Molecular Marker Development for the Rapid Differentiation of Black Rot Causing Xanthomonas campestris pv. campestris Race 7
Article information
Abstract
Xanthomonas campestris pv. campestris (Xcc) is a plant pathogen of Brassica crops that causes black rot disease throughout the world. At present, 11 physiological races of Xcc (races 1–11) have been reported. The conventional method of using differential cultivars for Xcc race detection is not accurate and it is laborious and time-consuming. Therefore, the development of specific molecular markers has been used as a substitute tool because it offers an accurate and reliable result, particularly a quick diagnosis of Xcc races. Previously, our laboratory has successfully developed race-specific molecular markers for Xcc races 1–6. In this study, specific molecular markers to identify Xcc race 7 have been developed. In the course of study, whole genome sequences of several Xcc races, X. campestris pv. incanae, X. campestris pv. raphani, and X. campestris pv. vesicatoria were aligned to identify variable regions like sequence-characterized amplified regions and insertions and deletions specific to race 7. Primer pairs were designed targeting these regions and validated against 22 samples. The polymerase chain reaction analysis revealed that three primer pairs specifically amplified the DNA fragment corresponding to race 7. The obtained finding clearly demonstrates the efficiency of the newly developed markers in accurately detecting Xcc race 7 among the other races. These results indicated that the newly developed marker can successfully and rapidly detect Xcc race 7 from other races. This study represents the first report on the successful development of specific molecular markers for Xcc race 7.
Brassicas, part of the Brassicaceae family, are globally important crop species that generate high economic and export value, such as edible oils, protein meals, vegetables, and condiments (Friedt et al., 2018). Cabbage production worldwide reached a reported 10.6 million metric tons (Food and Agriculture Organization of the United Nations, 2021). Black rot, which is primarily induced by the bacterium Xanthomonas campestris pv. campestris (Xcc), holds the utmost significance as a highly destructive disease for cabbage (Vicente and Holub, 2013; Williams, 1980). The most significant host for Xcc is Brassica oleracea, which includes economically important crops, cabbage, cauliflower, broccoli, brussels sprouts, and kale (Vicente and Holub, 2013). Black rot disease has emerged as a major constraint on cabbage production, and impacting the cabbage-growing area in Korea since the 1970s (Kim, 1986; Park, 2006). The disease affects production, notably of the vegetable Brassica, by inducing lesions on the leaves leading to a diminution in market value (Lema et al., 2012). Necrotic, darkening leaf veins and a chlorotic lesion in the shape of a V that begins at the leaf margin are typical signs of black rot (Cook et al., 1952; Lee et al., 2015; Vicente and Holub, 2013). The vascular pathogen Xcc, which infects plants through wounds, seeds, and insect transmission, is born in the seed (Cook et al., 1952; Williams, 1980).
Based on the gene-for-gene model, interactions between Xcc strains and various Brassica cultivars have led to the identification of 11 races of the Xcc pathogen (Fargier and Manceau, 2007; Kamoun et al., 1992; Vicente et al., 2001). So far, 11 Xcc races have been identified around the world using a set of differential cultivars. For example: in the United Kingdom, 6 races (races 1–6) have been reported by Vicente et al. (2001). In northwestern Spain, 7 races (races 1, 4, 5, 6, 7, 8, and 9) have been identified by Lema et al. (2012). Jensen et al. (2010), 5 races (races 1, 4, 5, 6, and 7) have been detected in Nepal. In south Africa, 4 races of Xcc (races 1, 3, 4, and 6) have been isolated by Chidamba and Bezuidenhout (2012). The newly identified races 10 and 11 have been reported in Portugal (Cruz et al., 2017). Among Xcc races, races 1 and 4 are predominant in Brassica oleracea crops throughout the world, however, race 6 is common in Brassica rapa (Lema et al., 2012; Vicente and Holub, 2013). As of present, the occurrence of Xcc race 7 has been reported in Nepal, Spain and Portugal along with the other Xcc races (Cruz et al., 2017; Jensen et al., 2010; Lema et al., 2012). Despite its constrained distribution, it is important to discriminate Xcc race 7 from the remaining races, since the frequent long-distance trade of contaminated seeds around the world can be associated with the successful dissemination of pathogen (Lema et al., 2012). Xcc exhibits diversity across many nations or regions of the same nation (Popović et al., 2013). Management of the disease includes cultivation of disease-free seeds and most of the time the phytopathogens spreads through the import and export of seeds materials (Lema et al., 2012). Therefore, there are many quarantine policies for prevention of the movement of different pests and pathogens during import and export (Martin et al., 2000).
