Biological Control of Fusarium oxysporum, the Causal Agent of Fusarium Basal Rot in Onion by Bacillus spp.
Article information
Abstract
Fusarium oxysporum is the main pathogen causing Fusarium basal rot in onion (Allium cepa L.), which incurs significant yield losses before and after harvest. Among management strategies, biological control is an environmentally safe and sustainable alternative to chemical control. In this study, we isolated and screened bacteria for antifungal activity against the basal rot pathogen F. oxysporum. Isolates 23-045, 23-046, 23-052, 23-055, and 23-056 significantly inhibited F. oxysporum mycelial growth and conidial germination. Isolates 23-045, 23-046, 23-052, and 23-056 suppressed the development of Fusarium basal rot in both onion seedlings and bulbs in pot and spray inoculation assays. Isolate 23-055 was effective in onion seedlings but exhibited weak inhibitory effect on onion bulbs. Based on analyses of the 16S rRNA and rpoB gene sequences together with morphological analysis, isolates 23-045, 23-046, 23-052, and 23-055 were identified as Bacillus thuringiensis, and isolate 23-056 as Bacillus toyonensis. All five bacterial isolates exhibited cellulolytic, proteolytic, and phosphate-solubilizing activity, which may contribute to their antagonistic activity against onion basal rot disease. Taken together B. thuringiensis 23-045, 23-046, 23-052, and 23-055 and B. toyonensis 23-056 have potential for the biological control of Fusarium basal rot in onion.
Genus Allium is among the most important genera in family Amaryllidaceae (Asparagales). It includes valuable crops such as the bulb onion (Allium cepa L.), garlic (Allium sativum L.), leek (Allium ampeloprasum var. porrum L.), chive (Allium schoenoprasum L.), and shallot (A. cepa var. ascalonicum L. and A. cepa var. aggregatum L.) (Sengupta and Bhattacharjee, 2004). The bulb onion is the most widely cultivated Allium species; it is a biennial crop, normally producing a bulb in the first year of growth and flowers and seed in the second year after vernalization by winter cold (Griffiths et al., 2002; Lyngkhoi et al., 2019). Onions are eaten fresh, roasted, grilled, and pickled, and are widely used to flavor sauces, soups, salads, and other snacks (Mahmood et al., 2021; Ren et al., 2018). Onions are rich in phytochemicals, including saponins, minerals, phenolics, oligosaccharides, and organosulfur compounds (Roldán-Marín et al., 2009; Shabir et al., 2022; Slimestad et al., 2007). Their bioactive compounds have numerous health benefits, such as antioxidant, antimicrobial, antiviral, anti-inflammatory, and anticancer activities.
Onion plants have many fungal pathogens worldwide, including Fusarium oxysporum, Botrytis aclada, Puccinia allii, Aspergillus niger, Sclerotium cepivorum, Rhizoctonia solani, and Peronospora destructor (Back et al., 2017; Kim et al., 2002; Moon et al., 2007; Park et al., 1995; Parthasarathy et al., 2016; Shin et al., 2023). Among these, F. oxysporum f. sp. cepae is the most common cause of Fusarium basal rot (FBR) in onion, but pathogenic complexes of several Fusarium species are also associated with FBR, including F. oxysporum, Fusarium commune, Fusarium proliferatum, and Fusarium redolens (Galván et al., 2008; Ghanbarzadeh et al., 2014; Haapalainen et al., 2016; Taylor et al., 2016). FBR infection occurs through onion roots and stem plates at any stage of onion plant development (Southwood et al., 2015). Rotting starts from the basal plate of the onion bulb, and then spreads to the outer bulb scales, followed by the inner bulb scales (Holz and Knox-Davies, 1985; Lager, 2011).
FBR is sometimes called damping-off in onion seedlings because the disease causes pre- and post-emergence damping-off in the early stages of onion plant development (Degani et al., 2022; Kintega et al., 2020; Le et al., 2021; Shin et al., 2023). Infected seedlings at the early stage, especially at transplantation, die or were severely stunted, but the symptoms were reduced or did not appear when infection occurs at later stages (Retig et al., 1970; Stadnik and Dhingra, 1997). However, Stadnik and Dhingra (1997) recovered the pathogen from asymptomatic onion plants, indicating that the fungus can remain a latent pathogen. The disease can develop in latently infected bulbs during storage, which can lead to huge onion losses (Labanska et al., 2022; Le et al., 2021).
