 |
 |
| Plant Pathol J > Volume 39(3); 2023 > Article |
|
Abstract
The plant microbiota plays a crucial role in promoting plant health by facilitating the nutrient acquisition, abiotic stress tolerance, biotic stress resilience, and host immune regulation. Despite decades of research efforts, the precise relationship and function between plants and microorganisms remain unclear. Kiwifruit (Actinidia spp.) is a widely cultivated horticultural crop known for its high vitamin C, potassium, and phytochemical content. In this study, we investigated the microbial communities of kiwifruit across different cultivars (cvs. Deliwoong and Sweetgold) and tissues at various developmental stages. Our results showed that the microbiota community similarity was confirmed between the cultivars using principal coordinates analysis. Network analysis using both degree and eigenvector centrality indicated similar network forms between the cultivars. Furthermore, Streptomycetaceae was identified in the endosphere of cv. Deliwoong by analyzing amplicon sequence variants corresponding to tissues with an eigenvector centrality value of 0.6 or higher. Our findings provide a foundation for maintaining kiwifruit health through the analysis of its microbial community.
Plants in their natural environment are in constant interaction with diverse microorganisms. This consortium of microorganisms, including bacteria, fungi, protists, and viruses, colonizes the plant and coevolves with the host plant to form a plant microbiome (Kaul et al., 2021). The phytobiome, which comprises the plant and its environment, is vital to natural ecosystems. Over the past few decades, researchers have recognized the crucial role of microorganisms in the natural world (Abdelfattah et al., 2022; Bettenfeld et al., 2022; Hardoim et al., 2015). The plant microbiome plays a pivotal role in nutrient acquisition, abiotic/biotic stress tolerance, and plant immune regulation (Turner et al., 2013; Vandenkoornhuyse et al., 2015).
The plant is known to harbor a diverse microbial community in the soil surrounding its roots, particularly in the rhizosphere (Qu et al., 2020). This region of soil, affected by root exudates, is composed of the highest abundance and species diversity of microbiomes (Mendes et al., 2013). The microbiome in the rhizosphere has been increasingly recognized to play a crucial role in determining plant health (Berendsen et al., 2012, Kim et al., 2019). In addition to the rhizosphere, specialized microorganisms can also colonize the inside of plant roots or shoots, which is known as the endosphere (Wang and Haney, 2020). The majority of microorganisms forming the plant microbiome originate from surrounding sources such as horizontal transmission from soil or air, and vertical transmission from seeds of the parent generation (Abdelfattah et al., 2021). Understanding the influence of these microorganisms on plant growth is crucial due to their growth-promoting attributes in the endosphere (Adeleke and Babalola, 2021).
Kiwifruit (Actinidia spp.) is a highly nutritious fruit that is cultivated globally due to its rich content of essential nutrients such as vitamin C, potassium, folate, and phytochemicals (Richardson et al., 2018). Kiwifruit is rapidly gaining popularity as a commercial crop in countries such as Australia, Italy, Japan, New Zealand, and the United States (Heatherbell et al., 1980). However, the cultivation of kiwifruit is hampered by various plant pathogens such as Botrytis cinerea (Michailides and Elmer, 2000), Alternaria alternata (Kwon et al., 2011), and Pseudomonas syringae (McCann et al., 2013), which can cause a range of diseases that adversely affect the yield and quality of kiwifruit. Hence, the management of these diseases is crucial for the sustainable production of kiwifruit. The microbiome has been recognized as a crucial component of plant health, impacting various plant tissues. Recent studies have shed light on the microbial ecology of fruits such as grapes (Chou et al., 2018), mangoes (Diskin et al., 2017), and olives (Abdelfattah et al., 2018), based on improvements in next-generation sequencing techniques. Despite the many investigations into the microbiome of fruit over the decades, little is known about the microbiota community structures in different tissues of kiwifruit. Thus, this research aims to investigate the microbiota of the rhizosphere and endosphere of kiwifruit, providing insights into the plant ecology of different tissues and cultivars (cv. Deliwoong, cv. Sweetgold).
