 |
 |
| Plant Pathol J > Volume 40(1); 2024 > Article |
|
Abstract
The conservation of the endangered Korean fir, Abies koreana, is of critical ecological importance. In our previous study, a yeast-like fungus identified as Aureobasidium pullulans AK10, was isolated and shown to enhance drought tolerance in A. koreana seedlings. In this study, the effectiveness of Au. pullulans AK10 treatment in enhancing drought tolerance in A. koreana was confirmed. Furthermore, using transcriptome analysis, we compared A. koreana seedlings treated with Au. pullulans AK10 to untreated controls under drought conditions to elucidate the molecular responses involved in increased drought tolerance. Our findings revealed a predominance of downregulated genes in the treated seedlings, suggesting a strategic reallocation of resources to enhance stress defense. Further exploration of enriched Kyoto Encyclopedia of Genes and Genomes pathways and protein-protein interaction networks revealed significant alterations in functional systems known to fortify drought tolerance, including the terpenoid backbone biosynthesis, calcium signaling pathway, pyruvate metabolism, brassinosteroid biosynthesis, and, crucially, flavonoid biosynthesis, renowned for enhancing plant drought resistance. These findings deepen our comprehension of how AK10 biostimulation enhances the resilience of A. koreana to drought stress, marking a substantial advancement in the effort to conserve this endangered tree species through environmentally sustainable treatment.
The Korean fir, Abies koreana, is a conifer native to South Korea, mainly concentrated on the slopes of Halla Mountain on Jeju Island. This species, enduring millennia on the Korean Peninsula, has been recognized for its distinctive ornamental shape and phytochemical constituents with medicinal value, such as anti-cancer properties and cognitive function enhancement (Kim et al., 2001, 2006). Despite these valuable attributes, A. koreana has experienced a severe population decline, leading to its classification as an endangered species by the International Union for Conservation of Nature (IUCN) in 2011 (Kim et al., 2011).
Climate change has exacerbated environmental stressors such as drought, which have been implicated in the decline of Korean fir populations (Kim et al., 2017). Previous research revealed a correlation between environmental stress and gene expression linked to heat stress within these trees, suggesting an intricate relationship between the declining numbers of A. koreana and abiotic stress factors (Kim et al., 2020). In our previous study (Mannaa et al., 2023), we isolated the yeast-like fungus Aurobasidium pullulans AK10 and observed its potential as a foliar treatment to enhance drought-stress tolerance in A. koreana seedlings, correlating with beneficial shifts in rhizosphere microbiome composition. Au. pullulans is known for its adaptability across various environmental matrices and utility in biotechnological applications because it produces metabolites, such as pullulan, and a suite of hydrolytic enzymes, which could be beneficial for plant health and soil ecology (Chi et al., 2009; Mannaa et al., 2023).
Recent advancements in high-throughput sequencing technologies, particularly transcriptome analysis, have revolutionized our understanding of complex biological interactions (Mandlik et al., 2011). These approaches, including in planta transcriptome analysis, allow for in-depth exploration of the dynamic interplay between biostimulants, and host plants. High-throughput transcriptome analysis enables a comprehensive, unbiased quantification of transcripts, thereby facilitating a high-resolution insight into genome-wide expression profiles under specific conditions. Such analyses are important in unraveling the molecular mechanisms underlying plant responses to external treatments, allowing for dissecting the intricate molecular networks activated in plants, shedding light on key defense-related pathways and genes involved in plant-pathogen interactions (Park et al., 2020; Shukla et al., 2018).
This study aimed to confirm the efficacy of Au. pullulans AK10 in enhancing drought tolerance in A. koreana. Moreover, we explored the underlying mechanisms using transcriptome analysis and investigated the key pathways affected by AK10 treatment. This will offer insights into the mechanisms through which Au. pullulans AK10 alleviates drought stress, thereby aiding in the conservation of this endangered tree species.