The cultivation of resistant cultivars remains the most effective, economical, and ecologically sustainable approach for reducing the damage caused by biotic stresses (Yerasu et al., 2019). Subsequently, an alternative strategy including the development and application of resistant cultivars has long been accepted, but in practice has had only modest success (Taylor et al., 2002). Various Brassica cultivars have shown resistance to the black rot disease, but these cultivars have not been substantiated to be resistant to all races of Xcc (Afrin et al., 2019; Vicente et al., 2001). It is essential that one considers the presence of different races and the genetic diversity of the disease (Jensen et al., 2010). Differentiating races of a pathogen is important for disease management since it allows for targeted and effective control strategies. By identifying and understanding the specific races of a pathogen, such as Xcc in the case of black rot in Brassica oleracea, researchers can develop and implement control measures that are tailored to the specific races present (Ignatov et al., 1998). It also allows in monitoring the prevalence and distribution of specific races and track their spread to implement appropriate management strategies. Therefore, accurate and immediate identification of the pathogen Xcc is necessary for effective black rot control of the pathogen. Xcc needs to be immediately and accurately identified in order to control. To facilitate disease control and breeding programs, it is essential to establish a quick and accurate method for detecting Xcc in Brassica seeds and plants (Eichmeier et al., 2019). A real-time polymerase chain reaction (PCR)-based method using the hrpF gene has been developed to distinguish Xcc-infected Brassica seeds from other bacteria (Berg et al., 2006). Additionally, the Xcc genomic structure has been classified and distinguished using repetitive DNA PCR (rep-PCR) based on repetitive DNA sequence elements such as Xcc, X. campestris pv. vesicatoria (Xcv), X. oryzae pv. oryzae (Xoo), and X. campestris pv. musacearum (Aritua et al., 2007; Louws et al., 1995; Vera Cruz et al., 1996). However, these approaches are time-consuming and demanding (Singh et al., 2014). PCR is incredibly effective at diagnosing plant diseases (Zaccardelli et al., 2007). PCR-based technique has been described as a powerful alternative tool for the rapid detection and identification of bacterial strains causing plant disease (Song et al., 2014).
Our research team has made significant advancements in the development of molecular marker-based PCR for race-specific identification of Xcc races 1–6. These newly developed markers offer rapid detection within a few hours, while requiring less labor and providing higher reliability and sensitivity compared to the traditional method that utilizes differential cultivars of the Brassicaceae family for Xcc race determination (Afrin et al., 2018, 2019, 2020; Rubel et al., 2017, 2019b). In continuation of this work, our objective was to develop a robust and specific molecular marker, designed through the alignment and re-sequencing of whole genome sequences of different Xcc races, capable of accurately detecting Xcc race 7 using a PCR-based approach, thereby distinguishing it from other bacterial strains.