Recently, FBR incidence in onion plants in South Korea has increased significantly in both the field and storage with the increasing use of mechanical seedling transplantation. It is thought that reusing plug trays in transplanters leads to contamination by FBR pathogens. No fungicides or biological control agents are registered for the control of FBR in onion plants in South Korea, increasing the need to find effective chemical or biological control agents against FBR. In this study, we isolated bacterial strains from soil samples from onion fields and evaluated their antagonistic properties against the FBR pathogen F. oxysporum.
Materials and Methods
Fungal strain and bacteria isolation
We screened antagonistic bacterial strains using Fusarium oxysporum strain 19–385 isolated from an onion infected with FBR in Hamyang, South Korea in 2019 (Shin et al., 2023). Soil samples were collected from onion fields in Hamyang, South Korea in 2022, brought to the laboratory in plastic bags, and stored in a refrigerator at 4°C before use. For bacterial isolation, 5 g of each soil sample was mixed with 45 ml sterile distilled water (SDW) in a 50-ml conical tube (SPL, Pocheon, Korea) and shaken at 100 rpm for 15 min. The suspensions were filtered through two layers of cheesecloth, diluted to 1:100 with SDW, and spread on Luria-Bertani (LB) agar (Difco Laboratories, Franklin Lakes, NJ, USA). The plates were incubated for 16 h at 25°C, after which single bacterial colonies were subcultured onto new LB plates. The isolates were mixed with 25% glycerol and preserved in a deep freezer at −75°C before use. F. oxysporum 19–385 and bacterial isolates were routinely grown on potato dextrose agar (PDA) and LB agar at 25°C in the dark.
In vitro inhibition of mycelial growth
Inhibition of F. oxysporum mycelial growth by bacterial isolates was tested on LPA (LB 50% + PDA 50%) medium. A mycelial agar plug of fungal pathogen from 7-day-old PDA culture was inoculated onto the center of the LPA, and three holes were punched with a sterile cork borer. Each hole was filled with 10 μl of bacterial suspension at an optical density at 600 nm (OD600) of 1.0. SDW was used as the control. After 3 days of incubation in the dark, inhibition of mycelial growth was measured in comparison with the control according to the following formula: Inhibition of growth (%) = (1 – T/C) × 100, where T and C are the average length of mycelium between the mycelial agar plugs and holes in the treatment and control, respectively. This experiment was repeated three times in triplicate.
In vitro inhibition of conidial germination
For the conidial germination assay, F. oxysporum was incubated in potato dextrose broth (PDB) containing carnation leaf pieces for 2 days with shaking at 150 rpm in the dark. Conidia were harvested from the PDB culture by filtering through two layers of Miracloth (Millipore, Burlington, MA, USA). The conidia were pelleted by centrifugation at 4,000 rpm for 10 min, washed twice with SDW, and adjusted to 1 × 105 conidia/mL using a hemocytometer. Bacterial isolates were incubated in LB broth, adjusted to an OD600 of 1.0, and serially diluted in SDW. The conidial suspension was mixed with the bacterial suspension in a 1:1 ratio to attain a final concentration of 5 × 104 conidia/mL and 10 to 1,000-fold dilutions of bacterial suspension from an initial OD600 of 1.0. Drops of the mixture, each 20 μl, were placed on a coverslip and incubated for 4 h in a moistened plastic box. Inhibition of conidial germination was measured in comparison with SDW-treated control conidia according to the following formula: Inhibition of conidial germination (%) = (1 – T/C) × 100, where T and C are the respective germination rates of the treatment and control. The experiment was conducted in triplicate and repeated three times (n ≥ 100 conidia per sample).
Molecular identification of bacterial isolates
Genomic DNA was extracted from overnight cultures of bacterial isolates in LB broth using the HiGene Genomic DNA Prep Kit (BIOFACT, Daejeon, Korea). The 16S rRNA and RNA polymerase beta subunit (rpoB) genes were amplified with the primer pairs 9F (5′-GAGTTTGATCCTGGCTCAG-3′)/1512R (5′-ACGGHTACCTTGTTACGACTT-3′) and BA-RF (5′-GACGATCATYTWGGAAACCG-3′)/BA-RR (5′-GGNGTYTCRATYGGACACAT-3′), respectively (Ko et al., 2003; Weisburg et al., 1991), and sequenced by BIOFACT. The sequences were analyzed using the National Center for Biotechnology Information BLAST program (http://blast.ncbi.nlm.nih.gov/). The obtained sequences were aligned using ClustalW in MEGA 6.0 and phylogenetic trees were generated using the maximum-likelihood method, with 500 bootstrap replicates (Tamura et al., 2011; Thompson et al., 1997).
