Identification and Characterization of Diplodia parva and Diplodia crataegicola Causing Black Rot of Chinese Quince
Article information
Abstract
Fungal isolates from infected Chinese quince trees were found to cause black rot in Yeongcheon, Gyeongsangbuk Province, Korea. The quince leaves withered and turned reddish-brown and fruits underwent black mummification. To elucidate the cause of these symptoms, the pathogen was isolated from infected leaf and fruit tissues on potato dextrose agar and Levan media. Several fungal colonies forming a fluffy white or dark gray mycelium and two types of fungi forming an aerial white mycelium, growing widely at the edges, were isolated. Microscopic observations, investigation of fungal growth characteristics on various media, and molecular identification using an internal transcribed spacer, β-tubulin, and translation elongation factor 1-α genes were performed. The fungal pathogens were identified as Diplodia parva and Diplodia crataegicola. Pathogenicity tests revealed that the pathogen-inoculated fruits exhibited a layered pattern, turning brown rotting; leaves showed circular brown necrotic lesions. The developed symptoms were similar to those observed in the field. Fungal pathogens were reisolated to fulfill Koch’s postulates. Apples were inoculated with fungal pathogens to investigate the host range. Strong pathogenicity was evident in the fruits, with browning and rotting symptoms 3 days after inoculation. To determine pathogen control, a fungicidal sensitivity test was conducted using four registered fungicides. Thiophanate-methyl, propineb, and tebuconazole inhibited the mycelial growth of pathogens. To the best of our knowledge, this is the first report on the isolation and identification of the fungal pathogens D. parva and D. crataegicola from infected fruits and leaves of Chinese quince, causing black rot disease in Korea.
The Chinese quince (Pseudocydonia sinensis), belonging to the family Rosaceae, is a tree commonly found in all regions of South Korea and other Asian countries. Ripened yellow fruits contain medicinally active compounds (Hamauzu et al., 2005). The fruit is consumed as tea and candies and is used in traditional medicine for treating asthma, cough, influenza, harsh throat, and tuberculosis, as well as liver protection (Chun et al., 2012). Ripened yellow fruits have been used in traditional therapy for respiratory ailments and as an additive in health products (Sawai et al., 2008). Quince pomes have a hard flesh with a highly acidic flavor and are astringent and sour when eaten raw (Alesiani et al., 2010). Furthermore, quince is used to make jams, marmalades, fresh fruit compotes, jellies, dried slices, and wines (Laureiro et al., 2009). Owing to its aromatic and functional properties, quince is used to fortify various products, such as beer and yogurt. In addition, quince seed mucilage and hydrocolloids can be used as thickeners and bulking agents in several food products, making them a possible choice in the food industry (Hanan et al., 2020). Despite these health benefits, fruits can be infected by several bacterial and fungal diseases. Bacterial diseases include fire blight caused by Erwinia amylovora (Myung et al., 2016), and fungal diseases include fruit canker caused by Nothophoma quercina (Kazerooni et al., 2021), brown spot infection caused by Didymosphaeria rubi-ulmifolii (Kazerooni et al., 2020), and leaf spot caused by Botrytis cinerea (Chen et al., 2019).
Chinese quince trees are often subjected to various pathogenic attacks during cultivation. Among them, Diplodia species, which cause black rot in Chinese quince, has become an important disease that reduction of crop production. The symptoms of black rot in Chinese quince appear as yellow halos around brown irregular spots on the leaves, and later, reddish-brown lesions spread on the entire leaf, which then withers. On the fruit, the symptoms start from the calyx as a black lesion and later the entire fruit becomes black as the symptoms enlarge.
Mierina et al. (2011) found that compared to other pome fruits, quince fruit was a major source of minerals. Baranowska-Bosiacka et al. (2017) reported that the quince fruit is rich in valuable nutrients, ascorbic acid, phenols, and fiber, and has low oxalate content. Quince fruits are considered a prospective source for use in medicine, cosmetics, and the food industry (Banaś and Korus, 2016; Nahorska et al., 2014). Norin and Rumpunen (2003) concluded that significant pest damage does not occur in quince; however, if the areas under quince cultivation increase, some fungi common to other crops from the family Rosaceae could attack the Japanese quince. Fungal diseases dominate Chaenomeles spp. and may cause fruit spots, fruit rot, and dieback of shoots and plants (Norin and Rumpunen, 2003). Among several Diplodia species, Diplodia seriata is a cosmopolitan and plurivorous fungal species occurring on woody hosts belonging to many plant genera and families (Phillips et al., 2007; Slippers and Wingfield, 2007). The fungus is encountered in many habitats but has a primarily temperate distribution and is present on most continents. Additionally, D. seriata causes canker, dieback, fruit rot, and leaf spot diseases in economically important crops (Farr and Rossman, 2020). Reports on the virulence of this pathogen vary depending on the crop, varieties, and hosts involved and are often regarded as a stress-related pathogen that takes advantage of weak or stressed plants. Similar to other members of the Botryosphaeriaceae, D. seriata can live inside plants (Crous et al., 2006; Slippers and Wingfield, 2007), and latent infections of fruits can result in storage rot. The pathogen is dispersed through both pycnidia and ascospores, and conidia are regarded as the most important inoculum source for short-distance spread.
The present study is the first to report the association between Diplodia species and black-rot symptoms in the fruits and leaves of Chinese quince in Korea. The objective of this study was to isolate and identify Diplodia species that cause black rot in quince obtained from disease-infected Chinese quince trees in Gyeongbuk Province, Korea. The results of this study can help further elucidate the etiology and epidemiology of this disease.
Materials and Methods
Sample collection and pathogen isolation
In July 2021, while investigating disease symptoms in quince orchard trees in Yeongcheon, Gyeongsangbuk Province, Korea, nearly 20 trees were found to be infected severely in a village with a disease incidence of 80–90%. The disease symptoms on quince were observed as withering of leaves with a reddish-brown color, forming a brown circular layer pattern, and black mummification. To determine the cause, leaves and fruits showing symptoms were collected as disease-infected samples from quince trees. Symptomatic tissues were cut into squares using a sterilized scalpel, surface-sterilized by immersion in 1% NaOCl solution for 1 min and rinsed 2–3 times in sterilized distilled water (SDW), and then dried on a sterilized filter paper. Thus, surface-sterilized samples were placed onto potato dextrose agar (PDA) plates and incubated at 25–28°C for 3 days. The fungal colonies were collected and transferred to freshly prepared PDA plates to obtain purified fungal colonies. The isolates were stored at −80°C in 20% glycerol solution for the long term.