Samples were collected three times from different tissues of kiwifruit in the Gardening Research Institute (Namhae, Republic of Korea; 34°48′56.8″N 127°55′42.7″E) to analyze microbial communities in kiwifruit orchards. Two different cultivars (cv. Deliwoong, cv. Sweetgold) were compared, and three kiwifruit trees were randomly selected for each cultivar. Samples were collected from various tissues, including rhizosphere, sap, leaf, flower bud, flower, and fruit, corresponding to the three periods (Supplementary Table 1). All samples were collected in 50 ml conical tubes, with rhizosphere samples collected from 1-3 mm below the root surface (n = 3) and the rest of the samples (leaf, flower bud, flower, fruit) collected from 3 trees (n = 3).
To isolate endosphere bacteria, the surface of the samples (excluding rhizosphere and sap) was sterilized. The samples were filled with 50 ml of 1× phosphate buffered saline (PBS) buffer (10× PBS: 80 g NaCl, 2 g KCl, 1.44 g Na2HPO4, 2.4 g KH2PO4, adjusted to a pH of 7.4, and a final volume of 1 liter) for 10 min. After sonication, the samples were sterilized with 70% ethanol for 30 s, 1% NaOCl for 30 s, and ddH2O for 2 times. The sterilized samples were then dried for 1 h on a clean bench and ground using 10 ml of 1× PBS buffer.
A modified NycoDenz density gradient centrifugation method was utilized for microbial isolation from all samples. In a 15 ml conical tube, 1 g of all samples and 9 ml of bacterial cell extraction (BCE) buffer (composed of 20 ml/l Triton X-100, 10 ml/l Tris-HCl pH 7.5, mass up to 1 liter with ddH2O, with 2 mM 2-mercaptoethanol added immediately before use) were vortexed. The mixtures were then filtered using sterilized Miracloth (Merck Millipore, Billercia, MA, USA) in a 50 ml conical tube. The filtered solution was subjected to centrifugation in a 15 ml conical tube at 10,000 ×g for 10 min using a LaboGene 1736R centrifuge (Lillerod, Denmark), and the supernatant was discarded. This process was repeated three times. The samples were then washed with BCE buffer and dissolved in 6 ml of 50 mM Tris-HCl (pH 7.5). Subsequently, 4 ml of NycoDenz (8 g NycoDenz dissolved in 10 ml of 50 mM Tris-HCl, pH 7.5; Axis-Shield, Dundee, Scotland, UK) was carefully poured onto the sample mixture, which was then centrifuged at 10,000 ×g for 90 min. After centrifugation, it was confirmed that a thin layer composed of bacterial cells was located between the NycoDenz solution and 50 mM Tris-HCl. Bacterial cells were then transferred to a microcentrifuge tube, and 1 ml of the resulting mixture was used for DNA extraction.
The DNA extraction from bacterial cells was carried out using the cetyltrimethylammonium bromide (CTAB) method described by Graham et al. (2003). Briefly, 100 μl of NycoDenz was added to 500 μl of CTAB solution in a microcentrifuge tube and incubated at 65°C for 30 min to decompose the cell wall and protein. After heat treatment, 700 μl of phenol:chloroform:isoamyl alcohol (25:24:1) was added to the mixture, which was then centrifuged at 12,470 ×g for 10 min. The supernatant containing DNA was carefully transferred to another microcentrifuge tube and mixed with 400 μl of isoprophanol. The mixture was then centrifuged at 12,470 ×g for 5 min at 4°C, and the supernatant was discarded. Next, 700 μl of 70% ethanol was added, and the mixture was centrifuged again at 12,470 ×g for 5 min at 4°C. The supernatant was removed, and the remaining DNA pellet was dried completely at room temperature. After drying, 20 μl of TE buffer (5 ml of 1 M Tris pH 8, 1 ml of 0.5 M EDTA pH 8, 496 ml of ddH2O) was added to melt the DNA. The concentration and purity of DNA were measured using a NanoDrop 2000C spectrophotometer (Thermo Scientific, Waltham, MA, USA) and stored at −20°C after extraction.
