Re-identification of Colletotrichum acutatum Species Complex in Korea and Their Host Plants
Article information
Abstract
Colletotrichum acutatum species complex is one of the most important groups in the genus Colletotrichum with a high species diversity and a wide range of host plants. C. acutatum and related species have been collected from different plants and locations in Korea and deposited into the Korean Agricultural Culture Collection (KACC), National Institute of Agricultural Sciences since the 1990s. These fungal isolates were previously identified based mainly on morphological characteristics, and a limitation of molecular data was provided. To confirm the identification of species, 64 C. acutatum species complex isolates in KACC were used in this study for DNA sequence analyses of six loci: nuclear ribosomal internal transcribed spacers (ITS), beta-tubulin 2 (TUB2), histone-3 (HIS3), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), chitin synthase 1 (CHS-1), and actin (ACT). The molecular analysis revealed that they were identified in six different species of C. fioriniae (24 isolates), C. nymphaeae (21 isolates), C. scovillei (12 isolates), C. chrysanthemi (three isolates), C. lupini (two isolates), and C. godetiae (one isolate), and a novel species candidate. We compared the hosts of KACC isolates with “The List of Plant Diseases in Korea”, previous reports in Korea and global reports and found that 23 combinations between hosts and pathogens could be newly reported in Korea after pathogenicity tests, and 12 of these have not been recorded in the world.
The fungal family Glomerellaceae contains only one genus Colletotrichum which consists of many phytopathogenic species with a wide range of hosts. Colletotrichum species were commonly reported as causal agents of anthracnose diseases and infected economically important crops such as apple, strawberry, pepper, citrus, peach, mango, avocado, banana, coffee and cereals (Cannon et al., 2012; Crouch and Beirn, 2009; González et al., 2006; Kim et al., 2008; Lenné, 2002; Nguyen et al., 2010; Oo et al., 2018; Peres et al., 2008; Sanders and Korsten, 2003; Tan et al., 2022).
The identification of Colletotrichum species was primarily based on morphological characteristics with 11 species recognized by von Arx (1957), 22 species by Sutton (1980), and 39 species by Sutton (1992). Later, 66 species were accepted by Hyde et al. (2009) based on the morphology and/or molecular analysis. However, morphological characteristics and the lack of molecular data cannot be used to identify Colletotrichum species accurately (Cai et al., 2011; Sato and Moriwaki, 2013). Therefore, Jayawardena et al. (2016) divided the genus Colletotrichum (189 species) into 11 species complexes and 23 single species, then Liu et al. (2022) updated to 280 accepted species with 16 species complexes and 15 singleton species, based on multi-locus phylogeny. The nuclear ribosomal internal transcribed spacer (ITS) region was used to determine Colletotrichum species complexes (Cannon et al., 2012). While, most members of the C. acutatum, C. dematium, C. destructivum, C. orchidearum, C. spaethianum, C. dracaenophilum, C. magnum, and C. truncatum species complexes were identified at species level by the combination of six loci: ITS, beta-tubulin 2 (TUB2), histone-3 (HIS3), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), chitin synthase 1 (CHS-1), and actin (ACT) (Damm et al., 2009, 2012, 2014, 2019; Jayawardena et al., 2016; Liu et al., 2022).
Species in the C. acutatum species complex are known as destructive plant pathogens worldwide, including Korea (Baroncelli et al., 2015; Cho et al., 2021; Garrido et al., 2009; Kim et al., 2022; Kwon and Kim, 2011; Lee et al., 2007; Oh, 1995; Peres et al., 2008; Talhinhas et al., 2011). To date, this complex was one of the biggest groups in the genus Colletotrichum with 43 accepted species and contained species with a wide or narrow range of host plants (Jayawardena et al., 2021; Liu et al., 2022). The variation in the conidial shape of many species in the complex often led to incorrectly identify based on morphology. For example, 39 Colletotrichum isolates, morphologically identified as C. gloeosporioides or Glomerella cingulata, were re-identified as 14 species in or closely related to the C. acutatum species complex based on the multi-locus analysis (Damm et al., 2012).
Isolates of C. acutatum and related species have been deposited into the Korean Agricultural Culture Collection (KACC), National Institute of Agricultural Sciences since the 1990s. These species were originally identified mainly based on morphological characteristics. In earlier studies, several KACC isolates in the C. acutatum species complex were re-identified using a single locus or the multi-locus combination. Nevertheless, some of these were ambiguous species because of insufficient molecular data (Han et al., 2014; Kim and Kim, 2020; Kim et al., 2006, 2008, 2020; Noh et al., 2014; Park et al., 2020; Xu et al., 2018). Accurate species identification within C. acutatum species complex plays an important role to understand species diversity and the host plants of this complex in Korea. Hence, this study aims to: (1) re-identify the isolates of C. acutatum species complex in KACC using the combined analysis of six loci (ITS, TUB2, HIS3, GAPDH, CHS-1, and ACT); (2) rearrange combination between host plants and re-identified species under C. acutatum species complex in Korea.