DNA extraction and polymerase chain reaction amplification
The selected pathogenic isolates, such as GYUN-10736 and GYUN-10738 were subjected to molecular identification based on the sequence homology of internal transcribed spacer (ITS), β-tubulin (TUB2), and translation elongation factor 1-α (TEF1-α) genes. For the molecular identification of isolates obtained from quince, DNA extraction, and polymerase chain reaction (PCR) was performed using ITS, TUB2, and TEF1-α gene sequences. The genomic DNA of all isolates was extracted using a HiGeneTM Genomic DNA Prep Kit (Biofact Co., Seoul, Korea) following the manufacturer’s instructions. The ITS, TUB2, and TEF1-α genes were amplified by PCR using Taq DNA polymerase and the primers listed in Table 1. The PCR was performed in a total reaction volume of 25 μl containing 1 μl template DNA, 1 μl forward primer, 1 μl reverse primer, 16 μl SDW, 0.5 μl Taq DNA polymerase (Biofact Co.), 0.5 μl dNTP mix, and 5 μl of 10× Taq Reaction buffer (25 mM MgCl2 mixed). The thermal cycling conditions were as follows: denaturation at 95°C for 2 min followed by 35 cycles at 95°C for 30 s, annealing at 52°C for 40 s and an extension at 72°C for 1 min. At the end of the cycle, the reaction mixture was held at 72°C for 10 min and then cooled to 4°C. The amplified PCR product was electrophoresed on 1% agarose gel to confirm the band and the PCR product was purified using a PCR purification kit (Biofact Co.). The PCR product obtained was sequenced by an automated sequencer (Genetic Analyzer 3130, Applied Biosystems, Carlsbad, CA, USA). This sequencing was performed by a company (Solgent, Daejeon, Korea). The same primers as above were used for this step. The sequences were compared with the homologs of the strains contained in the genomic database using the NCBI BLAST tool. Sequence alignment and phylogenetic tree construction were performed using the DNA Star software (version 5.02, DNAStar Inc., Madison, WI, USA) and MEGA 4.0, respectively (Biodesign Institute, Tempe, AZ, USA).
Phylogenetic analysis
Partial gene coding sequences of ITS, TUB2, and TEF1-α, were amplified using the primers listed in Table 1 (Hlaiem et al., 2020). The PCR was performed in a total reaction volume of 20 μl containing Taq reaction buffer 5 μl, 1.0 μl of template DNA, 1.0 μl of each primer (100 pmol), 1.0 μl of each deoxynucleotide triphosphate (200 μM), 2.0 mM MgCl2, and 1.0 U of Taq DNA polymerase (Bioneer Inc., Daejeon, Korea). PCR amplifications were performed with an initial denaturation at 95°C for 2 min; 35 cycles of denaturation at 95°C for 30 s, annealing at 54°C, 55°C, and 52°C for ITS, TUB2, and TEF1-α, respectively for 30 s, and extension at 72°C for 1 min; and a final extension at 72°C for 10 min. PCR products were purified and sequenced. The forward and reverse sequences of each nucleotide were combined using a SeqMan Pro version 7.1.0 (44.1) of the DNASTAR laser gene software (DNAStar Inc.). To analyze the molecular phylogenetic relationship, ITS, TUB2, and TEF1-α gene sequences of the isolates were compared with those of other Diplodia species collected from the NCBI GenBank database. The phylogenetic analyses inferred the data were constructed using MEGA-X software version 10.1.8 (Kumar et al., 2018).
Media optimization for the development of fungal pathogenic mycelia
To determine the effect of solid media on the growth and morphological characteristics of the isolated fungal pathogens, the selected fungal pathogenic isolates (GYUN-10736 and GYUN-10738) were inoculated on different solid media, including malt extract agar (MEA; 12.75 g, dextrin 2.75 g, peptone 0.78 g, agar 15 g, distilled water 1,000 ml), V8 agar (V8 juice 200 ml, CaCO3 6 g, distilled water 1,000 ml), and PDA plates. The plates were incubated at various temperatures (20, 25, 30, and 35°C) for 5–7 days. The colony diameters were measured at daily intervals (mm/day) at each temperature. The experiment was repeated at least once and each treatment consisted of three replicates (Petri dishes).
Morphological identification of GYUN-10736 and GYUN-10738
Morphological characterization of the fungal pathogenic mycelia was performed based on conidial characteristics as described by Phillips et al. (2013). After mycelia were fully grown on PDA plates, the formation of pycnidium was induced by incubation at 28°C under a 12/12 photoperiod for 3 weeks. Colony diameters were measured along two perpendicular axes when colonies covered almost the entire diameter of the plate, and the data were converted to daily radial growth (mm/day). For microscopic analysis, pycnidia were cut and mounted in 100% lactic acid, and the morphological characteristics of the conidia and conidiogenesis were observed under a ProgRes SpeedXT core3 Imager microscope. Images were captured using an AxioCam MRc5 camera (Zeiss, Oberkochen, Germany) and measurements were performed using AxioVision 4.6. Conidial dimensions are presented as the mean values of a minimum of 50 conidia, with extreme values in parentheses. At least 20 measurements were performed on the other structures.
Carbon utilization measurement using the Biolog FF MicroPlate
To analyze carbon source utilization by the isolated fungal pathogens, a Biolog FF Microplate system was used. The Biolog FF MicroPlate database allows rapid identification of filamentous fungi, such as Aspergillus, Penicillium, and yeast based on the utilization profile of 95 carbon sources. Color development and turbidity of each well were measured for quick and easy identification. The mycelia of the isolates cultured on PDA plates at 25°C for 7 days were collected using sterile cotton swabs. The collected mycelia were placed in a tube containing the IF solution and ground well, and the absorbance of the mycelium suspensions was adjusted to 72–75 using a turbidity meter at 490 nm. Then, 100 μl of the mycelial suspension was dispensed to each well of the Biolog FF MicroPlate. Biolog FF MicroPlates were incubated at 25°C for 96 h and then measured at 490 nm (mitochondrial activity) and 750 mm (mycelial growth) using a MicroLog Automated MicroStation System.