Microscopic examination of parasporal inclusion morphology
To observe the endospores and parasporal inclusions of bacterial isolates, the isolates were grown on Nutrient agar (Difco Laboratories) for 2 days at 30°C in the dark. Bacterial samples were suspended in SDW on a microscope slide, covered with a coverslip, and observed under a phase-contrast microscopy (Axio Image A2, Carl Zeiss, Jena, Germany) at 1,000× magnification with immersion oil (Kil et al., 2008; Kim et al., 1995).
Disease suppression assays
Onion bulb scales were used to evaluate the biocontrol efficiency of the bacterial isolates. Fusarium oxysporum was incubated in PDB containing carnation leaf pieces for 2 days with shaking (150 rpm). Then, the conidial suspension was filtered through two layers of Miracloth and adjusted to 5 × 105 conidia/mL in SDW using a hemocytometer. Bacterial isolates were incubated in LB broth with shaking overnight and adjusted to an OD600 of 1.0. Bacterial and conidial suspensions of F. oxysporum were sprayed onto bulb scales and incubated at 25°C in a moistened plastic box. SDW treatment was used as a negative control, and the F. oxysporum conidial suspension was sprayed on bulb scales as a positive control. Four days after inoculation, the percent diseased area was determined using ImageJ v1.53c (National Institutes of Health, Bethesda, MD, USA) (Kim et al., 2020). Control efficiency was calculated according to the following formula: Control efficiency (%) = (1 – T/C) × 100, where T and C are the diseased area in the treatment and positive control, respectively. The experiment was repeated three times in triplicate.
To evaluate the biocontrol efficiency of the bacterial isolates on onion seedlings, onion seeds (A. cepa L. cv. ‘Sunpower’) were surface-sterilized for 3 min in 1% sodium hypochlorite solution, rinsed thoroughly with SDW, and incubated for 2 days in sterile Petri dishes with wet paper towels in the dark. After incubation, the germinated seeds were soaked in a 10-ml mixture of conidial (7 × 105 conidia/ml) and bacterial (OD600 = 1.0) suspensions for 1 h. The seeds were then dried for 1 h and 50 seeds per treatment were planted in a pot. The seedling survival rate (%) was compared to that of the uninfected control after 14 days of incubation at 25°C under a 12-h light/12-h dark light cycle. This experiment was repeated three times in triplicate.
Analyses of proteolytic, cellulolytic, and phosphate-solubilizing activity
Cellulolytic bacteria were screened on carboxymethyl cellulose (CMC) agar (1.0 g KH2PO4, 0.5 g MgSO4·7H2O, 0.5 g NaCl, 0.01 g FeSO4·7H2O, 0.01 g MnSO4·H2O, 0.3 g NH4NO3, 10 g CMC, and 12 g agar/l) (Han et al., 2015b). After 12 days of incubation at 25°C in the dark, the plates were stained with 0.1% Congo red solution for 15 min and washed with 1 M NaCl solution. Proteolytic activity was tested on 3% skim milk agar (30 g skim milk and 15 g agar/l) (Sokol et al., 1979). A 4-mm-diameter hole was formed on each plate using a cork borer, and 10 μl of bacterial suspension (OD600 = 1.0) was dropped into each hole. Clear zone formation was observed after incubation for 20 h at 25°C in the dark. Phosphate-solubilizing activity was tested on Pikovskaya agar (10 g glucose, 5 g Ca3(PO4)2, 0.5 g (NH4)2SO4, 0.2 g NaCl, 0.1 g MgSO4·7H2O, 0.2 g KCl, 0.5 g yeast extract, 0.002 g MnSO4·H2O, and 0.002 g FeSO4·7H2O, and 15 g agar/l) (Nautiyal, 1999). After incubation for 14 days at 25°C in the dark, phosphate-solubilizing activity was confirmed by the formation of a clear zone. All experiments were conducted in triplicate and repeated three times.