Materials and Methods
Bacterial strains and media
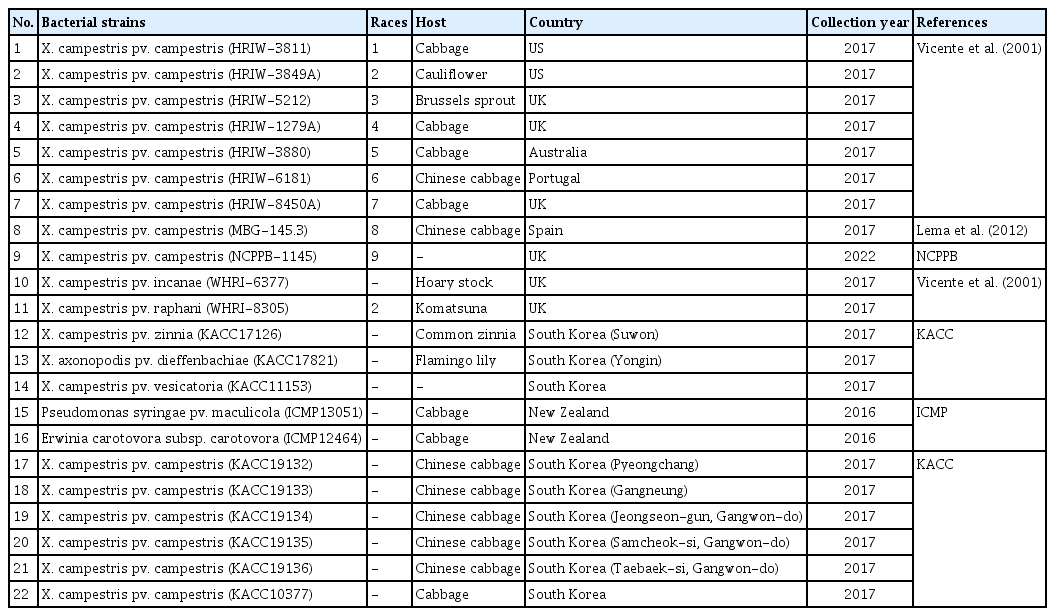
A group of 22 bacterial strains were employed for the analysis. This included nine standard reference races of Xcc (races 1–9), three X. campestris (Xc) pathovars, two Xc subspecies, two plant-infected bacteria and another six Xcc strains in Korea (race undetermined) (Table 1). The bacterial strains were cultured on King’s Medium B for 48 h at 30°C (King et al., 1954).
Extraction of genomic DNA
Genomic DNA of all bacterial isolates was extracted using QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. The concentration and purity of the extracted DNA were measured using a Nanodrop ND-1000 spectrophotometer (NanoDrop, Wilmington, DE, USA) and then stored at −20°C until study.
Identification of variable genomic regions
The complete sequences of Xcc races 1, 3, 4, and 9 as well as X. campestris pv. incanae (Xci), X. campestris pv. raphani (Xcr), and Xcv were obtained from the National Center for Biotechnology Information (NCBI) database. Meanwhile, DNA fragments from Xcc races 2, 5, 6, 7, and 8 (unpublished) were sequenced and compared with the aforementioned genomes using Integrative Genomics Viewer (IGV) (https://software.broadinstitute.org/software/igv/). Through this comparison, specific regions of insertion/deletion (InDel) were identified, which were unique to Xcc race 7 (Fig. 1). Thereafter, these regions were used to design molecular markers unambiguously targeting Xcc race 7.
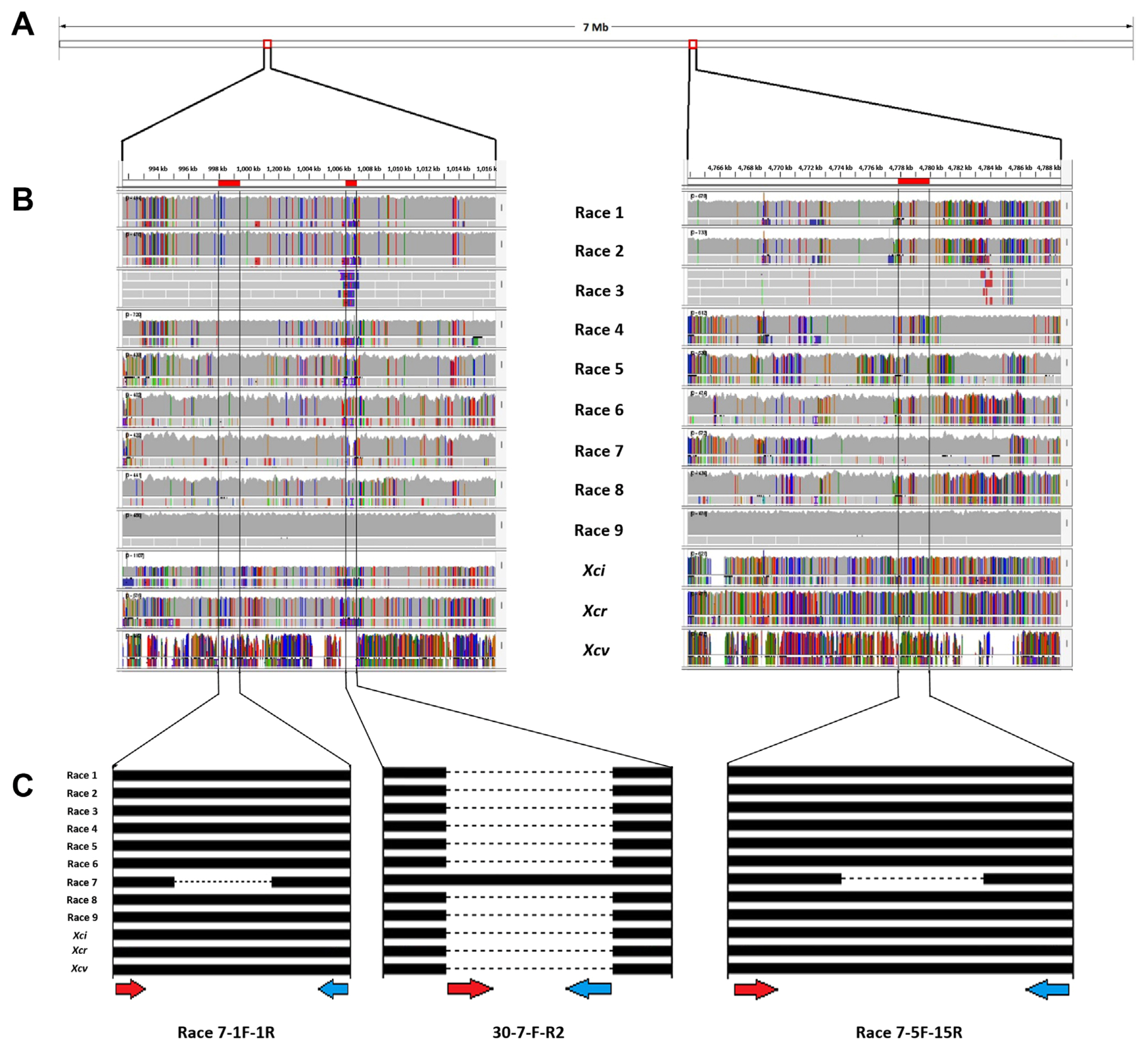