Pathogenicity test for quince fruits and leaves
Healthy quince fruits of similar size were selected for the pathogenicity test. The fruits were surface-sterilized with 70% ethanol followed by 2% NaOCl for 5 min, rinsed 2–3 times with SDW, and air-dried. The surface-sterilized fruits were wounded using a sterile needle. Mycelial plugs (5 mm in diameter) from the pathogenic fungal isolates (D. crataegicola GYUN-10738 and D. parva GYUN-10736) cultured on PDA plates for 5 days were inoculated onto the wounded or non-wounded sites of fruits. Fruits inoculated with only PDA plugs served as the untreated control group. Similar symptoms were observed in apples during our field survey. For the pathogenicity test on quince leaves, healthy leaves from a 3-year-old tree were detached and surface-disinfected with 70% ethanol for 30 s, followed by rinsing twice with SDW and then drying. Leaves were wounded at four sites using a sterile needle. Mycelial plugs (5 mm) of the two pathogens were inoculated onto wounded or non-wounded sites on the leaves. Leaves inoculated with PDA plugs alone served as the untreated control group. All inoculated leaves were placed on a square plate (40 × 40 cm) containing moist paper to maintain 70–80% relative humidity (RH). The disease lesion area (in diameter) was recorded 7 days after incubating at 25°C and compared with the untreated control group.
Pathogenicity test for apples
For the pathogenicity test on apples, the surface-sterilized apple fruits were wounded as were the quince fruits. Mycelial plugs (5 mm diameter) of the two pathogens were inoculated onto the wounded or non-wounded sites of the apple, as was done for the quince fruits. Apples inoculated with PDA plugs alone served as the untreated control group. The diameter of the lesion area was recorded 7 days after incubation at 25°C under moist conditions with 70–80% RH. The results were compared with those of the untreated control group. The experiment was performed twice with three replicates, and each replicate consisted of six fruits.
Fungicide sensitivity test on the fungal pathogen growth
The two fungal pathogens, D. parva GYUN-10736 and D. crataegicola GYUN-10738, isolated from quince fruits were tested for fungicide sensitivity following the method described by Arrigoni et al. (2019). The four fungicides used in our study were thiophanate-methyl, propineb, tebuconazole, and pyraclostrobin. These fungicides are available in the market with commercial-grade names, such as topsin M, anthracol, silvaco, and cabrio, respectively. They were dissolved in SDW at various concentrations as follows: thiophanate-methyl (350, 700, and 1,400 μg/ml), propineb (700, 1,400, and 2,800 μg/ml), tebuconazole (125, 250, and 500 μg/ml), and pyraclostrobin (28.625/57.25/114.5 μg/ml), and mixed in autoclaved PDA (cooled to approximately 50°C). Before solidification, the fungicide-containing medium was poured onto the Petri plates. Control plates contained PDA without fungicides. A mycelium plug (5 mm diameter) of the two fungal pathogens cultured for 7 days was placed at the center of PDA plates with or without fungicides, and the plates were incubated at 25°C for 7 days. Mycelial growth was observed and compared with that of mycelia grown on PDA plates without fungicides. When the colonies on the control plates (without fungicide) covered 75% of the agar surface, the diameters of all colonies of the microorganism were measured. The resistance or sensitivity of the two pathogens to the fungicides was determined based on their inhibition zones. The experiment was performed twice with three replicates per treatment. The percentage (%) inhibition of mycelial growth was calculated as follows:
Statistical analysis
All data were analyzed for variance using the SAS software (1989, SAS Institute Inc., Cary, NC, USA). Significant differences between the treatment means were determined at a significance level of at P < 0.05. The data for each experiment were analyzed separately. All experiments were repeated at least once.
Results
Observation of black rot on Chinese quince in orchards in Korea
Black rot in Chinese quince plants affects both the fruits and leaves (Fig. 1). The symptoms of black rot in Chinese quince appeared as yellow halos around brown irregular spots on the leaves (Fig. 1A–C), and later, the lesions spread as reddish-brown areas on the entire leaf and the leaves withered (Fig. 1D). On the fruits, the symptoms started from the calyx as a black lesion (Fig. 1E) and later stage, the entire fruit became black as the symptoms enlarged (Fig. 1F).

Observation of symptoms on quince trees in Yeongcheon, Gyeongsangbuk Province. (A–D) Leaves exhibit round or irregular brown spots and yellowing symptoms around the spots that progress and merge with other lesions, causing the leaves to turn reddish-brown and die. (E) Black rot starts from the calyx of the fruit and enlarges as the disease progresses. (F) The quince fruit is blackened and mummified.
Isolation of pathogenic fungi from the diseased parts of quince trees
Typical Diplodia colonies were isolated from the diseased leaves and fruits of a 3-year-old Chinese quince tree in July 2021. On inoculating the infected fruits and leaf samples from Chinese quince trees collected from Yeongcheon, Gyeongsangbuk Province, Korea onto different solid media, the formation of fungal colonies with brown spots was observed. A total of 29 bacterial and fungal strains were isolated, of which 10 strains were identified as D. parva and four strains were identified as D. crataegicola (Supplementary Table 1). Among the several Diplodia species, only two strains, D. parva GYUN-10736, and D. crataegicola GYUN-10738 were used in further studies. These two pathogens were deposited at Korean Agricultural Culture Collection, Korea, with the names GYUN-10736 (KACC 410047) and GYUN-10738 (KACC 410049).
Molecular identification of fungal pathogens
The two isolates, GYUN-10736 and GYUN-10738, were further characterized by ITS, TUB2, and TEF1-α sequencing. The gene sequences of the ITS (GenBank accession no. ON705843), TUB2 (OL588334), and TEF1-α (OP700709) for the isolate GYUN-10736; and the gene sequences of the ITS (GenBank accession no. ON705781), TUB2 (OP700706), and TEF1-α (OP700708) for the isolate GYUN-10738 suggested that both isolates belonged to Diplodia genus and had the highest homology with D. parva GYUN-10736 and D. crataegicola GYUN-10738 (98%), respectively. Thus, both morphological and molecular characteristics confirmed that these species were D. parva and D. crataegicola (Fig. 2). The molecular phylogenetic relationship was analyzed by combining the analyzed nucleotide sequences of GYUN-10736 and GYUN-10738 with sequences of ITS, TUB2, and TEF1-α genes of other Diplodia species collected from GenBank. The combined dataset consisted of 1,093 sequences. Botryosphaeria dothidea CMW8000 was included as the outgroup.

The phylogenetic tree was constructed using a concatenated internal transcribed spacer (ITS), β-tubulin (TUB2), and translation elongation factor 1-α (TEF1-α) sequences of 21 isolates of Diplodia species from NCBI was inferred based on the maximum likelihood method using MEGA 7.0. The number below the branches indicates the percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (100 replicates). The scale bar indicates 0.02 substitutions per nucleotide position.