The 16S rRNA V4 region was amplified by diluting DNA to a 5-8 ng/μl and using the 515F-806R primer set, including overhang adapter sequences. The polymerase chain reaction reaction was carried out using a total of 40 μl of reaction mixture containing 2.5 μl of genomic DNA, 1 μl of each primer (10 pmol), 20 μl of 2× PCR buffer for KOD FX Neo, 4 μl of dNTPs (2 mM), and 0.3 μl of KOD FX Neo (1.0 U/μl; Toyobo, Tokyo, Japan), with ddH2O added to a final volume of 40 μl. The thermal cycling conditions consisted of an initial denaturation step at 95°C for 3 min, followed by 25 cycles of denaturation at 95°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 30 s, using a T100 thermal cycler (Thermo Fisher Scientific, Waltham, MA, USA).
The PCR product was loaded onto a 1% agarose gel in an electrophoresis machine (Mupid ExU, Japan) for 20 min at 100 V and a 100 bp DNA Ladder Marker (MGmed, Seoul, Korea). The band of interest was excised using a sterilized scalpel and purified using the ExpinTM Gel SV kit (GeneAll, Seoul, Korea). The purified band was then transferred to a microcentrifuge tube, and 300 μl of GB buffer was added, followed by incubation at 50°C for 10 min. The tube was then centrifuged at 12,470 ×g for 1 min, and the entire solution was transferred to a column tube, which was then centrifuged at 12,470 ×g for 1 min. The flow-through was discarded, and 500 μl of 80% ethanol was added to the column tube, which was then centrifuged at 12,470 ×g for 3 min. Finally, the column was air-dried for 30 min, and the genomic DNA was eluted from the column. Sequencing was performed by Illumina Miseq (Macrogen Co., Seoul, Korea), and the raw sequences were processed to generate amplicon sequence variant (ASV) using DADA2 version 1.8 (Callahan et al., 2015) following the guidelines provided at https://benjjneb.github.io/dada2/tutorial_1_8.html in R version 4.0. The libraries were truncated at 300 bp, with a minimum quality score of 30, and the forward and reverse reads were merged and checked for chimeras. The taxonomic classification was assigned using the Silva release 138 database (https://www.arb-silva.de/) and IDTAXA (Murali et al., 2018), with a minimum similarity of 95%. The resulting data were visualized using ggplot2 version 3.3.3. Rarefaction curve, alpha diversity, and beta diversity analyses were conducted using the ggiNEXT version 2.0.20 and pyloseq version 3.12 packages. Statistical analysis was performed using the Kruskal-Wallis rank sum test (Kruskal and Wallis, 1952), followed by the Conover-Iman post-hoc test (Dinno and Dinno, 2017). The principal coordinates analysis (PCoA) was performed based on Bray-Curtis distance. Finally, co-occurrence network analysis was conducted using SparCC (Sparse correlations for compositional data) for degree centrality and eigenvector centrality (Friedman and Alm, 2012; Ruhnau, 2000). GenBank accession information such as BioProject, BioSample, and SAR were presented in Supplementary Table 2.