Alignment of whole genome sequences and identification of variable regions. (A) Line diagram representation of the whole genome length of Xanthomonas campestris pv. campestris (Xcc) race 7. (B) Whole genome alignment of Xcc races 1 to 9 and subspecies to identify the variant regions using Integrative Genomics Viewer (IGV) software with two parallel lines indicating the region where variants are identified. (C) A line diagram representation of the variable region identified; red and blue arrow represents forward and reverse primers respectively. Xci, X. campestris pv. incanae; Xcr, X. campestris pv. raphanin; Xcv, X. campestris pv. vesicatoria.
PCR amplification and primer development
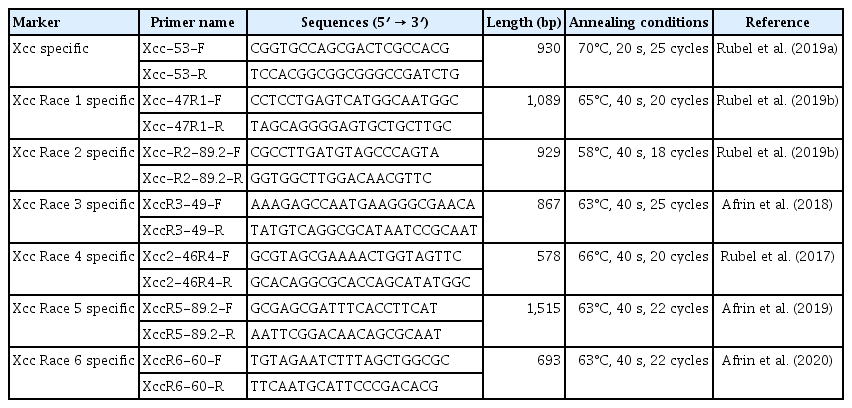
A seven-set of Xcc race-specific molecular markers previously reported from our laboratory was used to amplify DNA fragments (Table 2). Race 7 specific primers were designed using Primer 3 online program (https://primer3.ut.ee/) and the blast tool was used to check the specificity of the primers. In this study, eight primer pairs were designed for the detection of race 7 (Table 3). Thereafter, all these primer pairs were validated with all the bacteria in Table 1. Using TaKaRa TP600 Thermal cycler system (TaKaRa, Tokyo, Japan) race 7 primers were amplified with the following conditions: denaturation at 95°C for 2 min followed by 30 cycles (95°C for 15 s, 70°C for 15 s or 30 s and 72°C, 15 s) and terminated by a final elongation at 72°C for 2 min (Table 3). PCR reaction was prepared for a 10 μl reaction containing 1 μl (50 ng) of DNA, 5 μl of 2× Prime Taq Premix (GenetBio, Daejeon, Korea), 0.5 μl of each forward and reverse primers (0.2 mM) and 3 μl of sterile distilled water. The PCR products were resolved on 2.5% agarose gel for 30 min on an Agaro-Power electrophoresis unit (Bioneer, Daejeon, Korea), and visualized with a GD-1000 gel documentation system (Axygen, Union City, CA, USA) under UV light at 320 nm. All the PCR was conducted with two biological replicates and repeated three times independently.