Pairwise sequence comparison of Diplodia spp
Sequence data were used to construct a phylogenetic tree and the same data were used for pairwise comparisons. Sequence similarity analysis revealed that GYUN-10736 had the highest similarity with D. parva KNU16-007 (99.86%), followed by D. sapinea and D. intermedia (99.28% and 99.14%, respectively). GYUN-10738 showed the highest similarity to D. crataegicola MFLU 15-1311 (99.71%), followed by D. intermedia and D. rosacearum (99.57%) (Fig. 3).
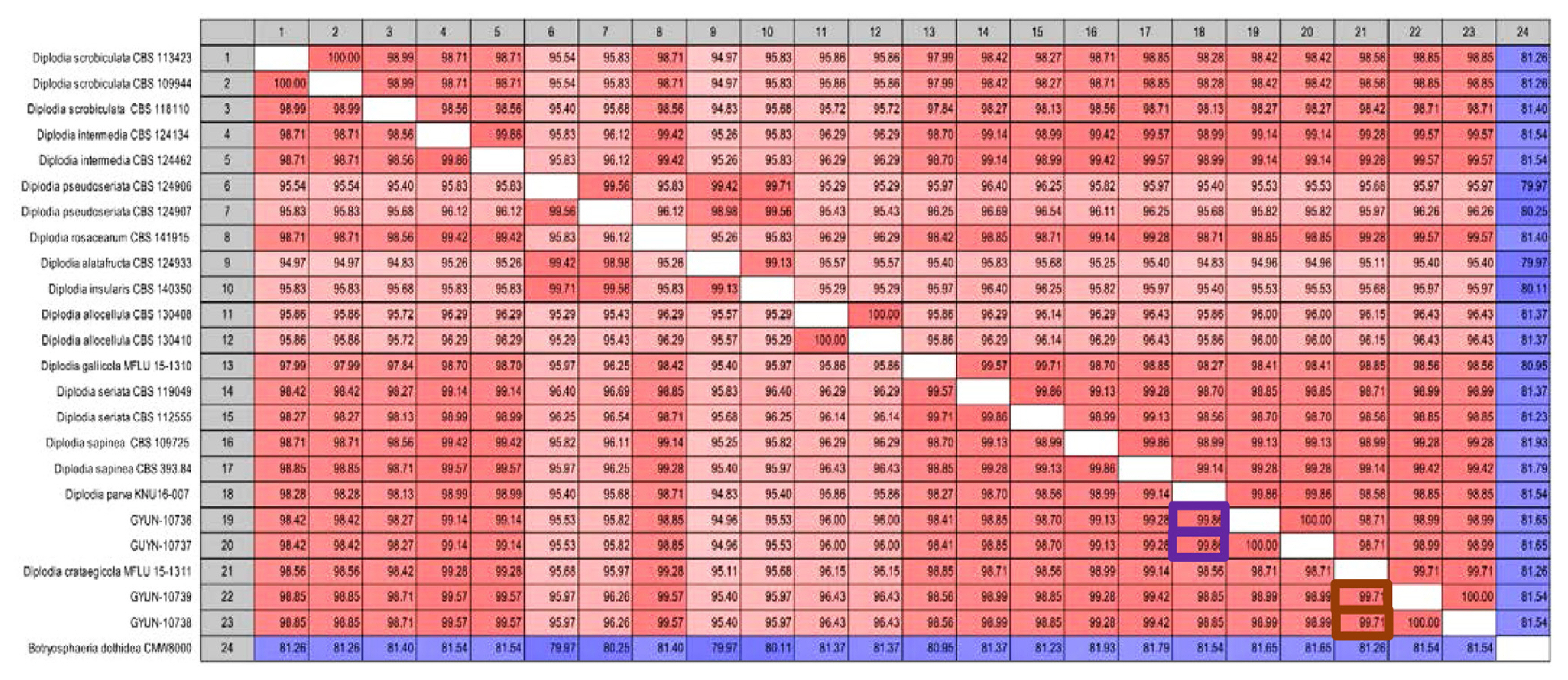
Pairwise identity matrix of all the pathogens based on internal transcribed spacer (ITS), β-tubulin (TUB2), and translation elongation factor 1-α (TEF1-α) gene sequences. Based on the ITS, TUB2, and TEF1-α gene sequence binding data, the nucleotide sequence similarity of GYUN-10736, GYUN-10738 and other Diplodia species registered with NCBI was analyzed. The similarity between GYUN-10736 and Diplodia parva was highest at 99.71%, and GYUN-10738 showed a similarity of 99.84% with Diplodia crataegicola. Red color highlights the isolate pairs with relatively high similarity and white color indicates isolate pairs with low similarity.
Media optimization and temperature conditions for growth of pathogenic fungi
Colony diameters on different media were measured to determine the growth rates of the two fungal pathogens, GYUN-10736 and GYUN-10738. When these two isolates were cultured on different media (V8, MEA, and PDA) under various incubation temperatures (20, 25, 30, and 35°C), the mycelial growth of GYUN-10736 was observed to be greater at 25°C than at other temperatures (Fig. 4A). The mycelial growth diameter was observed as 85, 70, and 53 mm in V8, MEA, and PDA media, respectively, at 25°C. At 30°C and 35°C, the colony grew very slowly or did not grow at all. The fungal colony appeared as a dark gray colony on the V8 medium and a wrinkled white downy colony on PDA and MEA medium; later, it became dark gray (Fig. 4A). GYUN-10738 also showed a rapid growth rate in the V8 medium at 20°C, but in the case of MEA and PDA medium, the diameter was 77.3 and 60 mm, respectively, and the growth rate was faster at 20°C than at 25°C (Fig. 4B). The fungal colonies appeared as dark gray or black colonies with white aerial mycelia on V8 medium and white colonies with irregular and wrinkled shapes on MEA and PDA media, which turned dark gray from the edge. The conidia of GYUN-10736 and GYUN-10738 were not well formed at different incubation temperatures or in the three media types (Fig. 4B).