To assess the richness and evenness of bacterial communities in kiwifruit, we sequenced the V4 region of 16S rRNA using synthesis for sequencing. A total of 2,099,746 reads were identified in this study. The number of reads confirmed in the second and third samplings was 995,142 and 619,301, respectively (Supplementary Table 3). In the first sampling, clusters of 485,303 taxonomic bacteria were identified. ASVs were assigned by IDTAXA for sequence-based bacterial taxonomic classification. In the rhizosphere of cvs. Deliwoong and Sweetgold, the number of ASVs ranged from 263 to 456, regardless of sampling times (Fig. 1). In cv. Deliwoong, the Observed index values showed statistical differences and were differently grouped according to the sampling times (P = 0.02732). The observed values for cv. Deliwoong ranged from 62 to 194 in the endosphere and were lower than those in the rhizosphere (Fig. 1A). No statistical difference was found in the endosphere of cv. Deliwoong (P = 0.1341) during stages. Similarly, the number of ASVs in cv. Sweetgold was higher than in cv. Deliwoong, ranging from 103 to 287, but there was no significant difference (Fig. 1B). The Shannon index, which indicates the evenness of bacterial communities, showed no significant difference according to the sampling period or tissue within the cultivar (range, 3.7343-5.4588). However, the endosphere of cv. Sweetgold cultivar showed a significant difference (P = 0.0462).
The relative abundance of bacterial communities was determined at the order and family levels, with the top 10 taxa presented in Fig. 2. At the order level, Pseudomonadales, Enterobacterales, Xanthomonadales, Rhizobiales, Streptomycetales, Propionibacteriales, Burkholderiales, Flavobacteriales, Micrococcales, and Corynebacteriales were found to be abundant in all tissues (Supplementary Fig. 1A). No significant difference was observed in the rhizosphere between cultivars, with Pseudomonadales, Enterobacterales, and Xanthomonadales being the most abundant, accounting for 7.61-7.28%, 1.86-35.81%, and 2.27-25.27%, respectively. In contrast, bacterial communities in the endosphere showed differences between cultivars. In cv. Deliwoong, the sap was dominated by Enterobacterales (38.8-78.61%), while in cv. Sweetgold, Pseudomonadales (16.6-46.44%) were more abundant. Streptomycetales increased from 0-1.38% to 11.76-32.61% in the flower compared to the sap. In addition, Streptomycetales accounted for 20.48-27.61% of the bacterial community in the leaf of cv. Deliwoong at the last sampling, whereas Enterobacterales increased to 60.58-69.22% in the leaf of cv. Sweetgold.
Pseudomonadaceae, Enterobacteriaceae, Xanthomonadaceae, Moraxellaceae, Streptomycetaceae, Yersiniaceae, Propionibacteriaceae, Rhizobiaceae, Weeksellaceae, and Oxalobacteraceae were identified as the top 10 families (Supplementary Fig. 1B). The results showed that the microbial communities in the rhizosphere were similar, with Pseudomonadaceae, Xanthomonadaceae, and Moraxellaceae dominating at 3.87-49.61%, 2.27-25.27%, and 0.36-68.85%, respectively. At the family level, differences were observed between the endosphere and rhizosphere, with cultivars playing a significant role. Yersiniaceae was the predominant family in cv. Deliwoong (12.02-77.99%), whereas Pseudomonadaceae dominated in cv. Sweetgold (17.44-46.35%). In other plant tissues, Pseudomonadaceae and Streptomycetaceae were present at 3.5-53.3% and 0-32.66%, respectively, in both cultivars. Enterobacteriaceae increased from 6.46% to 57.71% in the leaf and fruit samples of cv. Sweetgold during the final sampling period.
PCoA revealed dissimilarities in microbial community composition between tissues based on Bray-Curtis distance (Fig. 2). The statistical significance of these differences was confirmed by permutational analysis of variance (PERMANOVA), with a Padj value of 0.001 (Fig. 2A). The PCoA plot showed a clear separation of samples into two groups, rhizosphere, and endosphere. To investigate differences in microbial community composition between cultivars, we further analyzed the PCoA plot by dividing it into tissues and analyzing each tissue by cultivar (Fig. 2B). This analysis revealed that microbial communities were statistically similar across cultivars within each tissue. These results indicate that while there were significant differences in microbial community composition between tissues, there were no significant differences between cultivars.