Cloning and sequencing
The PCR amplicons of race 1 (801 bp) and race 7 (469 bp) from the ‘Race 7-1F-1R’ primer were excised from the agarose gel and purified using Wizard SV Gel and PCR Cleanup System (Promega, Madison, WI, USA). The purified fragments were cloned using TOPcloner Blunt Kit (Enzynomics, Daejeon, Korea) according to the protocol provided by the manufacturer. The positive clones were selected and further cultured in liquid LB media plate containing ampicillin. The purification of plasmid DNA was extracted using QIAprep Spin Miniprep Kit (Qiagen). Thereafter, the cloned sequences and alignment were performed in Multiple Sequence Alignment by the ClustalW program (https://www.genome.jp/tools-bin/clustalw).
Sensitivity test
The sensitivity and specificity of three primer pairs of race 7 were tested to check the level of detection by PCR amplification which was aforementioned, 20 ng/μl of purified DNA of race 7 was taken and diluted serially up to 10−4 dilution. Thereafter, 1 μl of DNA from each dilution (20 ng/μl, 2 ng/μl, 0.2 ng/μl, 0.02 ng/μl, and 0.002 ng/μl) was taken and used for PCR reaction. Sterile distilled water was used as a negative control.
Results
Xcc race 7 specific marker development and their specificity
To establish a PCR-based marker system specifically to detect Xcc race 7, we compared the complete genome sequences of Xcc races 1, 3, 4, 9, Xci, Xcr, and Xcv sequences obtained from the NCBI database. Additionally, we sequenced and aligned DNA fragments from races 2, 5, 6, 7, and 8 with the aforementioned genomes (Fig. 1), facilitating the design of primers specific to race 7.
By analyzing the whole-genome sequences, we were able to realign and compare them, thereby identifying variable regions that were present across the strains. Specifically, we identified specific InDel regions that served as targets for developing unique molecular markers specific to race 7. Ultimately, we designed eight primer pairs to differentiate race 7, from which three primer sets; one sequence-characterized amplified region (SCAR) marker (30-7-F-R2) and two InDel markers (race 7-1F-1R and Race 7-5F-5R) for specific detection of Xcc race 7 (Table 3) were able to specifically detect the race 7. To validate the efficacy of the newly developed primers, we performed PCR reactions using all the bacterial strains listed in Table 1.
The SCAR marker (30-7-F-R2) demonstrated the ability to specifically amplify a 320 bp amplicon solely with Xcc race 7 DNA, while all other strains’ DNA failed to produce any amplicons (Fig. 2A). In contrast, the two designed InDel markers, Race 7-1F-1R and Race 7-5F-5R, generated specific amplicons with Xcc race 7 DNA, measuring approximately 600 bp and 700 bp, respectively (Fig. 2B and C). Remarkably, all other strains yielded an amplicon of approximately 750 bp when utilizing either primer set, Race 7-1F-1R or Race 7-5F-5R (Fig. 2B and C).