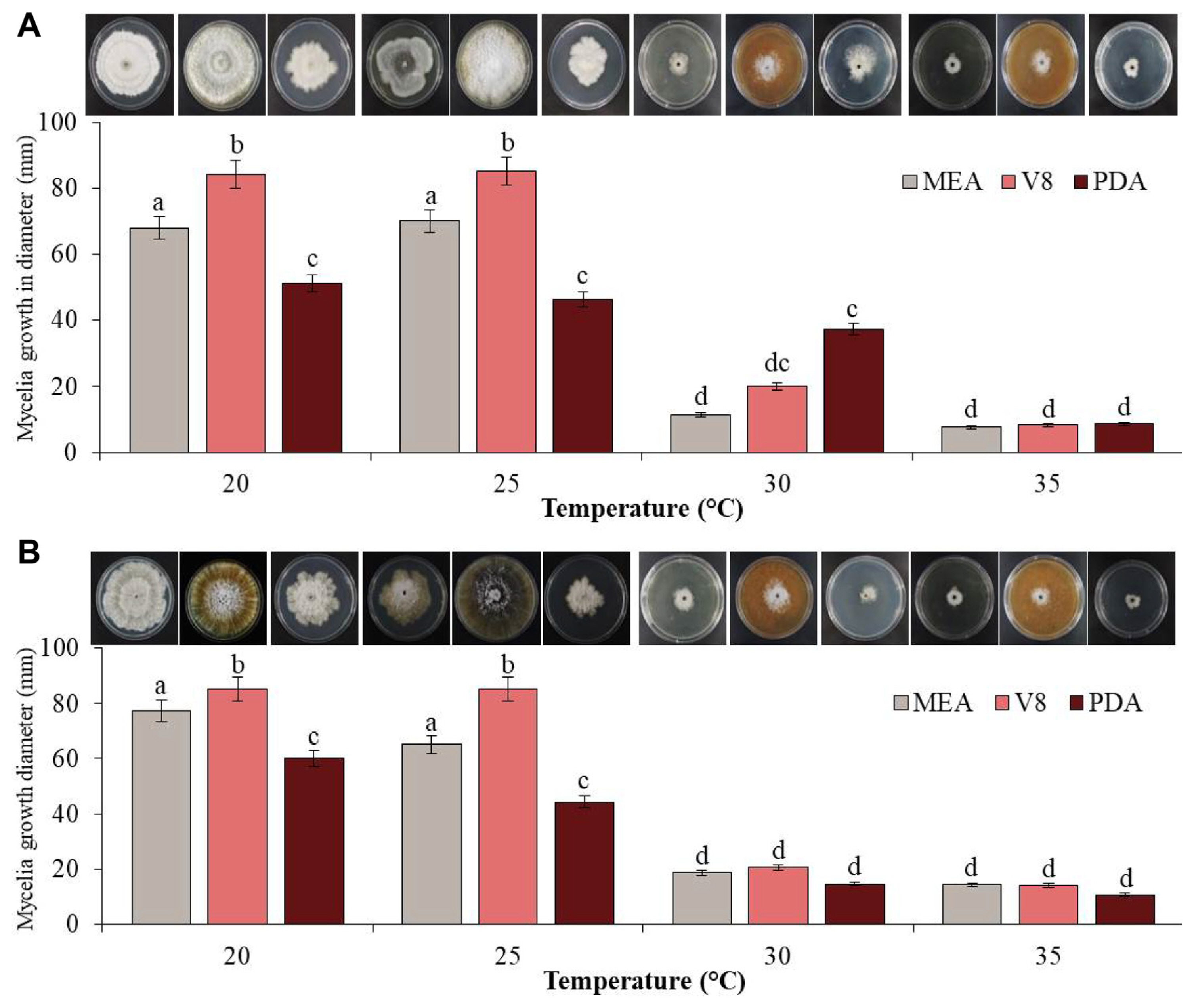
Effect of various media and temperature conditions on the mycelial growth of fungal pathogens GYUN-10736 (A) and GYUN-10738 (B) in vitro. The experiment was repeated at least once with three replicates per treatment. Bars with the same letters do not differ from each other according to the least significant difference at P < 0.05. Color and diameter of the mycelial colony were observed 7 days after incubation at various temperatures. MEA, malt extract agar; PDA, potato dextrose agar.
Morphological characteristics, microscopic observations, and Biolog analysis
The morphological characteristics of the two fungal pathogens (D. parva GYUN-10736 and D. crataegicola GYUN-10738) were observed and compared in PDA. Conidia of each strain were observed under a microscope (Table 2). Pycnidia were formed on the surface of 2% water agar medium and cultured at 25°C for 3 weeks. The conidia of D. parva GYUN-10736 were yellow or light yellow, elongated oval, without a septum, and with a blunt tip (Fig. 5A). The size of conidia was 18.22–27.17 × 8.25–11 (length × width) μm. D. crataegicola GYUN-10738 had yellow or brown circular conidia with no septa. The size of conidia was 12.28–17.55 × 8.59–12.34 (length × width) μm and oval or transparent spores were observed (Table 2, Fig. 5B).

Morphological characteristics of Diplodia parva GYUN-10736 and Diplodia crataegicola GYUN-10738 and compared with domestic and foreign strains

(A) Morphological characteristics and microscopic observations of fungal pathogen Diplodia parva GYUN-10736. a, culture growing on potato dextrose agar (PDA); b, pycnidia developing in culture (scale bar = 500 μm); c, d, conidiogenous cells with developing conidia (scale bars = 10 μm); e, f, the conidia are yellow or brown oval without a septum with blunt tip. Conidia were 18.22–27.17 × 8.25–11 (length × width) μm in size (scale bars = 10 μm). (B) Morphological characteristics and microscopic observations of fungal pathogen Diplodia crataegicola GYUN-10738. a, culture growing on PDA; b, pycnidia developing needles in culture (scale bar = 500 μm); c, d, conidiogenous cells with developing conidia (scale bars = 10 μm); e, conidia are yellow or brown oval without a septum with blunt tip (scale bar = 10 μm); f, conidia later become dark brown oval and were 12.28–17.55 × 8.59–12.34 (length × width) μm in size (scale bar = 5 μm).
The Biolog results for the two fungal pathogens were determined based on the utilization of 95 carbon sources (Table 3). D. parva GYUN-10736 responded positively to 90 carbon sources. Three carbon sources, such as L-fucose, β-hydroxy-butyric acid, and L-lactic acid were not utilized. In the case of the other fungal pathogen D. crataegicola GYUN-10738, nearly 16 carbon sources were not utilized. A comparison of these traits with those in the Biolog database revealed that these strains had a match probability of 94% with the above two pathogens.