Correlations were estimated to examine interactions between ASVs in the network using SparCC (Fig. 3). The network was analyzed using the sum of samples cvs. Deliwoong and Sweetgold because there were no significant differences between tissues. Degree centrality and eigenvector centrality were calculated to determine the network’s properties. Analysis based on degree centrality revealed that the network was divided into three groups. Dots with a small number of adjacent nodes indicated a weak relationship with other microorganisms, while high-degree centrality was indicative of a stronger relationship with other ASVs (Fig. 3A). The co-occurrence network using eigenvector centrality showed similar interactions to those observed with degree centrality (Fig. 3B). The network was also divided into three groups. ASVs with an eigenvalue of 0.6 or higher were selected from the rhizosphere and endosphere of each cultivar. Corynebacteriaceae, Enterobacteriaceae, Streptococcaceae, Nitrososphaeraceae, Moraxellaceae, and Pseudomonadaceae had high eigenvector centrality (0.940443-1) in the rhizosphere of cv. Deliwoong (Table 1). In the rhizosphere of cv. Sweetgold, Enterobacteriaceae, and Xanthomonadaceae had eigenvector centrality values ranging from 0.612183-0.955234 and 0.692647-1, respectively (Table 2). Enterobacteriaceae was common in the rhizosphere with an eigenvector value of 0.6 or more in both cultivars. Streptomycetaceae had a high relationship with other ASVs (0.920587-1) in the endosphere of cv. Deliwoong (Table 3). However, in the endosphere of Sweetgold, Enterobacteriaceae, Isosphaeraceae, Rubinisphaeraceae, Moraxellaceae, Pseudomonadaceae, and Xanthomonadaceae showed high relationships (0.626701-0.879513) with other bacteria (Table 4).
The structure of microbial communities and their diversity contributes to the overall health of plants (Bettenfeld et al., 2022). The study of plant-microbes relationships and functions has been ongoing for decades. Despite its significance, the precise nature of the relationship between plants and microbiota remains unclear. In this study, we examined the differences in microbial communities present in different cultivars and tissues of kiwifruit.
The endosphere tissues exhibited differences in microbial communities within cultivars compared to the rhizosphere. cv. Deliwoong showed 0.69-77.99% occupancy by Yersiniaceae and Streptomycetaceae, while cv. Sweetgold was dominated by Pseudomonadaceae at 3.5-46.35%, and Enterobacteriaceae increased to 0.2-57.71%, respectively. However, there was no significant difference observed between cultivars within the tissues, as determined by PERMANOVA. While the relative abundance of the top 10 taxa showed some differences, the PCoA analysis revealed no significant variation between the cultivars. Therefore, these results suggest that the PCoA analysis demonstrated a similarity in the entire environmental microorganisms, while the relative abundance analysis highlighted differences only in the top 10 taxa. To explore the interaction between ASVs, a co-correlation network was estimated using SparCC. To determine the similarity between cultivars, the cvs. Deliwoong and Sweetgold cultivars were both used to analyze the network. Degree centrality and eigenvector centrality were used to examine the relationships between ASVs. The analysis based on degree centrality revealed that the network was divided into three distinct groups. Eigenvector centrality measures the importance of a node based on the microbes in its neighborhood, and it focuses on the overall pattern of the network (Bonacich and Bailey, 1971; Grassi et al., 2007). The eigenvector-based network exhibited a similar structure to that revealed by the degree centrality-based network. It was suggested that degree centrality and eigenvector centrality are highly correlated, regardless of the type of network (He and Meghanathan, 2016). These findings were supported by previous studies that