Polymerase chain reaction amplifications using 3 primer sets tested against all 16 bacterial strains. (A) 30-7-F-R2, 320-bp. (B) Race 7-1F-1R, 600-bp. (C) Race 7-5F-5R, 700-bp. Lane M, DNA ladder (100 bp); lane 1–9, Xanthomonas campestris pv. campestris (Xcc) races 1–9; lane 10, WHRI-6377 (X. campestris pv. incanae [Xci]); lane 11, WHRI-8305 (X. campestris pv. raphani [Xcr]); lane 12, KACC17126 (X. campestris pv. zinnia [Xcz]); lane 13, KACC17821 X. axonopodis pv. dieffenbachiae [Xad]; lane 14, KACC11153 (X. campestris pv. vesicatoria [Xcv]); lane 15, ICMP13051 (Pseudomonas syringae pv. maculicola [Psm]); lane 16, ICMP12464 (Erwinia carotovora subsp. carotovora [Ecc]).
The PCR products showed that: a SCAR marker ‘30-7-F-R2’ was amplified with an approximate size of 320 bp fragment specific to Xcc race 7 only while the other sample was unamplified (Fig. 2A). Thereafter, InDel marker ‘Race 7-1F-1R’ showed polymorphic amplification to race 7 with an amplicon size of 600 bp and 850 bp for race 7 and Xcc races and two Xc pathovars (Xci and Xcr), respectively (Fig. 2B). Another InDel primer pair ‘Race 7-5F-5R’ also gave the polymorphic amplification to race 7 in amplicon size of 700 bp for race 7 only and Xcc races and Xci with 800 bp size, while other samples were not detected (Fig. 2C). Therefore, these 3 primer pairs can successfully and rapidly detect Xcc race 7 from other Xcc.
Race determination for Xcc unknown KACC strains
In this study, the three primer pairs were also effective in detecting the race of unknown Xcc strains obtained from the Korean Agriculture Centre Collection (KACC), namely KACC19132, KACC19133, KACC19134, KACC19135, and KACC19136. These strains exhibited the same specific amplicons as Xcc race 7, confirming their classification as race 7, with one exception-strain KACC10377 (Table 1, Fig. 3A–J). Notably, previous research conducted by Rubel et al. (2019b) identified KACC10377 as race 1. Thus, our findings demonstrate that the race-unknown strains obtained from KACC are predominantly race 7, except for KACC10377, which corresponds to race 1 based on the aforementioned study.

Polymerase chain reaction amplification of Xanthomonas campestris pv. campestris (Xcc) races 1–6 and Xcc race 7-specific using different 10 primer sets including 3 designed for race 7 in this study. (A) Xcc specific (Xcc-53, 930 bp). (B) Xcc race 1-specific (Xcc-47R1, 1,089 bp). (C) Xcc race 2-specific (Xcc-R2-89-2, 929 bp). (D) Xcc race 3-specific (XccR3-49, 867 bp). (E) Xcc race 4-specific (Xcc2-46R4, 578 bp). (F) Xcc race 5-specific (XccR5-89.2, 1,515 bp). (G) Xcc race 6-specific (XccR6-60, 693 bp). (H) Xcc race 7-specific (30-7-F-R2, 320 bp). (I) Xcc race 7-specific (race 7-1F-1R, 600-bp). (J) Xcc race 7-specific (race 7-5F-5R, 700 bp). All these primers were validated with genomic DNA listed in Table 1. Lane M, DNA ladder: 100 bp; lanes 1–9, Xcc races (1–9); lane 17, Xcc (KACC19132); lane 18, Xcc (KACC19133); lane 19, Xcc (KACC19134); lane 20, Xcc (KACC19135); lane 21, Xcc (KACC19136); lane 22, Xcc (KACC10377).
Sensitivity of the developed markers
To assess the detection sensitivity of the developed markers, PCR amplifications were conducted using five different concentrations of race 7 genomic DNA: 20 ng/μl, 2 ng/μl, 0.2 ng/μl, 0.02 ng/μl, and 0.002 ng/μl. Sterile distilled water was used as a negative control. All three markers successfully amplified DNA at a minimum concentration of 2 ng/μl (Fig. 4A–C). In contrast, ‘Race 7-5F-5R’ marker demonstrated the ability to amplify DNA even at lower concentrations, specifically as low as 0.02 ng/μl (Fig. 4B).