Materials and Methods
Fungal isolates
Sixty-four cultures of Colletotrichum from many host plants and different locations in Korea have been deposited and preserved in liquid nitrogen at the KACC. Details of these isolates such as sources, locations, collected date and previous re-identifications were documented. Fresh cultures were recovered on potato dextrose agar (PDA) and used in this study.
DNA extraction, polymerase chain reaction amplification, and sequencing
Fungal mycelia were scraped from 7-day-old cultures on PDA plates. Around 50 mg of fresh mycelia was used for DNA extraction using the DNeasy plant mini kit (Qiagen, Hilden, Germany), following the manufacturer’s instructions. DNA templates were checked by a NanoDrop Spectrophotometer (Thermo Scientific, Waltham, MA, USA).
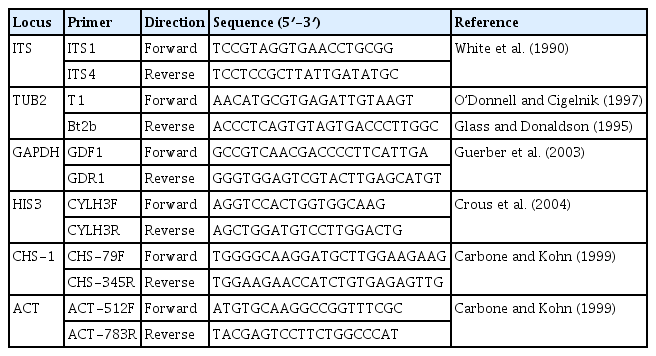
The primer pairs, including ITS1/ITS4 (White et al., 1990), T1/BT2b (Glass and Donaldson, 1995; O’Donnell and Cigelnik, 1997), GDF1/GDR1 (Guerber et al., 2003), CYLH3F/CYLH3R (Crous et al., 2004), CHS-79F/CHS-345R (Carbone and Kohn, 1999), and ACT-512F/ACT-783R (Carbone and Kohn, 1999), were used for the amplification of ITS, TUB2, GAPDH, HIS3, CHS-1, and ACT, respectively (Table 1). Each polymerase chain reaction (PCR) volume (25 μl) consisted of 12.5 μl MyTaq HS Mix, 1 μl (4.5 pMol) of each primer, 8.5 μl nuclease-free water and 2 μl DNA template (100 ng/μl). PCR reactions were performed in a MJ Research PTC-200 Thermal Cycler (MJ Research, Ramsey, MN, USA) with an initial denaturation step at 94°C for 5 min, followed by 30 cycles: denaturation at 94°C for 30 s; annealing at 58°C (ITS), 61°C (TUB2 and GAPDH) and 61.5°C (HIS3, CHS-1, and ACT) for 30 s, extension at 72°C for 1 min and final extension at 72°C for 10 min. PCR products were checked by gel electrophoresis before sending to the Macrogen (Seoul, Korea) for sequencing with the amplifying primer pairs.
Phylogenetic analysis
Raw sequences obtained in this study were assembled by MEGA 11 (Tamura et al., 2021) and deposited to RDA-GeneBank (http://genebank.rda.go.kr) with accession numbers in Table 2. The sequence datasets contained sequences of 64 KACC isolates, 43 reference species (Supplementary Table 1) in the C. acutatum species complex, and C. orchidophilum (outgroup) (Liu et al., 2022). The multiple sequence alignment of each locus was separately performed using the ClustalW program in MEGA 11 and concatenated afterward. A maximum likelihood (ML) phylogenetic tree of six loci was inferred using IQ-TREE with the best-fit model “TIM2+F+I+G4”, and 1,000 ultrafast bootstrap replicates. The phylogenetic tree was viewed in MEGA11 and depicted in Adobe Illustrator.
