to P. capsici zoospores. The reduction in zoospore germination rate and germ tube length of P. capsici by both treatments might imply the presence of bacterial secondary metabolites antagonistic to the pathogen that are more distinct in zoospores rather than mycelial growth of P. capsici. Similarly, De Vrieze et al. (2018) found that biocontrol P. ferederikbergensis strain S19 efficiently inhibited zoospore production more than mycelial growth of P. infestans.
The successful establishment of biocontrol agents depends significantly on the sustainable survival of bacterial populations in the rhizospheres. Primary colonization and subsequent establishment in rhizoplanes or rhizospheres (Herrera et al., 2020) are the major traits of microbial survival and function. Previous studies (Sheoran et al., 2015; Sun et al., 2017) have demonstrated that P. putida successfully colonizes root surfaces and interiors of plants, including pepper, Arabidopsis, ginger, pepper, and tomato. In our study, P. plecoglossicida YJR13 and P. putida YJR92 maintained a population of more than 106 cells/cm root on pepper roots grown in a square dish system without exogenous nutrient supplementation for up to 7 days. This bacterial colonization of plant roots could be enhanced by competitive characteristics, including swimming and swarming activities, and biofilm formation. Swimming and swarming motilities of Pseudomonas strains, such as P. chlororaphis M71, Pseudomonas corrugata CCR04 and CCR08, and P. putida M71 have been previously reported (Dutta and Lee, 2022; Raio et al., 2020; Sang and Kim, 2014). Furthermore, Gao et al. (2016) revealed that the swarming motility of rhizobacteria could play a crucial role in tomato root colonization by rhizobacteria. Similarly, both swimming and swarming activities along with biofilm formation were strongly detected in the strains YJR13 and YJR92 tested in our study. Furthermore, strains YJR13 and YJR92 had significant chemotactic ability towards several components, including amino acids (aspartic acid, glutamic acid, glycine, and threonine), organic acids (citric acid, fumaric acid, oxalic acid, and succinic acid), and sugars (arabinose, fructose, glucose, and maltose) of pepper root exudates (Kamilova et al., 2006; Vančura and Hovadik, 1965). Bacterial chemotaxis is an important trait for root colonization. Van de Broek et al. (1998) exhibited that Azospirillum brasilense non-chemotactic mutants reduced the colonization ability of wheat roots. More recently, Sun et al. (2017) reported that efficient bacterial wilt biocontrol was achieved by the strong chemotaxis of P. putida A1 in plant extracts. Therefore, the chemotaxis and biofilm-forming activity of the tested strains, YJR13 and YJR92, can promote root colonization observed in our square dish system.
Collectively, our results suggest that P. plecoglossicida YJR13 and P. putida YJR92 inhibit infection of the soilborne oomycete P. capsici on pepper plants by inhibiting mycelial growth, zoospore germination, and germ tube elongation of the oomycete pathogen, protecting plant roots through bacterial motility (swimming and swarming activities), colonization, and biofilm formation on pepper roots, and stimulating bacterial motility with root exudate compounds. Therefore, strains YJR13 and YJR92 are promising biocontrol agents for controlling Phytophthora blight of pepper caused by P. capsici. To our knowledge, this is the first report of P. plecoglossicida as a biocontrol candidate for the pepper disease management.
Acknowledgment
Elena Volynchikova was supported by the Korean Government Scholarship Program (KGSP) during her PhD study at the Korea University, Seoul, Korea.
Notes
Conflicts of Interest
No potential conflict of interest relevant to this article was reported.
Electronic Supplementary Material
Supplementary materials are available at The Plant Pathology Journal website (http://www.ppjonline.org/).