Polymerase chain reaction amplification of endochitinase (chit) and exochitinase (chi36) genes
The polymerase chain reaction (PCR) for chit and chi36 genes was performed according to the method of Djenane et al. (2017). The chit and chi36 genes were amplified with the primer pairs chit(f) (5′-ATTCACACTGCTATTACTATC-3′)/chit(r) (5′-TGACGGCATTTAAAAGTTCGGC-3′) and chi36(f) (5′-GATGTTAAACAGGTTCAA-3′)/chi36(r) (5′-TTATTTTTGCAAGGAAAG-3′), respectively (Arora et al., 2003; Djenane et al., 2017; Raddadi et al., 2009).
Analytical Profile Index 50 CHB test
The biochemical profiles of the bacterial isolates were determined using the Analytical Profile Index (API) 50 CHB kit, a ready-to-use medium that tests the fermentation of 49 carbohydrates on an API 50 CH strip, according to the manufacturer’s instructions (BioMérieux, Marcy l’Étoile, France). Bacterial suspensions of each isolate were prepared in an ampule of API 50 CHB medium, which equals 1.0 McFarland standard. Tubes with the API CH strip were filled with the inoculated API 50 CHB medium and covered with mineral oil. The results were recorded after 24 h at 30°C in the dark and interpreted using the API Web database system (http://apiweb.biomerieux.com).
Data analyses
The data were analyzed statistically using IBM SPSS Statistics v27.0 (IBM Corp., Armonk, NY, USA). Inhibition of mycelial growth and conidial germination, diseased area on bulb scales, and seedling survival rate were evaluated using Duncan’s multiple range test at a significance level of P < 0.05.
Results
In vitro antifungal activity of bacterial isolates
In total, 400 bacterial isolates were obtained from soil samples collected from onion fields in Hamyang, South Korea, and screened for antifungal activity against the onion basal rot pathogen F. oxysporum using a dual culture method. Ultimately, five bacterial isolates, 23-045, 23-046, 23-052, 23-055, and 23-056, strongly inhibited the mycelial growth of F. oxysporum (Fig. 1A and B). Isolates 23-045, 23-046, 23-052, 23-055, and 23-056 inhibited the mycelial growth of F. oxysporum by 43%, 44%, 27%, 37%, and 35%, respectively. These five isolates also exhibited antifungal activity on the conidial germination of F. oxysporum (Fig. 2). Ten-fold diluted cultures of each isolate inhibited > 70% of conidial germination compared to the SDW-treated control conidia. Strains 23-046, 23-052, and 23-056 were more effective at inhibiting conidial germination than 23-045 and 23-055 at 10 to 1,000-fold dilutions. The efficacy of the five isolates at inhibiting conidial germination dropped significantly at 1,000-fold dilution, with strains 23-046 and 23-056 inhibiting less than 40% of conidial germination, and strain 23-045 only inhibiting 5% of conidial germination.

Inhibition of mycelial growth of Fusarium oxysporum by bacterial isolates. (A) Mycelial agar plugs of F. oxysporum from 7-day-old potato dextrose agar (PDA) cultures were placed in the center of LPA (Luria-Bertani [LB] + PDA) medium. Bacterial isolates were inoculated in holes in the LPA medium. Photographs were taken after 3 days of co-culture at 25°C. (B) Measurement of mycelial growth inhibition (%) of F. oxysporum by bacterial isolates in comparison with the control (0% mycelial growth inhibition). Error bars represent standard deviations. Different letters on bars indicate significant differences (P < 0.05, Duncan’s multiple range test).

Inhibitory effect of bacterial isolates on conidial germination of Fusarium oxysporum. A conidial suspension was mixed with each bacterial isolate in a 1:1 ratio to a final concentration of 5 × 104 conidia/ml and 10 to 1,000-fold dilutions of bacterial suspension from an OD600 of 1.0 and placed on a coverslip. Inhibition of conidial germination was measured after 4 h of incubation in comparison with the sterile distilled water (SDW)-treated control (90% conidial germination). Error bars represent standard deviations. Different letters on bars indicate significant differences (P < 0.05, Duncan’s multiple range test).