To evaluate the effects of Au. pullulans AK10 on drought resistance in A. koreana, a seedling assay was conducted as described previously (Mannaa et al., 2023). Briefly, three-year-old A. koreana seedlings were obtained from International Horticultural Seedlings, Seoul, and Republic of Korea. Seedlings were replanted into plastic pots (11.5 à 16 cm) filled with soil from Punong, Gyeongju, Republic of Korea. They were then maintained in a greenhouse, with conditions set at a temperature of 25 ¹ 5°C and a relative humidity of 50 ¹ 10%, along with daily watering. The AK10 treatment was performed by culturing in yeast extract peptone dextrose (YPD) broth for 24 h at 25°C under constant agitation. The optical density of the culture was adjusted to OD600 = 0.8. Tween-20 was added to a final concentration of 250 Οg/ml. This mixture was foliar-sprayed onto seedlings three times: 7 days before, 4 days before, and 4 days after the initiation of drought stress. A control treatment was YPD broth with 250 Οg/ml Tween-20. Drought stress was simulated by withholding water. Three weeks after stopping irrigation, the seedlings were evaluated for drought symptoms based on a predefined scale. The evaluation scale employed for assessing seedling health ranged from 0 to 5. A score of 0 indicated healthy seedlings, characterized by vibrant green needles. A score of 1 was assigned to seedlings with less than 20% of the needles exhibiting a brown, wilted appearance. Scores of 2, 3, and 4 corresponded to seedlings with 20-39%, 40-59%, and 60-79% brown, wilted needles, respectively, with the latter also displaying bending in the terminal shoots. A score of 5 was given to seedlings where 80-100% of the needles were brown and wilted, signifying complete wilting of the seedling. This seedling assay was replicated twice, each with three replicates per treatment.
For the transcriptome analysis, RNAs were extracted from Korean fir seedlings using a modified CTAB method (Azevedo et al., 2003; Park et al., 2020), 3 weeks after stopping irrigation. After surface sterilization with 70% ethanol and rinsing, 1 g of leaf tissue was ground in liquid nitrogen and homogenized in 15 ml of extraction buffer (100 mM Tris-HCl, 2% CTAB, 30 mM ethylenediaminetetraacetic acid, 2 M NaCl, 0.05% spermidine, 2% polyvinylpolypyrrolidinone, 2% 2-mercaptoethanol, and 1.5 mg/ml proteinase K). After incubation at 42°C for 90 min and chloroform-isoamyl alcohol extraction, RNAs were precipitated with 10 M LiCl, washed with ethanol, and resuspended in diethylpyrocarbonate-treated water. RNA quality was confirmed using gel electrophoresis and quantified using a NanoDrop2000 spectrophotometer (Thermo Scientific, Barrington, IL, USA). Three replicates of drought-affected seedlings treated with Au. pullulans AK10 and three replicates of untreated drought-affected seedlings were conducted for RNA extraction and subsequent RNA sequencing (RNA-seq) analysis.
RNA integrity was evaluated using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). Using the TruSeq RNA Sample Preparation Kit (Illumina, San Diego, CA, USA), 1 Îźg of total RNAs was processed to produce cDNA libraries. These libraries were sequenced on a HiSeq 2000 platform (Illumina) by Macrogen (Seoul, Korea), generating 101-bp paired-end reads. Raw read quality was checked using FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/), and sequencing data were deposited in the NCBI Gene Expression Omnibus under accession no. GSE248350.
The Douglas fir (Pseudotsuga menziesii) reference genome was obtained from the Pine RefSeq database (https://treegenesdb.org/FTP/Genomes/Psme/v1.0/). Genes, exons, and coding sequences were identified in the GTF file. Duplicate gene predictions were removed from Augustus, GenomeThreader, and Gmap using a custom Python script.
Raw FASTQ reads were refined using the FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/). Raw FASTQ reads were refined by eliminating sequences with suboptimal quality scores. A bespoke Python script ensured the alignment of the forward and reverse reads. These curated reads were mapped to the reference genome using the Burrows-Wheeler Aligner (BWA), employing the maximal exact match approach (Li and Durbin, 2009). Using SAMtools, we converted the mapping outcome from SAM to BAM format and sorted it according to genomic coordinates (Li et al., 2009). The Subread package and featureCounts tool (Liao et al., 2014) tallied the reads for each coding sequence. Gene transcript levels were assessed using the RPKM (reads per kilobase million) metric (Mortazavi et al., 2008).
For AK10-specific differential expressed genes (DEGs) in drought-stressed A. koreana seedlings, we used the DEGseq package in R (Wang et al., 2010). The MARS model within this package, with random sampling, was used to identify and visualize intensity-dependent shifts in the transcriptomic dataset (Yang et al., 2002). Using the RPKM values as our dataset, we assessed the impact of AK10 treatment on genomic expression in the control. To account for multiple tests, individual gene P-values were corrected to false discovery rate (FDR) values using the Benjamini-Hochberg approach (Benjamini and Hochberg, 1995). Expressions with a change met the criteria of |log2-fold change| ⼠1 and FDR < 0.05. DEGs were functionally annotated using BlastKOALA, focusing on eukaryotic classification (Kanehisa et al., 2016). We developed BRITE functional outlines and associated pathways using the designated Kyoto Encyclopedia of Genes and Genomes (KEGG) orthologs.