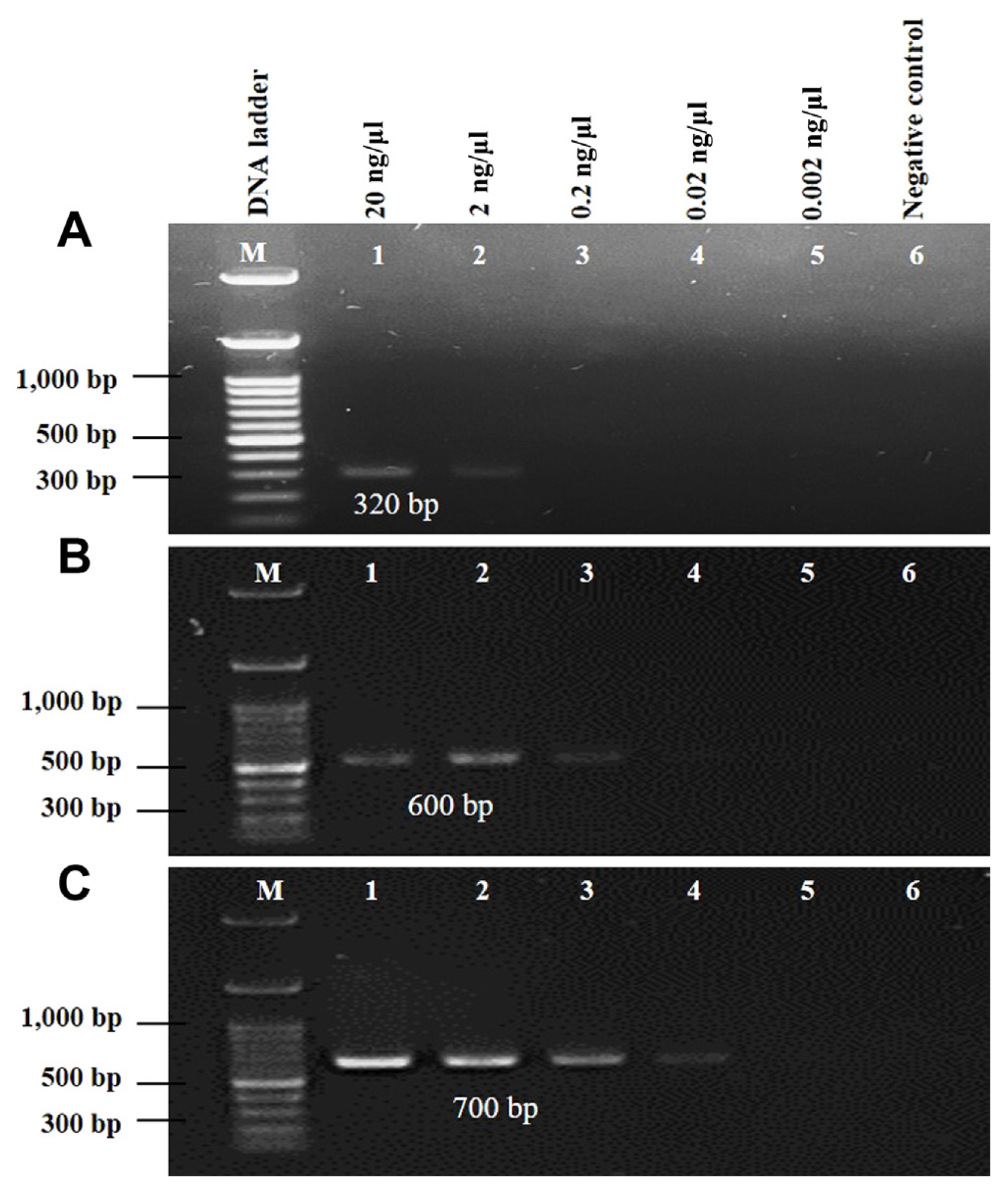
Limits of detection of Xanthomonas campestris pv. campestris (Xcc) race 7 from genomic DNA: (A) 30-7-F-R2, (B) race 7-1F-1R, and (C) race 7-5F-5R. Lane M, DNA ladder (100 bp); lane 1, 20 ng/μl; lane 2, 2 ng/μl; lane 3, 0.2 ng/μl; lane 4, 0.02 ng/μl; lane 5, 0.002 ng/μl; lane 6, negative control. For the polymerase chain reaction amplification, 1 μl DNA of Xcc race 7 from each other dilution was used as a template.