Molecular identification of bacterial isolates
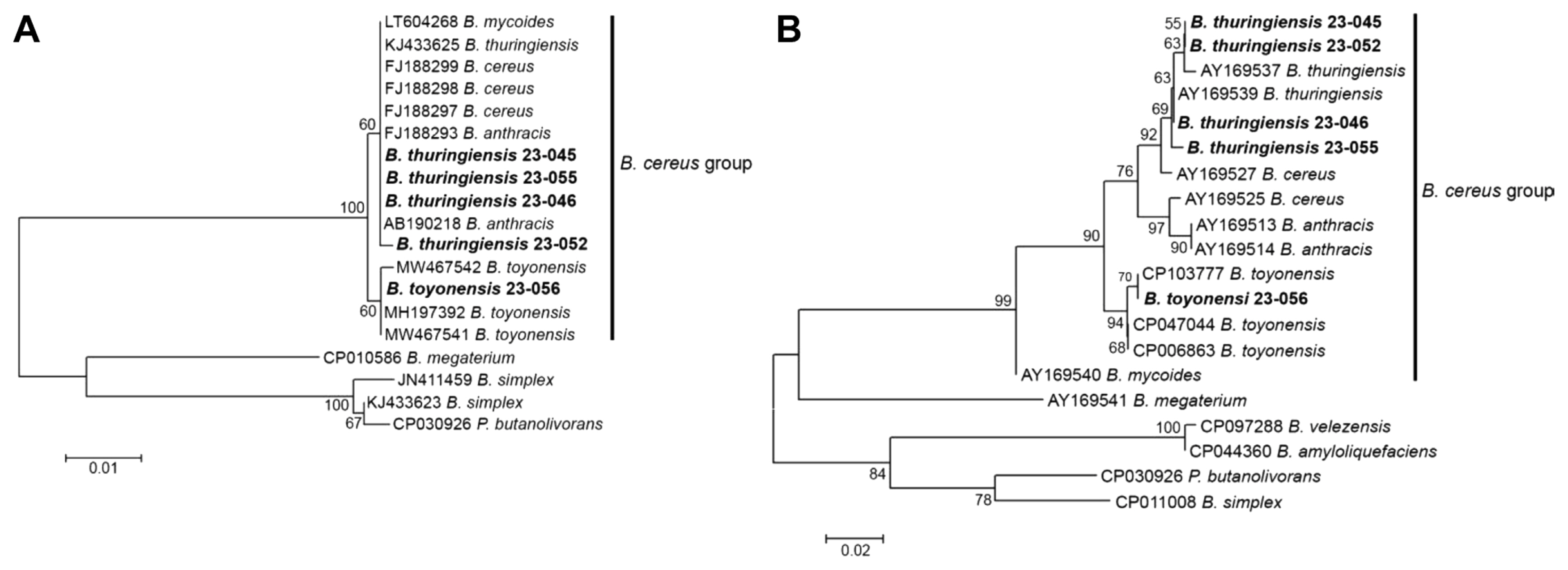
To identify the five isolates exhibiting antifungal activity, 16S rRNA and rpoB gene sequences were analyzed. After NCBI BLAST searches of the 16S rRNA and rpoB gene sequences of each isolate, isolates 23-045, 23-046, 23-052, and 23-055 were identified as Bacillus thuringiensis, whereas isolate 23-056 was identified as Bacillus toyonensis (Table 1). To analyze the genetic relationships of the Bacillus isolates, phylogenetic trees were generated using the maximum-likelihood method based on the 16S rRNA and rpoB genes (Fig. 3A and B). The 16S rRNA gene analysis failed to discriminate closely related Bacillus species (Fig. 3A). Strains of the Bacillus cereus group were separated into two subgroups, with isolates 23-045, 23-046, 23-052, and 23-055 assigned to the subgroup with Bacillus mycoides, B. thuringiensis, B. cereus, and Bacillus anthracis, and isolate 23-056 was assigned to another subgroup with B. toyonensis. The rpoB gene analysis discriminated B. anthracis, B. toyonensis, and B. mycoides from B. cereus and B. thuringiensis, with isolates 23-045, 23-046, 23-052, and 23-055 closely grouped with B. thuringiensis (Fig. 3B).