Results
Multi-locus phylogeny
The ITS, TUB2, GAPDH, HIS3, CHS-1, and ACT alignments contained 545, 497, 247, 378, 251, and 234 characters including gaps, respectively. Concatenated alignment of six loci included the members in the C. acutatum species complex in this study and references (43 accepted species, and C. orchidophilum as an outgroup). A combined phylogenetic tree (Fig. 1) showed that 64 KACC isolates were composed of six different species and a novel species candidate. Of these, 24 KACC isolates were grouped with the ex-type strain of C. fioriniae (CBS 128517), supported by a 100% ML bootstrap value. Twenty-one KACC isolates were clustered together with the ex-type strain of C. nymphaeae (CBS 515.78) with a 100% ML bootstrap value. Twelve KACC isolates and the ex-type strain of C. scovillei (CBS 126529) formed a single clade with a 99% ML bootstrap value. Three KACC isolates (KACC 40700, KACC 40801, and KACC 47036) were segregated into a separate group with C. chrysanthemi (IMI 364540, authentic strain), well supported by a 100% ML bootstrap value. KACC 47255 and KACC 47254 were in a group with the ex-type strain of C. lupini (CBS 109225) with a 97% ML bootstrap value. A single clade was generated by KACC 42911 and the ex-type strain of C. godetiae (CBS 133.44), well supported with a bootstrap value of 98%. The isolate KACC 40014 formed a single clade and had a distant genetic relationship with others. The GAPDH sequence of KACC 40014 had highest similarities with C. sloanei (IMI 364297, ex-type strain, 97.37%), C. paxtonii (IMI 165753, ex-type strain, 96.93%) and had 93.86% similarity with C. simmondsii (CBS 122122, ex-type strain). The loci of KACC 40014 were lower than 99% similarity with the closely related species such as ACT (98.7% with C. simmondsii and C. paxtonii), HIS3 (98.66% with C. simmondsii and C. sloanei), TUB2 (98.98% with C. paxtonii and 98.57% with C. sloanei), and ITS (98.9% with C. simmondsii). The data suggested that Colletotrichum sp. (KACC 40014) could be considered as a novel species candidate of the genus Colletotrichum.
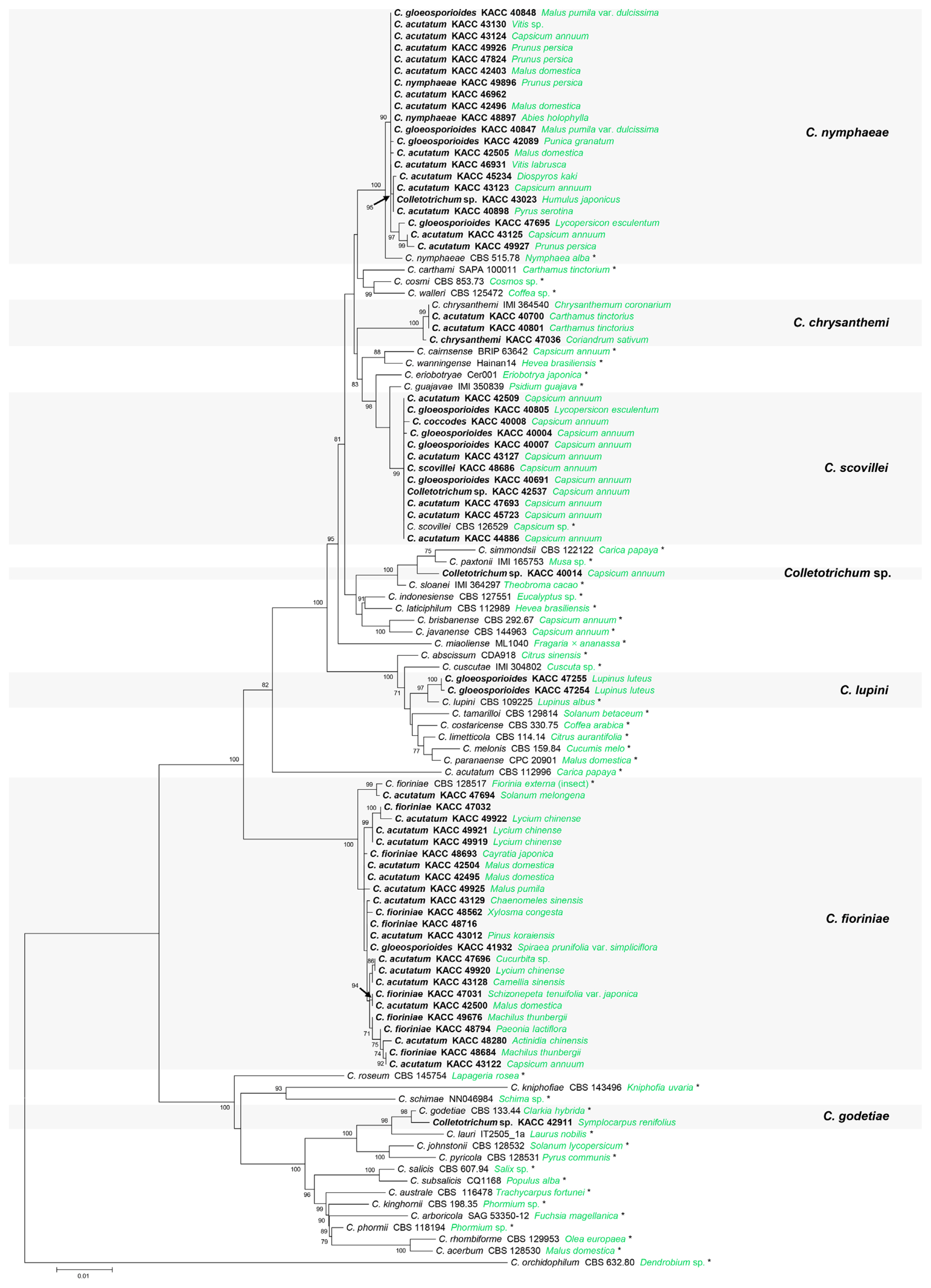
A maximum likelihood tree was generated based on the analysis of multi-locus sequences of internal transcribed spacers (ITS), beta-tubulin 2 (TUB2), histone-3 (HIS3), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), chitin synthase 1 (CHS-1), and actin (ACT). Species names are followed by isolate numbers and their hosts (green). Isolates in this study are in bold (original species names and isolate numbers). Ex-type strains are noted by *. Colletotrichum orchidophilum was used as the outgroup.
Host plants of C. acutatum species complex in this study