Pathogenicity test on quince fruits, quince leaves, and apples
The pathogenicity of two pathogens was tested on detached fruits and leaves of a 3-year-old quince tree (Fig. 6). Disease symptoms were observed in quince fruits 7 days after inoculation with the two pathogens. Diseased lesions appear as circular brown or dark brown spots. The diseased lesions progressed by 7 and 4 mm/day in wounded fruits inoculated with GYUN-10738 and GYUN-10736, respectively (Fig. 6A), whereas the lesions were 4 and 2 mm in non-wounded fruits inoculated with GYUN-10738 and GYUN-10736, respectively. More diseased lesions were found on wounded fruits than in non-wounded fruits, whereas no disease symptoms were observed in non-inoculated fruits.
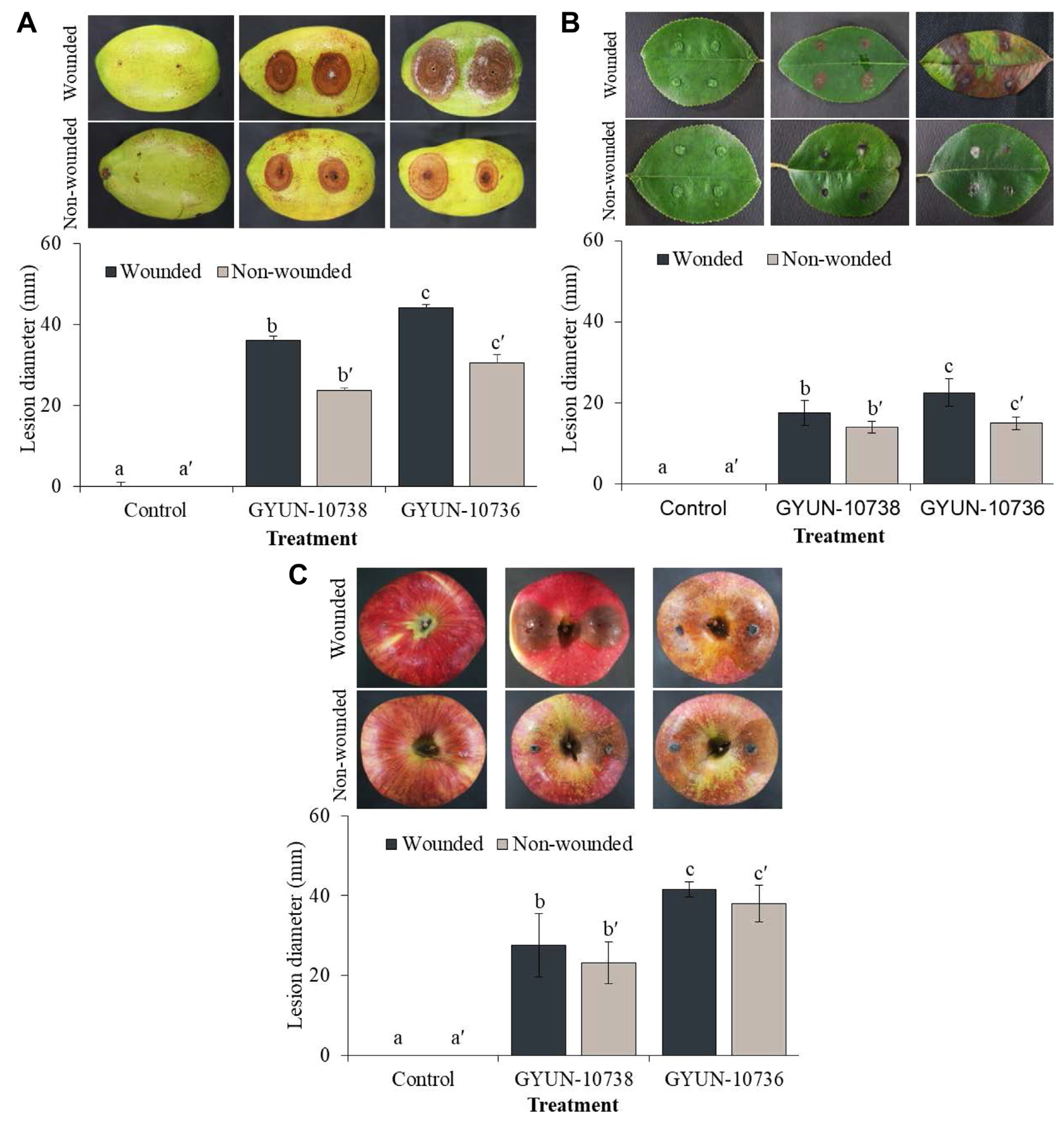
(A) Pathogenicity test on quince fruits. Immature quince fruits were artificially inoculated with mycelial plugs of two fungal pathogens on wounded and non-wounded fruits. The fruits inoculated with potato dextrose agar (PDA) plugs alone were considered as the non-treated control group. The disease symptoms were observed 7 days after incubation at 25°C. (B) Pathogenicity test on quince leaves. Healthy quince leaves after surface sterilization were artificially inoculated. Development of disease symptoms on wounded leaves one-week after incubation at 25°C with halos around the black spots, which later enlarged at 3 weeks. No disease symptoms were developed on non-inoculated leaves and non-wounded leaves. (C) Pathogenicity test on apples. Immature apples were artificially inoculated with mycelial plugs of two fungal pathogens on wounded and non-wounded fruits. The fruits inoculated with PDA plugs alone were considered as the non-treated control group. The disease symptoms were observed 7 days after incubation at 25°C. The experiment was repeated at least once with three replicates (fruits/leaves) per treatment. Bars with the same letters do not differ from each other according to the least significant difference at P < 0.05.
In the case of quince leaves, the symptoms initially appeared as yellow-brown spots, which later developed into circular lesions with yellowing of the lateral veins and appeared light brown in the center. In the later stages, the lesions merged, causing the leaves to wither to form reddish-brown and black pycnidium on the surface in the wounded leaves inoculated with GYUN-10738; however, such symptoms were not observed, except for small lesions on inoculation sites, in non-wounded leaves (Fig. 6B). Leaves inoculated with GYUN-10736 formed light brown spots initially during the 1st week and later started spreading across the leaf by the 2nd week. The leaves completely turned dark brown by the 3rd week in wounded leaves, whereas such symptoms were not observed, except for small lesions on inoculation sites, in non-wounded leaves, and no symptoms developed in non-inoculated wounded or non-wounded leaves.
In the case of pathogenicity on ripened apples; symptoms were observed on wounded apples 3 d after inoculation with mycelial plugs. The symptoms manifested as browning of the wounded fruits (Fig. 6C). The diseased lesions progressed to 41.5 mm and 27.5 mm in GYUN-10736- and GYUN-10738-inoculated apple fruits, respectively, on the 7th day. In contrast, in non-wounded apple fruits, 7 days after inoculation with pathogenic mycelial plugs, disease symptoms were also observed in apples inoculated with GYUN-10736 and GYUN-10738 fungal pathogens (Fig. 6C), whereas no disease symptoms were observed in non-inoculated apple fruits.