Cloning and sequencing analysis
Additionally, the PCR amplicons from race 1 and race 7 using the ‘Race 7-1F-1R’ primer were verified through cloning and sequencing. The size of race 1 amplicon was 801 bp (Supplementary Fig. 1A), but the sequencing result showed an amplicon size of 753 bp (Supplementary Fig. 1B). The size of the race 7 amplicon was 469 bp (Supplementary Fig. 1C). This may be due to the partial sequencing. These sequences were then aligned with the sequences of race 7 amplicon using ClustalW. The alignment revealed a deletion of 284 bp in race 7 compared to race 1 (Supplementary Fig. 2).
Discussion
The pathogen Xcc is a significant plant pathogen that causes devastating black rot disease in the Brassicaceae family, particularly affecting cabbage crops worldwide. In order to effectively manage this disease accurate diagnosis is crucial. Traditionally, race identification has relied on the use of differential cultivars from the Brassicaceae family, which is a time-consuming process requiring fieldwork over at least one cropping season. Therefore, it is essential to employ proper and reliable techniques for the detection of infected crops.
PCR-based markers have been successfully utilized for identifying leaf and seed infections caused by bacterial and fungal pathogens (Thangavelu et al., 2022; Wang et al., 2010). Molecular markers have also been developed for the detection of Xcc races 1 to 6 using whole genome realignment (Afrin et al., 2018, 2019, 2020; Rubel et al., 2017, 2019b). These markers have proven to be effective in rapidly and accurately distinguishing between Xcc races within a few hours.
Similar PCR-based marker approaches have been applied in the diagnosis of other plant diseases. For instance, Thangavelu et al. (2022) developed race-specific markers for the differentiation of Indian Fusarium oxysporum f. sp. cubense, the causal agent of Fusarium wilt in bananas. Wang et al. (2010) utilized SCAR markers to reliably detect two races (CYR32 and CYR33) of wheat stripe rust caused by Puccinia striiformis f. sp. tritici. Pasquali et al. (2007) successfully employed an inter-retrotransposon sequence-characterized amplified region marker to identify race 1 of F. oxysporum f. sp. lactucae on lettuce. Cho et al. (2011) developed sensitive and specific primers for the detection of bacterial leaf blight caused by Xoo in rice, utilizing real-time and conventional PCR approaches based on an rhs family gene.
In this study, we developed novel PCR amplification primers for the specific detection of Xcc race 7. The availability of genome sequences of Xcc races (1, 3, 4, and 9), as well as other pathovars Xci, Xcr, and Xcv and their alignment, facilitated the discovery of highly variable genomic regions and the development of unique markers specific to Xcc race 7. The SCAR marker (30-7-F-R2) and two InDel markers (Race 7-1F-1R and Race 7-5F-5R) developed in this study reliably and effectively detect Xcc race 7, differentiating it from other Xcc races, Xc subspecies, and other plant-infecting bacteria. Additionally, the five strains of Xcc unknown have amplified with the newly developed race 7-specific markers and this indicates that five unknown KACC strains may belong to race 7 (Fig. 3H–J). Therefore, this finding provides a reliable diagnostic tool for the accurate and rapid detection of Xcc race 7, offering an alternative to the time-consuming and laborious method of using differential cultivars for race determination.
Notes
Conflicts of Interest
No potential conflict of interest relevant to this article was reported.
Acknowledgments
This work was supported by grants from Ministry of Agriculture, Food and Rural Affairs (MAFRA) (322059-03-2-HD050). We would thank Dr. Pilar Soengas, Department of Plant Genetics, Spain for providing Xcc race 8. We also thank Dr. Joana G. Vicente, University of Warwick, UK for providing Xcc races (races 1–7) and Xc pathovars. We thank the Korean Agriculture Culture Collection (KACC), Korea for providing Xcc species and ICMP collection, New Zealand for providing isolates of plant pathogenic bacteria.
Electronic Supplementary Material
Supplementary materials are available at The Plant Pathology Journal website (http://www.ppjonline.org/).