showed a strong correlation between eigenvector centrality and degree centrality (Valente et al., 2008). Upon conducting a PCoA, a statistical difference was observed between the microbial communities of the rhizosphere and endosphere. The rhizosphere, influenced by root exudates, exhibited a higher microbiome abundance and species diversity, as previously reported (Mendes et al., 2013). In contrast, the plant endosphere harbored highly specific microbial communities that differed significantly from those observed in the rhizosphere (Vandenkoornhuyse et al., 2015). These findings suggest that the observed differences in microbial community composition between the rhizosphere and endosphere are biologically meaningful and not merely due to chance
In our systematic analysis of network patterns, we observed that eigenvector centrality values of 0.6 or higher were present in the rhizosphere and endosphere, depending on the cultivar. Notably, Streptomycetaceae dominated the endosphere of the Deliwoong cultivar, suggesting an important role in this plant compartment. In a recent study, Kim et al. (2022) reported the appearance of Streptomyces venezuelae 1-1 9D in the rhizosphere of kiwifruits, where it demonstrated biocontrol activity against Pseudomonas syringae pv. actinidiae. Additionally, we observed that cv. Sweetgold tree died during the third sampling time, suggesting a potential role for Streptomyces in promoting plant health in the endosphere of cv. Deliwoong. To our knowledge, this research represents the first attempt to investigate microbiota community structures across different tissues and developmental stages in kiwifruit. Our findings contribute to the growing body of knowledge on fruit microbiota research and provide further insights for future studies.
Acknowledgments
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MIST) [2020R1A2C20041777].
Electronic Supplementary Material
Supplementary materials are available at The Plant Pathology Journal website (http://www.ppjonline.org/).
Fig. 1
Species richness and evenness of kiwifruit. Average sequence variant was assigned using IDTAXA based on a 97% or greater similarity in 16S rRNA sequence. Alpha diversity was assessed using the Observed, Shannon, and Simpson indices. Kiwifruit trees of different varieties (cv. Deliwoong, cv. Sweetgold; n = 3) were randomly selected for comparison. Samples were collected from the rhizosphere, sap, leaf, flower bud, flower, and fruit in each of the three periods. All samples showed statistically significant differences based on the Kruskal-Wallis rank sum test. Post-hoc analysis was performed using the Conover-Iman test for multiple pairwise comparisons. (A) Alpha diversity of kiwifruit (cv. Deliwoong). (B) Alpha diversity of kiwifruit (cv. Sweetgold).
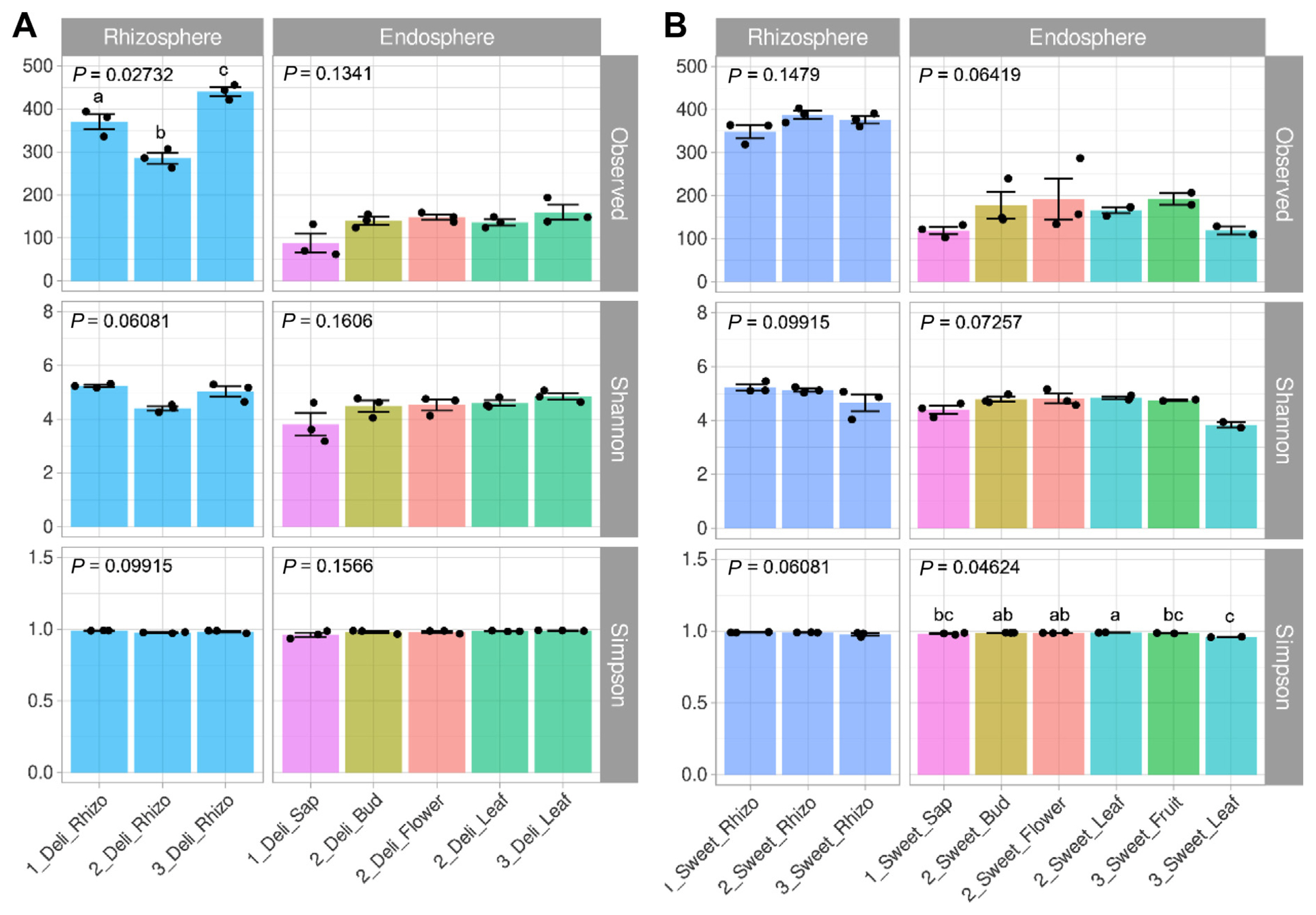
Fig. 2
Dissimilarity comparison of the microbial community. Principal coordinate analysis (PCoA) based on Bray-Curtis dissimilarity metrics revealed microbial community differences between cultivars (cv. Deliwoong, cv. Sweetgold). Statistical significance was analyzed by permutational multivariate analysis of variance (PERMANOVA). (A) The comparison of microbial communities corresponding to the entire dataset. (B) The dissimilarity of the microbial community according to the cultivar in kiwifruit tissues, where fruit tissue was only present in the Sweetgold cultivar.
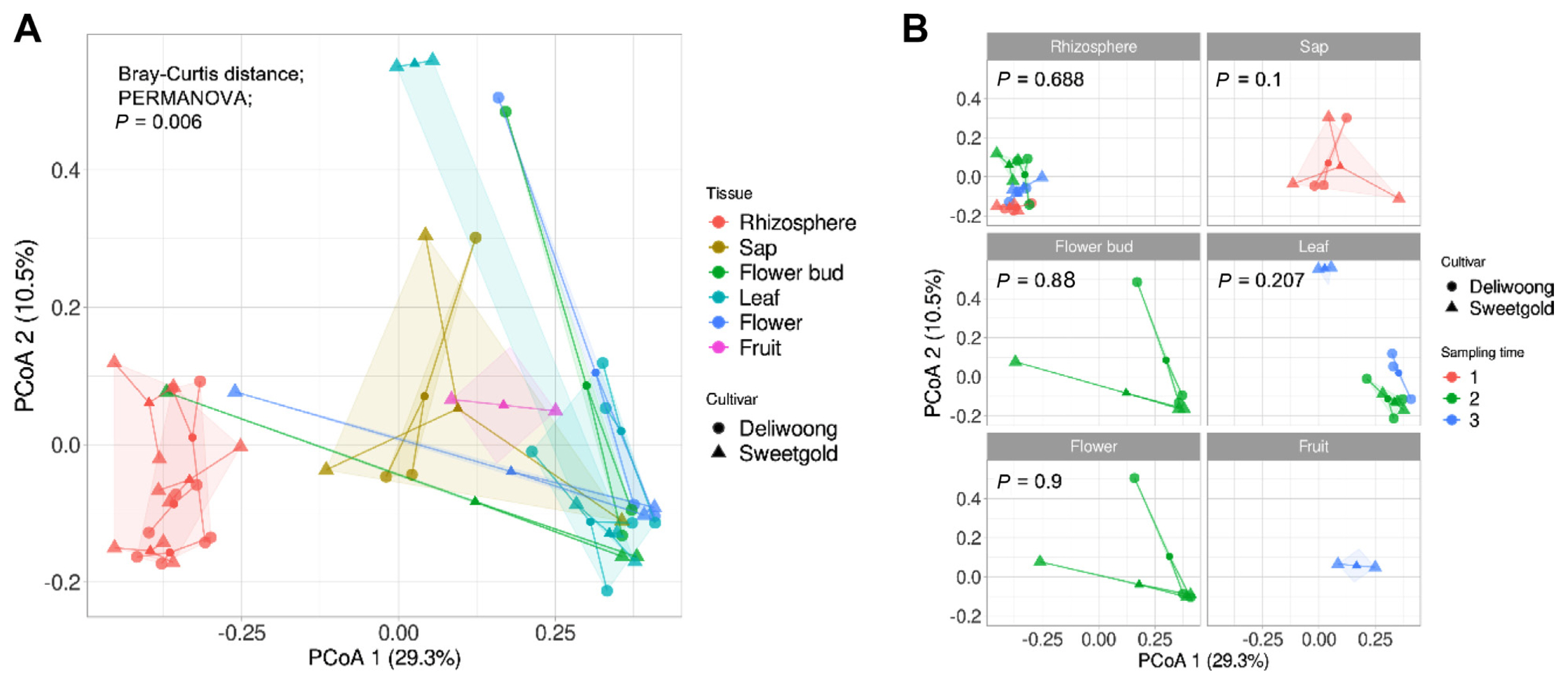
Fig. 3
Co-occurrence network of the microbiota in kiwifruit. The correlation of tissues in the amplicon sequence variant (ASV) table was analyzed using Sparse correlations for compositional data (SparCC). Blue lines indicate a positive relationship, while red lines indicate a negative relationship. Each dot represents an ASVs, with dot size increasing as centrality increases. Two centrality measures were used to construct, the co-occurrence network: (A) degree centrality, and (B) eigenvector centrality.