Phase-contrast microscopic examination
Bacillus thuringiensis is a gram-positive rod-shaped bacterium characterized by the production of parasporal inclusions, which differentiates B. thuringiensis from B. cereus (Kil et al., 2008; Whiteley and Schnepf, 1986). Therefore, the formation of endospores and parasporal inclusions of the bacterial isolates were observed by phase-contrast microscopy. After incubation on Nutrient agar for 2 days, all isolates produced white endospores within bacterial cells (Fig. 4). Isolates 23-045, 23-046, 23-052, and 23-055 also produced oval and cuboid parasporal inclusions together with the endospores, indicating that the strains belong to B. thuringiensis. The 23-056 isolate, which was identified as B. toyonensis by 16S rRNA and rpoB gene analysis, did not produce parasporal inclusions.
Biocontrol efficacy of bacterial isolates against FBR in onion bulbs and seedlings
Next, we assessed the biocontrol efficiency of bacterial isolates against FBR in onion bulbs. Because F. oxysporum can remain latent in the basal plate or roots of onion bulbs, we used onion bulb scales to generate the disease. Onion bulb scales were sprayed with bacterial suspension (OD600 = 1.0) followed by conidial suspension (5 × 105 conidia/ml) of F. oxysporum. As shown in Table 2 and Fig. 5A, the positive control (without bacterial isolates) had a disease area of 26.3 ± 14.7%, whereas the disease areas was significantly reduced by treatment with bacterial isolates 23-045, 23-046, 23-052, or 23-056, to 0.4 ± 0.2%, 0.9 ± 0.5%, 2.3 ± 1.9, and 2.1 ± 1.5%, respectively. However, strain 23-055 showed little reduction of disease development in onions, with a control efficiency of 7.2%.

Control efficiency of bacterial isolates on onion bulb scales and seedling survival rate against Fusarium oxysporum

Suppression of Fusarium basal rot caused by Fusarium oxysporum on onion bulb scales and seedlings by bacterial isolates. (A) Onion bulb scales were sprayed with bacterial suspension at an optical density at 600 nm (OD600) of 0.1 and conidial suspension of F. oxysporum (5 × 105 conidia/ml). Photographs were taken 4 days after inoculation. (B) Onion seeds were soaked in conidial suspension of F. oxysporum (7 × 105 conidia/ml) and bacterial suspension (OD600 = 1) for 1 h and sown in plastic pots. Photographs were taken 14 days after inoculation.
Next, germinated onion seeds were soaked in a 10-ml mixture of conidial (7 × 105 conidia/ml) and bacterial (OD600 = 1.0) suspensions for 1 h and incubated in a pot for 14 days. All bacterial isolates, including strain 23-055, which was ineffective on onion bulb scales, significantly reduced pre-emergence damping-off (Table 2, Fig. 5B). Seedling survival rates of onion seeds after treatment with isolates 23-045, 23-046, 23-052, 23-055, or 23-056 were 79.5%, 82.8%, 78.8%, 63.0%, and 80.2%, respectively, with only 26.8% of seedling survival rate in the positive control (without bacterial isolates). These data indicate that isolates 23-045, 23-046, 23-052, and 23-056 are effective for inhibiting FBR in onion bulbs and seedlings, whereas isolate 23-055 is effective in onion seedlings.
Antagonistic and phosphate-solubilizing activity of bacterial isolates
The antagonistic and phosphate-solubilizing activity of the bacterial isolates were tested on CMC, skim milk, and Pikovskaya agar media (Fig. 6). CMC agar was used to identify isolates with cellulase activity. On CMC agar, isolates 23-045, 23-046, 23-052, and 23-055 formed white clear zones around the colonies, whereas isolate 23-056 formed an orange clear zone that was distinguishable from CMC stained by Congo red. Skim milk agar was used to identify proteolytic activity. All isolates formed transparent clear zones around the colonies on skim milk agar. Phosphate-solubilizing activity were screened on Pikovskaya agar; all isolates formed transparent clear zones on Pikovskaya agar. Together, these results indicate that isolates 23-045, 23-046, 23-052, 23-055, and 23-056 have cellulolytic, proteolytic, and phosphate-solubilizing activity. In addition, we assessed the presence of endochitinase (chit) and exochitinase (chi36) genes in the bacterial isolates by PCR amplification (Fig. 7). We found that the chit gene was detected in the B. thuringiensis isolates, 23-045, 23-046, 23-052, and 23-055, but not in the B. toyonensis isolate 23-056 (Fig. 7). In contrast, the chi36 gene was not found in all bacterial isolates (Fig. 7B).