Functional enrichment analyses based on KEGG pathways and gene ontology (GO) terms were performed to identify significantly activated biological systems in A. koreana seedlings treated with AK10 under drought conditions. Gene annotations were obtained from the PineRefSeq database. We quantified the up-regulated DEGs by employing a hypergeometric distribution via the R phyper function to ascertain significantly enriched systems (Qureshi and Sacan, 2013). We used the up-regulated DEGs identified in the transcriptomic study as inputs for the STRING database to analyze gene/protein interactions (Szklarczyk et al., 2019). STRING v11 encompasses interactions among 24.6 million proteins across 5,090 organisms. In our study, the Tracheophyta group served as the background, and the interactions were filtered using a confidence threshold of 0.50.
The specifics of the bioinformatics analyses have been detailed in their respective sections. Regarding the seedling assay, data were subjected to statistical evaluation using the General Linear Model procedure within the SAS software framework (SAS Institute Inc., Cary, NC, USA). Differences in means related to drought severity were determined employing the least significant difference test, with a significance threshold set at P < 0.05.
In seedling assays, drought-stressed seedlings showed leaf browning and shoot wilting. However, seedlings treated with AK10 via foliar spray displayed enhanced drought resilience with visibly reduced browning and wilting symptoms compared with their untreated counterparts (Fig. 1A). The control seedlings were regularly watered, vibrated, and healthy. The drought severity index further verified these observations, indicating a significant (P < 0.05) reduction in the drought effects in AK10-treated seedlings (Fig. 1B). These findings are consistent with those of our previous study (Mannaa et al., 2023), reinforcing the significant role of Au. Pullulans AK10 treatment in enhancing the drought tolerance of A. koreana. This is consistent with the findings of numerous studies highlighting the various benefits of Au. pullulans. Known for its biotechnological versatility, Au. pullulans synthesizes pullulan, a biodegradable extracellular polysaccharide used in many applications, highlighting its importance in sustainable practices and biotechnology (Chi et al., 2009). In addition to pullulan production, this fungus secretes a spectrum of hydrolytic enzymes that contribute to soil health and nutrient cycling, potentially affecting plant health both directly and indirectly (Zalar et al., 2008). Furthermore, Au. pullulans aids plant growth and produces volatile organic compounds, contributing to its biocontrol efficacy (Di Francesco et al., 2021). These potential benefits of Au. pullulans are reflected in the results of this study, suggesting that A. pullulans AK10 plays a critical role in enhancing the drought-stress resilience of A. koreana.
After establishing the role of AK10 in enhancing drought resilience in A. koreana, we conducted A. koreana transcriptome analysis via RNA-seq. We obtained approximately 445 million paired-end reads from the six RNA-seq libraries, averaging 74 million reads per library. Reads with an average Phred score of 36 (Supplementary Table 1) were narrowed down to 396 million after quality filtering. Details of the sequences, read lengths, GC content, and raw and mapped reads are listed in Table 1 and Supplementary Table 1. The processed reads were aligned to the Douglas fir P. menziesii reference genome (Table 2).
While investigating the genomic expression changes due to AK10 treatment, we set a threshold of |log2-fold change| ⼠1 and FDR < 0.05 (Fig. 2). AK10-treated seedlings were distinctly separated in the transcriptome analysis, suggesting significant gene expression changes compared to those in the controls (Fig. 2A). Scatter and volcano plots revealed each A. koreana gene as a point, with those meeting the FDR criteria marked in red (Fig. 2B and C). We observed 3,032 DEGs under drought stress with AK10 treatment; 601 genes were up-regulated, and 2,431 were downregulated (Fig. 2D). The treatment seemed to modulate physiological responses to drought, with fewer genes up-regulated by AK10, potentially the key to enhancing drought tolerance in A. koreana seedlings.
The donut charts in Fig. 3 show the KEGG pathway enrichment analysis for the DEGs, with segment dimensions and color intensity corresponding to gene counts and enrichment levels. Up-regulated DEGs in AK10-treated A. koreana seedlings enriched pathways, such as terpenoid backbone biosynthesis and flavonoid biosynthesis, implicated in plant defense and stress adaptation (LaouĂŠ et al., 2002; Tiedge et al., 2022). Additionally, the AMPK signaling pathway, known for its essential role in energy homeostasis, suggests a potential regulatory mechanism for enhanced drought tolerance under AK10 treatment (Hardie, 2011) (Fig. 3A).