C. fioriniae (24 KACC isolates) were collected from 15 different plant species belonging to 15 genera and 11 families with an abundance of Rosaceae (six isolates) and Solanaceae (six isolates). Ten host species (Actinidia chinensis, Capsicum annuum, Camellia sinensis, Cayratia japonica, Chaenomeles sinensis, Machilus thunbergii, Pinus koraiensis, Schizonepeta tenuifolia var. japonica, Spiraea prunifolia var. simpliciflora, and Xylosma congesta) have not been reported in Korea and six of which, including C. japonica, C. sinensis, P. koraiensis, S. tenuifolia var. japonica, S. prunifolia var. simpliciflora and X. congesta, have not been reported in the world. Five species (Cucurbita sp., Lycium chinense, Malus domestica, Paeonia lactiflora, and Solanum melongena) were previously reported in Korea. C. nymphaeae (21 KACC isolates) were isolated from 10 different host species, from nine genera and seven families. Most of them are from the family Rosaceae (10 isolates) and Solanaceae (four isolates). Eight host species (Abies holophylla, Capsicum annuum, Humulus japonicas, Lycopersicon esculentum, Prunus persica, Punica granatum, Pyrus serotina, and Vitis labrusca) have not been reported in Korea, of which two species (A. holophylla and H. japonicus) have not been recorded in the world. Two other species (Diospyros kaki and Malus domestica) were previously reported in Korea. C. scovillei (12 KACC isolates) were obtained from only the family Solanaceae, including 11 isolates from Capsicum annuum (reported in Korea) and one isolate from Lycopersicon esculentum (unreported in the world). C. chrysanthemi (three KACC isolates) was isolated from Coriandrum sativum (unreported in the world) and Carthamus tinctorius (unreported in Korea). C. lupini (two KACC isolates) was found on Lupinus luteus (reported in Korea). C. godetiae (one KACC isolate) was from Symplocarpus renifolius (unreported in the world). A novel species candidate (KACC 40014) was collected from Capsicum annuum (Tables 2 and 3).
Discussion
Sixty-four Korean isolates in C. acutatum species complex were accurately identified into six different species (C. fioriniae, C. nymphaeae C. scovillei, C. chrysanthemi, C. lupini, and C. godetiae) and a novel species candidate, based on the combination of multi-locus sequences of ITS, TUB2, HIS3, GAPDH, CHS-1, and ACT. Forty-eight isolates changed their species names from the original names given by depositors. The present results also demonstrated that the identifications of the species in the C. acutatum species complex using a single ITS region and/or TUB2 gene in the previous publications (Han et al., 2014; Kim et al., 2006, 2008; Noh et al., 2014) were insufficient.