In vitro fungicide-sensitivity test of the two fungal pathogens
The two fungal pathogens, D. parva GYUN-10736 and D. crataegicola GYUN-10738 were tested for their sensitivity to chemical fungicides in vitro (Fig. 7). Among the four chemical fungicides tested, both fungal pathogens were sensitive to three fungicides (thiophanate-methyl, propineb, and tebuconazole) at 350 μg/ml, 700 μg/ml, and 125 μg/ml concentrations, respectively, and no mycelial growth was observed. However, minor mycelial growth of these two pathogens was observed when inoculated onto V8 agar plates supplemented with the chemical fungicide pyraclostrobin. The mycelial growth of pathogen GYUN-10736 was observed in approximately 35% of the plate and the mycelial growth of the fungal pathogen GYUN-10738 was observed in 30% of the plate. No further growth was observed at 28.625 μg/ml concentration of pyraclostrobin, whereas mycelial growth of the two fungal pathogens was observed in the non-treated control.
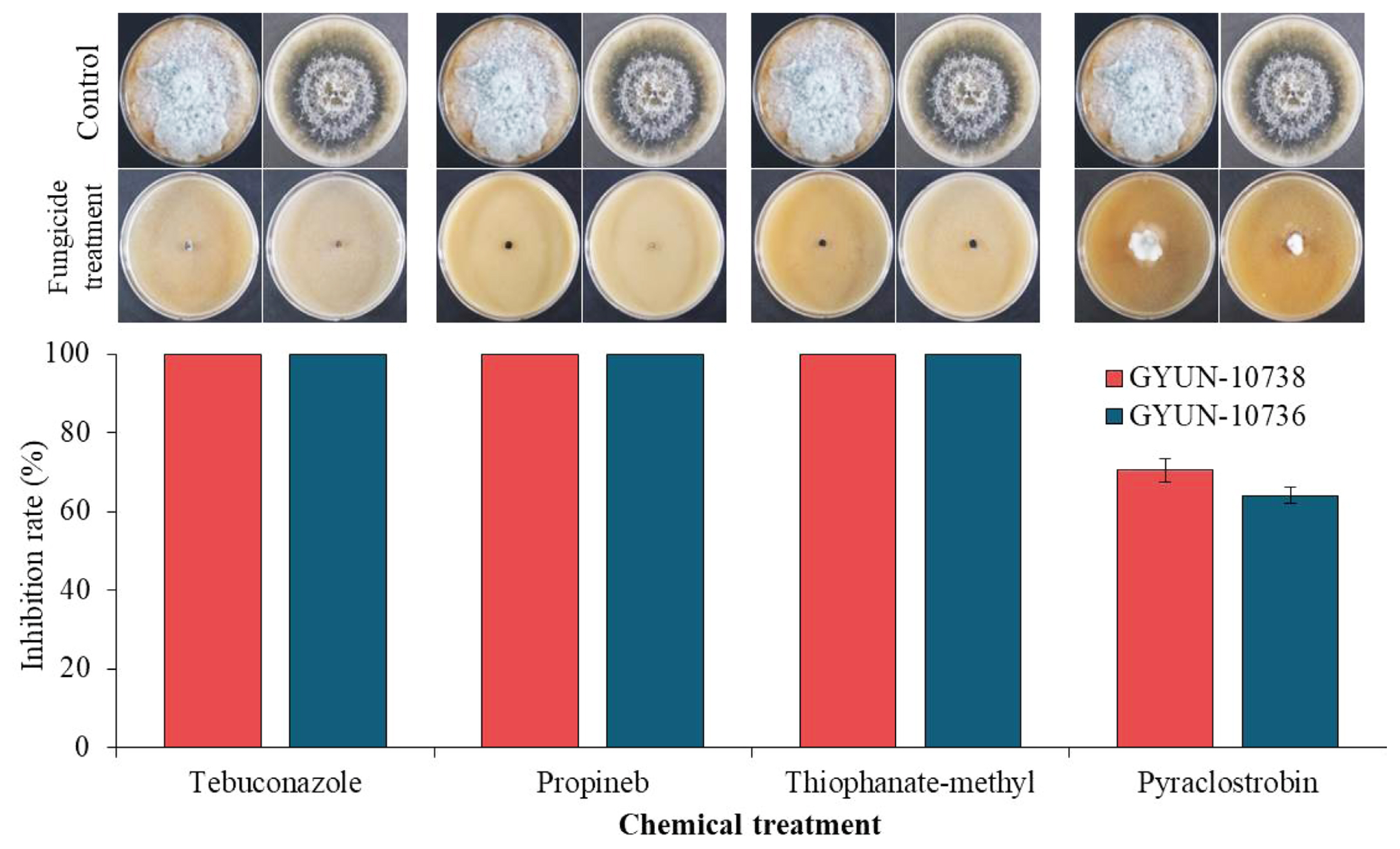
Fungicide-sensitivity to two fungal pathogens under in vitro conditions. Mycelial plugs of fungal pathogens were inoculated onto V8 agar plates supplemented with fungicides (pyraclostrobin, thiophanate-methyl, propineb, and tebuconazole) at a 0.5× concentration. A V8 agar plate without fungicide served the non-treated control. Mycelial growth was observed 7 days after incubation at 25°C and compared with the non-treated control. The experiment was performed twice with three replicates per treatment. Percentage (%) inhibition of mycelial growth rate was calculated.
Discussion
In this study, we identified and characterized two fungal pathogens that cause black rot of Chinese quince in South Korea. A survey of quince orchards showed yellow halos around brown irregular spots on the leaves, and later, reddish-brown lesions spread on the entire leaf, which then withered. On the fruit, symptoms started from the calyx with black lesions, and later, the entire fruit became black as the symptoms enlarged. Isolation of fungal pathogens from lesions and ITS, TUB2, and TEF1-α sequencing indicated that the isolates belonged to the black-rot pathogen Diplodia species, D. parva, and D. crataegicola. The molecular phylogenetic relationship was analyzed by combining the analyzed nucleotide sequences of GYUN-10736 and GYUN-10738 with the gene sequences of ITS, TUB2, and TEF1-α of other Diplodia species collected from GenBank. Diplodia species have been found in comparable numbers across several countries and are present in all forest inventory analyses (Adamson et al., 2021). Although observed in a wide range of locations, this pathogen showed low recovery rates across the region. When the recovered isolates were compared with those housed in various culture collections, the results revealed that many of the D. corticola isolates examined were not phylogenetically distinct based on the location or gene regions studied. Previous studies have indicated that D. corticola might have been inadvertently introduced into the United States from areas in Europe, where D. corticola has been associated with cork oak decline since 2000 (Lynch et al., 2010).