Table 1
ASV number present in the rhizosphere of Deliwoong according to eigenvector centrality (>0.6)
Table 2
ASV number present in the rhizosphere of cv. Sweetgold according to eigenvector centrality (>0.6)
Table 3
ASV number present in the endosphere of cv. Deliwoong according to eigenvector centrality (>0.6)
Table 4
ASV number present in the endosphere of cv. Sweetgold according to eigenvector centrality (>0.6)
References
Abdelfattah, A., Ruano-Rosa, D., Cacciola, S. O., Li Destri Nicosia, M. G and Schena, L. 2018. Impact of Bactrocera oleae on the fungal microbiota of ripe olive drupes. PLoS ONE 13:e0199403.



Abdelfattah, A., Tack, A. J. M., Wasserman, B., Liu, J., Berg, G., Norelli, J., Droby, S. and Wisniewski, M. 2022. Evidence for host-microbiome co-evolution in apple. New Phytol 234:2088-2100.



Abdelfattah, A., Wisniewski, M., Schena, L. and Tack, A. J. M 2021. Experimental evidence of microbial inheritance in plants and transmission routes from seed to phyllosphere and root. Environ. Microbiol 23:2199-2214.



Adeleke, B. S and Babalola, O. O 2021. Roles of plant endosphere microbes in agriculture: a review. J. Plant Growth Regul 41:1411-1428.