Analyses of cellulolytic, proteolytic, and phosphate-solubilizing activity. Cellulase production was examined on carboxymethyl cellulose agar stained with 0.1% Congo red for 15 min followed by decolorization with 1 M NaCl solution for 15 min. Protease production was examined on 3% skim mild agar. Phosphate-solubilization was examined on Pikovskaya agar.
Biochemical profiles of bacterial isolates with the API 50 CHB system
The five Bacillus isolates were examined using an API 50 CHB kit to test carbohydrate fermentation (Table 3). Although the isolates were identified as B. thuringiensis or B. toyonensis in our study, isolates 23-045, 23-046, 23-052, 23-055, and 23-056 were identified as B. cereus based on the API 50 CHB system, with 94.9%, 94.9%, 97.4%, 57.6%, and 99.0% similarity, respectively (weak reactions were scored as positive), because the API 50 CHB system cannot distinguish B. thuringiensis from B. cereus, and B. toyonensis is not included in the system (Logan and Berkeley, 1984). For carbohydrate fermentation, all five tested Bacillus isolates were able to utilize glycerol, ribose, d-glucose, d-fructose, N-acetylglucosamine, arbutin, esculin, salicin, cellobiose, maltose, trehalose, starch, and glycogen. The four B. thuringiensis isolates were unable to utilize d-mannose, whereas B. toyonensis 23-056 utilized it.
Discussion
Crop pests and pathogens, including bacteria, fungi, nematodes, viruses, and insects, significantly reduce annual global agricultural production by more than 20% (Pérez-García et al., 2011). To control diseases, growers have relied heavily on manufactured pesticides, which may result in quick effective control of diseases and increase yield. However, excessive and repeated use of chemical pesticides in agriculture have led to environmental pollution, reducing biodiversity, increased development of resistant pests and pathogens, and toxicity in animals including humans (Hassaan and El Nemr, 2020; Mesnage et al., 2014). The adoption of biological control is an important technique to reduce chemical pesticides, and genus Bacillus is among the most exploited microbial groups due to its high abundance, diversity, survival ability, and production of various bioactive compounds with direct and indirect antagonistic effects (Caulier et al., 2018; Han et al., 2015a; Saxena et al., 2020). Currently, several commercial Bacillus-based biological control agents are used in agriculture, including Bacillus amyloliquefaciens, Bacillus subtilis, B. thuringiensis, and Bacillus velezensis (Fira et al., 2018).
In this study, we isolated and identified five Bacillus isolates exhibiting high antifungal activity against the FBR pathogen F. oxysporum in onion. Our 16S rRNA and rpoB gene sequence analyses identified the isolate 23-056 as B. toyonensis. Other four isolates (23-045, 23-046, 23-052, and 23-055) were grouped with B. thuringiensis, but not clearly differentiated from B. cereus. The B. cereus group, also called B. cereus sensu lato, includes B. anthracis, B. cereus, Bacillus cytotoxicus, B. mycoides, Bacillus pseudomycoides, B. thuringiensis, B. toyonensis, and Bacillus weihenstephanensis, and is extremely close phylogenetically (Fiedler et al., 2019). Although rpoB sequence analysis can discriminate some strains in the B. cereus group, B. cereus and B. thuringiensis cannot be clearly differentiated (Ko et al., 2003). B. thuringiensis produces rounded (oval and cuboid) or diamond-shaped parasporal inclusion bodies during sporulation, which are used to differentiate it from B. cereus. Therefore, we also used phase-contrast microscopy to identify parasporal inclusions and confirmed that all four isolates produced parasporal inclusions. Together with the gene sequence analysis results, our morphological analysis showed that the four isolates were B. thuringiensis.