In a recent report, Vučković et al. (2022) stated that D. seriata is a fungal pathogen with a wide host range having more than 34 different hosts. In Serbia, it has been previously described in apple fruits and ornamental trees (Zlatković et al., 2016). D. seriata causes frog-eye leaf spots, canker, shoot dieback, and black rot in pome fruit (Phillips et al., 2007). It has been reported as a quince pathogen (Cydonia oblonga M.) in Canada, Greece, New Zealand, South Africa, and Spain (Farr and Rossman, 2017). The disease incidence has been reported to be low, but its symptoms are severe. The affected fruits exhibit large decayed brown areas with a concentric band towards the lesion margin. In 2021, the symptoms of black rot in apples (‘Fuji’) were observed in Yeongcheon, Korea. Fruit decay symptoms included purple pimple spots, black rot around the seed cavity (calyx end), and mummified fruits. Similarly, in a previous report, B. dothidea was reported causing Botryosphaeria canker in apple trees, whereas D. seriata was reported causing apple dieback in Canada and Uruguay (Delgado-Cerrone et al., 2016).
D. parva GYUN-10736 and D. crataegicola GYUN-10738 were characterized and identified as causing new diseases, in which the leaves of quince trees grown in Korea turned reddish-brown and the fruits turned black. Diplodia contains more than 1,000 species with a wide host range (Slippers et al., 2007). Diplodia species are difficult to identify morphologically because they have few interspecies morphological features. Therefore, these pathogens have long been identified and classified based on their association with host plants; as a result, many species of Diplodia are likely to be the same species (Phillips et al., 2012). With the development of molecular biology, Diplodia species have been classified according to differences in nucleotide sequences and morphological characteristics of the TUB2 and TEF1-α genes, including the ITS gene of the isolate (de Wet et al., 2003; Phillips et al., 2007). The present study isolated pathogens that cause new diseases in Chinese quince trees grown in Korea and performed phylogenetic analysis and morphological characterization based on nucleotide sequences of ITS, β-tubulin, and TEF1-α genes. The isolate GYUN-10738 was found to be similar to D. rosacearum and D. intermedia, but was most closely related to D. crataegicola, indicating that it was distinct from D. rosacearum, D. intermedia, and other Diplodia species (Dissanayake et al., 2016), whereas GYUN-10736 was found to show the closest relationship with D. parva, D. crataegicola, and D. galiicola (Lee et al., 2021).
The morphological characteristics of GYUN-10736 have been found to be similar to those of D. sapinea and D. intermedia, which are closely related phylogenetically. The length and width of the conidia were much shorter than those of the conidia described in a previous report (Lee et al., 2021). GYUN-10738 formed round or ovoid conidia, unlike D. rosacearum and D. intermedia, which have ovoid or oval conidia and are much shorter in length and width (Giambra et al., 2016; Phillips et al., 2012). It has been reported that the conidial length of all reported Diplodia species range from 21.5 to 52.5 μm and the width varies from 10 to 22 μm (Phillips et al., 2013). On the other hand, the conidia of D. crataegicola measures 11–16 × 6–10 (length × width) which clearly distinguishes this species from all other reported species (Ariyawansa et al., 2015). The utilization of carbon sources by these pathogens was determined using a Biolog FF MicroPlate. GYUN-10736 used 90 carbon sources, whereas GYUN-10738 did not use 18 carbon sources and used 76 carbon sources. The distinguishing of the two types of Diplodia based on their utilization of different carbon sources was confirmed.
Previously, Diplodia pseudoseriata was isolated from native Myrtaceae trees in Uruguay (Pérez et al., 2010), and D. alatafructa was isolated from Pterocarpus angolensis in South Africa (Mehl et al., 2011). In contrast, in another report by Phillips et al. (2012), the phylogeny constructed with the isolates of both species clustered in a single clade, suggesting that they represent a single phylogenetic species. Therefore, it seems that they should be regarded as synonyms, and given that D. pseudoseriata was the first name to be published, it takes priority over D. alatafructa. In the present study, the pathogenicity of GYUN-10736 and GYUN-10738 was assessed in quince and apple, which resulted in quince and apple fruits turning brown and rotting, and black pycnidium forming on the surface of the apple fruits. GYUN-10736 showed stronger pathogenicity than GYUN-10738 on both wounded and unwounded apples. According to Epstein et al. (2008), pathogenicity levels are dependent on the type of pathogen or strain and differ from host to host.
Currently, most fungal pathogens have developed resistance to various chemical fungicides because of their continuous application to crop plants (Kiiker et al., 2021; Kunova et al., 2021). D. citri and D. seriata, which are representative pathogens of Diplodia, are controlled by the application of fungicides, such as carbendazim, MBC, and QoI (Cerioni et al., 2017; Martínez-Diz et al., 2021). In our study, four fungicides were tested for the effective control of GYUN-10736 and GYUN-10738. The fungicides thiophanate-methyl, tebuconazole, and propineb completely inhibited the mycelial growth of two pathogens, whereas pyraclostrobin showed low control against D. parva GYUN-10736 and D. crataegicola GYUN-10738. To date, the resistance of various pathogens to pyraclostrobin fungicides has been reported (Fernández-Ortuño et al., 2012; He et al., 2021). However, this drawback is not limited to pyraclostrobin fungicides; resistance to pathogens, such as Colletotrichum gloeosporioides to thiophanate-methyl has also been reported (Ali et al., 2019; Isa and Kim, 2022). To solve this challenge, the continuous spraying of a particular fungicide system should be reduced and replaced instead with alternatively replacing the fungicides to establish a better systematic control system.
Notes
Conflicts of Interest
No potential conflict of interest relevant to this article was reported.
Acknowledgments
This work was supported by the Korea Institute of Planning and Evaluation for Technology in Food, Agriculture and Forestry (IPET) through the Technology Commercialization Support Program, funded by the Ministry of Agriculture, Food and Rural Affairs (MAFRA) (122039-02).
Electronic Supplementary Material
Supplementary materials are available at The Plant Pathology Journal website (http://www.ppjonline.org/).