C. fioriniae has been reported as an entomopathogenic, endophytic and phytopathogenic fungus (Damm et al., 2012; Marcelino et al., 2008). This species has been reported as the causal agent of anthracnose diseases on Cucurbita moschata, Lycium chinense, Malus domestica, Paeonia lactiflora, Solana melongena, Ilex integra, Prunus persica, Prunus salicina, Schisandra chinensis, and Vaccinium sect. Cyanococcus in Korea. Six of them (M. domestica, P. salicina, S. chinensis, and V. sect. Cyanococcus) were not listed in The List of Plant Diseases in Korea (http://genebank.rda.go.kr/english/plntDissInfo.do), and five species (I. integra, P. persica, P. salicina, S. chinensis, and V. sect. Cyanococcus) were not found in this work. Meanwhile, ten host species of C. fioriniae in this study have not been recorded in Korea or in the world. C. nymphaeae was associated with serious anthracnose diseases in a wide range of host plants, especially strawberries (Fragaria × ananassa) (Damm et al., 2012; Jayawardena et al., 2016). In the previous research, six host species of C. nymphaeae (Diospyros kaki, Malus domestica, Actinidia argute, Prunus salicina, Vaccinium sect. Cyanococcus, and Ziziphus jujube) were reported in Korea, three of that plant (M. domestica, V. sect. Cyanococcus, and Z. jujube) have not been updated in The List of Plant Diseases in Korea, and four species (A. argute, P. salicina, V. sect. Cyanococcus, and Z. jujube) were not recorded in this study. However, eight host species in the present study were not introduced in relationship with this fungal species before in Korea or in the world. C. scovillei has a narrow host range and was commonly reported as one of the highly aggressive diseases of Capsicum spp. in many countries such as Brazil (Giacomin et al., 2021), Korea (Oo et al., 2017) and Asia (de Silva et al., 2019). C. scovillei also infected Clausena lansium (Lin et al., 2020), Mangifera indica (Qin et al., 2019), Musa sp. (Zhou et al., 2017), and Pseudodracontium lacourii (Liu et al., 2022). In our findings, C. scovillei was isolated from Lycopersicon esculentum and this has not been reported in the world.
C. chrysanthemi was only reported in the family Asteraceae, including Carthamus tinctorius in Italy (Baroncelli et al., 2015), Glebionis carinata (vascular discoloration) in the Netherlands, Glebionis coronaria (leaf spot) in China (Damm et al., 2012), Chrysanthemum coronarium in Korea and Calendula officinalis in Japan (Sato and Moriwaki, 2013). In this study, C. chrysanthemi was not only collected from the Asteraceae (Carthamus tinctorius) but also obtained from an unrecorded family Apiaceae (Coriandrum sativum).
C. lupini was first reported to cause anthracnose on yellow lupin (Lupinus luteus) in Korea and Asia in 2013 (Han et al., 2014). This species has a narrow range of host plants, but it showed high virulence and globally widespread disease on some host species of the genus Lupinus (Alkemade et al., 2021). This fungal species was also found on Camellia sp., Cinnamomum verum, Manihot utilissima, and Olea europaea (Alkemade et al., 2021; Damm et al., 2012). C. godetiae was infected economically important crops and had a wide host range and global distribution (Alizadeh et al., 2015; Liu et al., 2022; Shivas et al., 2016; Tan et al., 2022; Tóth et al., 2017). However, this species was not commonly associated with valuable crops in Korea. In our study, this fungal species could be a new finding on Asian skunk cabbage (Symplocarpus renifolius).
On the other hand, considering species under C. acutatum species complex according to economic host plants in this study, C. scovillei (n = 11) was dominant on Capsicum annuum (pepper) and C. nymphaeae (n = 3), C. fioriniae (n = 1) followed. Five C. nymphaeae and four C. fioriniae isolates were identified from Malus spp. including apple. Only four and two isolates of C. nymphaeae were isolated from peach (Prunus persica) and grape (Vitis labrusca), respectively.
The most important finding in this study is that 23 new combinations could be suggested in Korea and 12 of these have not been reported in the world. Of which, C. fioriniae on pepper (Capsicum annuum) and kiwi (Actinidia chinensis), C. nymphaeae on peach (Prunus persica), pear (Pyrus serotina) and grape (Vitis labrusca), and C. scovillei on tomato (Lycopersicon esculentum), are meaningful information in the agricultural field of Korea. However, the pathogenicity of KACC isolates on hosts is not clear. KACC did not confirm the pathogenicity of deposited Colletotrichum isolates on host plants, but depended on only the depositor’s information. Therefore, new combinations suggested in this study need to be clarified via the pathogenicity tests in further studies.
Notes
Conflicts of Interest
No potential conflict of interest relevant to this article was reported.
Acknowledgments
This study was supported by a grant (PJ017286) from the National Institute of Agricultural Sciences and was a part of the “2023 KoRAA Long-term Training Program”, Rural Development Administration, Republic of Korea.
Electronic Supplementary Material
Supplementary materials are available at The Plant Pathology Journal website (http://www.ppjonline.org/).