Bacillus thuringiensis has been used as a biological control agent for insect pests in agriculture due to its parasporal inclusions, which often contain one or more insecticidal proteins (de Almeida Melo et al., 2016; Dulmage, 1970; Kil et al., 2008; Lacey et al., 2015; Wasano et al., 1998). In addition to its properties as a biopesticide, ongoing attention is being paid to the use of B. thuringiensis to control plant pathogenic fungi (Jouzani et al., 2017; Zhou et al., 2008). For example, B. thuringiensis BK4, C25, and var. israelensis showed antifungal activity against Fusarium wilt in tomato caused by F. oxysporum (Jung et al., 2005), lettuce drop caused by Sclerotinia minor (Shrestha et al., 2015), and Sclerotium rolfsii infecting soybean seedlings (Reyes-Ramírez et al., 2004), respectively. We tested the antifungal activity of the B. thuringiensis isolates 23-045, 23-046, 23-052, and 23-055 against the FBR pathogen F. oxysporum. Consistent with previous reports, B. thuringiensis isolates 23-045, 23-046, 23-052, and 23-055 significantly inhibited the mycelial growth and conidial germination of F. oxysporum. Moreover, isolates 23-045, 23-046, and 23-052 successfully reduced disease development on onion seedlings and bulbs. These results show that B. thuringiensis has great potential for use as a biological agent for FBR in onion. The B. thuringiensis isolate 23-055 also strongly inhibited pre-emergence damping-off in onion seedlings, but had weak inhibitory effect on onion bulb scales. B. cereus var. toyoi BCT-7112T, also called B. toyonensis BCT-7112T, is an active ingredient in Toyocerin, the first probiotic to receive European Union approval for use as a feed additive for domesticated animals such as swine, poultry, cattle, and rabbits (Cutting, 2011; Jiménez et al., 2013; Williams et al., 2009). Recently, B. toyonensis has been studied as a biological control agent against plant pathogens (Kim et al., 2018; Lopes et al., 2017; Rojas-Solis et al., 2020). For example, B. toyonensis CAB12243-2 was effective at suppressing soft rot in Chinese cabbage caused by Pectobacterium carotovorum subsp. carotovorum (Kim et al., 2018). B. toyonensis COPE52 had antifungal activity against the gray mold phytopathogen Botrytis cinerea (Rojas-Solis et al., 2020). Consistent with reports that B. toyonensis can suppress plant diseases, we demonstrated that B. toyonensis isolate 23-056 strongly inhibited FBR in onion seedlings and bulbs, inhibiting mycelial growth and conidial germination of F. oxysporum.
Many antagonistic bacteria secrete hydrolytic enzymes such as chitinase, glucanase, and protease, which plays a major role in fungal suppression (Fatima et al, 2023; Xu et al., 2016). For example, the proteolytic activity of antagonistic bacteria exhibits strong inhibitory effect on fungal mycelial growth and conidial germination (Gandhi Pragash et al., 2009; Ji et al., 2020; Ling et al., 2022). In the current study, a qualitative assay of antagonistic substances showed that the B. thuringiensis and B. toyonensis isolates exhibited proteolytic activity. We think that the ability of B. thuringiensis and B. toyonensis isolates to produce the hydrolytic enzymes contributes to their antifungal activity against the basal rot pathogen F. oxysporum. In addition, we found that B. thuringiensis isolates harbored the endochitinase chit gene that is a candidate gene to control plant pathogenic fungi (Djenane et al., 2017). Thus, chitinases could be involved in the antifungal activity of B. thuringiensis isolates. On the other hand, cellulolytic and phosphate-solubilizing activity were found in the bacterial isolates. Cellulolytic activity plays an important role for antagonistic bacteria to colonize the endosphere of plants (Santoyo et al., 2021). Microbial phosphate solubilization makes phosphorous available to plants, which promotes plant growth (Lee et al., 2012; Rodríguez and Fraga, 1999). Although we could not find plant growth-promoting effects of bacterial isolates in onion seedling test in pots (data not included), it is worth to investigate plant growth-promoting and endosphere colonizing abilities under field condition.
In this study, the B. thuringiensis and B. toyonensis isolates strongly inhibited fungal growth, conidial germination, and FBR disease development in onion bulbs and seedlings of F. oxysporum. Studies of suitable application methods and cost effectiveness are currently underway, including field experiments, which will contribute to the application of these bacteria as biological control agents.
Notes
Conflicts of Interest
No potential conflict of interest relevant to this article was reported.
Acknowledgments
This work was carried out with the support of the “Cooperative Research Program for Agriculture Science & Technology Development (Project No. PJ01667601), and supported by (2023) the RDA Fellowship Program of the National Institute of Horticultural & Herbal Science, Rural Development Administration, Republic of Korea.