Berendsen, R. L., Pieterse, C. M. J and Bakker, P. A. H. M 2012. The rhizosphere microbiome and plant health. Trends Plant Sci 17:478-486.


Bettenfeld, P., Canals, J. C., Jacquens, L., Fernandez, O., Fontaine, F., van Schaik, E., Courty, P.-E and Trouvelot, S. 2022. The microbiota of the grapevine holobiont: a key component of plant health. J. Adv. Res 40:1-15.


Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A and Holmes, S. P 2015. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13:581-583.


Chou, M.-Y., Vanden Heuvel, J., Bell, T. H., Panke-Buisse, K. and Kao-Kniffin, J. 2018. Vineyard under-vine floor management alters soil microbial composition, while the fruit microbiome shows no corresponding shifts. Sci. Rep 8:11039.




Dinno, A. and Dinno, M. A 2017. Conover-Iman test of multiple comparisons using rank sums. R Foundation for Statistical Computing, Vienna, Austria.
Diskin, S., Feygenberg, O., Maurer, D., Droby, S., Prusky, D. and Alkan, N. 2017. Microbiome alterations are correlated with occurrence of postharvest stem-end rot in mango fruit. Phytobiomes 1:117-127.

Friedman, J. and Alm, E. J 2012. Inferring correlation networks from genomic survey data. PLoS Comput. Biol 8:e1002687.



Graham, J., Marshall, B. and Squire, G. R 2003. Genetic differentiation over a spatial environmental gradient in wild Rubus ideaus populations. New Phytol 157:667-675.



Grassi, R., Stefani, S. and Torriero, A. 2007. Some new results on the eigenvector centrality. J. Math. Sociol 31:237-248.

Hardoim, P. R., Van Overbeek, L. S., Berg, G., Pirttilä, A. M., Compant, S., Campisano, A., Döring, M. and Sessitsch, A. 2015. The hidden world within plants: ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol. Mol. Biol. Rev 79:293-320.




He, X. and Meghanathan, N. 2016. Correlation of eigenvector centrality to other centrality measures: random, small-world and real-world networks. In: Proceedings of the 8th International Conference on Networks and Communications (NeCoM), CSITEC -2016, eds. by N Meghanathan and J Zizka, pp. 09-18. AIRCC Publishing Corporation, Chennai, India.

Heatherbell, D. A., Struebi, P., Eschenbruch, R. and Withy, L. M 1980. A new fruit wine from kiwifruit: a wine of unusual composition and Riesling Sylvaner character. Am. J. Enol. Vitic 31:114-121.

Kaul, S., Choudhary, M., Gupta, S. and Dhar, M. K 2021. Engineering host microbiome for crop improvement and sustainable agriculture. Front. Microbiol 12:635917.



Kim, D.-R., Cho, G., Jeon, C.-W., Weller, D. M., Thomashow, L. S., Paulitz, T. C and Kwak, Y.-S 2019. A mutualistic interaction between Streptomyces bacteria, strawberry plants and pollinating bees. Nat. Commun 10:4802.




Kim, S.-H., Kim, D.-R and Kwak, Y.-S 2022. Characteristics of Streptomyces venezuelae 1-1 9D strain against kiwifruit bacterial canker pathogen. Korean J. Pestic. Sci 26:9-15.

Kruskal, W. H and Wallis, W. A 1952. Use of ranks in one-criterion variance analysis. . J. Am. Stat. Assoc 47:583-621.

Kwon, J.-H., Cheon, M.-G., Kim, J. and Kwack, Y.-B 2011. Black rot of kiwifruit caused by Alternaria alternata in Korea. Plant Pathol. J 27:298.

McCann, H. C., Rikkerink, E. H. A., Bertels, F., Fiers, M., Lu, A., Rees-George, J., Anderson, M. T., Gleave, A. P., Haubold, B., Wohlers, M. W., Guttman, D. S., Wang, P. W., Straub, C., Vanneste, J., Rainey, P. B and Templeton, M. D 2013. Genomic analysis of the kiwifruit pathogen Pseudomonas syringae pv. actinidiae provides insight into the origins of an emergent plant disease. PLoS Pathog 9:e1003503.



Mendes, R., Garbeva, P. and Raaijmakers, J. M 2013. The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev 37:634-663.


Michailides, T. J and Elmer, P. A. G 2000. Botrytis gray mold of kiwifruit caused by Botrytis cinerea in the United States and New Zealand. Plant Dis 84:208-223.


Murali, A., Bhargava, A. and Wright, E. S 2018. IDTAXA: a novel approach for accurate taxonomic classification of microbiome sequences. Microbiome 6:140.




Qu, Q., Zhang, Z., Peijnenburg, W. J. G. M., Liu, W., Lu, T., Hu, B., Chen, J., Chen, J., Lin, Z. and Qian, H. 2020. Rhizosphere microbiome assembly and its impact on plant growth. J. Agric. Food Chem 68:5024-5038.


Richardson, D. P., Ansell, J. and Drummond, L. N 2018. The nutritional and health attributes of kiwifruit: a review. Eur. J. Nutr 57:2659-2676.




Valente, T. W., Coronges, K., Lakon, C. and Costenbader, E. 2008. How correlated are network centrality measures? Connect 28:16-26.
- TOOLS
-
METRICS

- ORCID iDs
-
Youn-Sig Kwak

https://orcid.org/0000-0003-2139-1808 - Related articles



 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Supplement1
Supplement1 Print
Print