In contrast, downregulated DEGs span a more extensive range of functions, with marked decreases in pathways such as starch and sucrose metabolism. This may reflect a strategic shift towards conserving resources under stress conditions (Smith et al., 2005). Similarly, the suppression of the phenylpropanoid biosynthesis, mitogen-activated protein kinase signaling, and plant hormone signal transduction pathways might indicate reprogramming of growth and developmental processes to prioritize drought survival strategies (Peleg and Blumwald, 2011; Rodriguez et al., 2010) (Fig. 3B). These findings suggest that the predominant effect of AK10 treatment could be the downregulation of specific biological processes, shifting from energy-consuming growth processes to survival modes under drought conditions. In addition to the upregulation of the AMPK signaling pathway, terpenoid backbone biosynthesis, and flavonoid biosynthesis, these results provide a possible explanation for the observed drought resistance. Future research must elucidate the precise molecular interactions by which AK10 mediates these effects.
Among the main observations of our study, treatment with Au. pullulans AK10 distinctly augmented the flavonoid biosynthesis pathway in A. koreana (Fig. 4). This pathway progression, depicted in the KEGG map (map00941), showed concerted upregulation in the expression of genes associated with key steps in flavonoid biosynthesis. PSME_05253, PSME_1376, and PSME_4268, which are responsible for the early stages of the pathway, showed a marked increase in expression, as indicated by normalized log2 fold-change values. Furthermore, the upregulation extends to genes PSME_2566 and PSME_3856, which are integral to synthesizing dihydroflavonols and flavonols such as kaempferol and quercetin. These flavonoid compounds have previously been shown to enhance drought tolerance in various plant species, offer protective functions against oxidative stress, and assist in stabilizing cell structures under water-deficient conditions (Baozhu et al., 2022; Yang et al., 2020). In addition to influencing stomatal movement, which is regulated by abscisic acid and hydrogen peroxide (Watkins et al., 2017). Our findings suggest that the application of Au. pullulans AK10 may stimulate flavonoid biosynthesis, thereby enhancing drought resilience in A. koreana seedlings. The upregulation of flavonoid pathway genes is correlated with an improved physiological state that enables plants to withstand water scarcity. However, the exact molecular interactions between AK10 and flavonoid pathway induction remain to be elucidated and require further investigation.
Our examination of the up-regulated DEGs revealed protein-protein interactions when Au. pullulans AK10 was applied, as illustrated in Fig. 5. Interaction network maps are instrumental in elucidating the dynamic biological processes that could be linked to enhancing drought tolerance. By applying an interaction cutoff value of 0.5, we identified a network of 36 protein nodes and 10 additional extended-interacting proteins. Integration of enriched GO terms for biological processes in this network map highlighted significant pathways associated with drought-stress response in A. koreana. These included the flavonoid biosynthesis pathway (PSME_42680 and PSME_38560), which is implicated in cellular antioxidant defense, and the brassinosteroid biosynthesis pathway (PSME_37926), which is known to modulate growth and stress responses (Ye et al., 2017). Additionally, genes related to calcium signaling (PSME_50714 and PSME_2774), which is essential for drought-stress signal transduction (Shao et al., 2008), and pyruvate metabolism (PSME_32155), which plays a role in energy homeostasis under stress conditions (Shen et al., 2017), were up-regulated. These interconnected pathways underscore the complex regulatory mechanism that enhances the ability of A. koreana to withstand drought.
In conclusion, this study confirmed the beneficial effects of foliar application of Au. pullulans AK10 on A. koreana under drought-stress conditions. The transcriptomic analyses conducted in this study shed light on the underlying mechanisms by which AK10 treatment confers drought resistance. We identified the upregulation of specific pathways and functional systems, including biological processes intrinsically associated with improved plant drought resistance. This study represents a step towards a deeper understanding of the strategies by which A. koreana can be enhanced against drought stress, thereby contributing to efforts to conserve this endangered species amidst escalating climatic challenges.
Acknowledgments
This study was supported by the Research and Development Program for Forest Science Technology (Project No. FTIS 2021334B10-2323-CD02) and was provided by the Korea Forest Service (Korea Forestry Promotion Institute).
Electronic Supplementary Material
Supplementary materials are available at The Plant Pathology Journal website (http://www.ppjonline.org/).
Fig. 1
Effect of Aureobasidium pullulans AK10 foliar spray on drought-stress response in Abies koreana seedlings. (A) The effects of drought and Au. pullulans AK10 treatment on A. koreana seedlings, captured three weeks post-treatment. (B) The severity of drought on the treated seedlings. The bars represent the average of three replicates, and different letters atop the error bars indicate a statistically significant difference at P < 0.05. Seedlings that were regularly watered served as control for the seedlings assay.
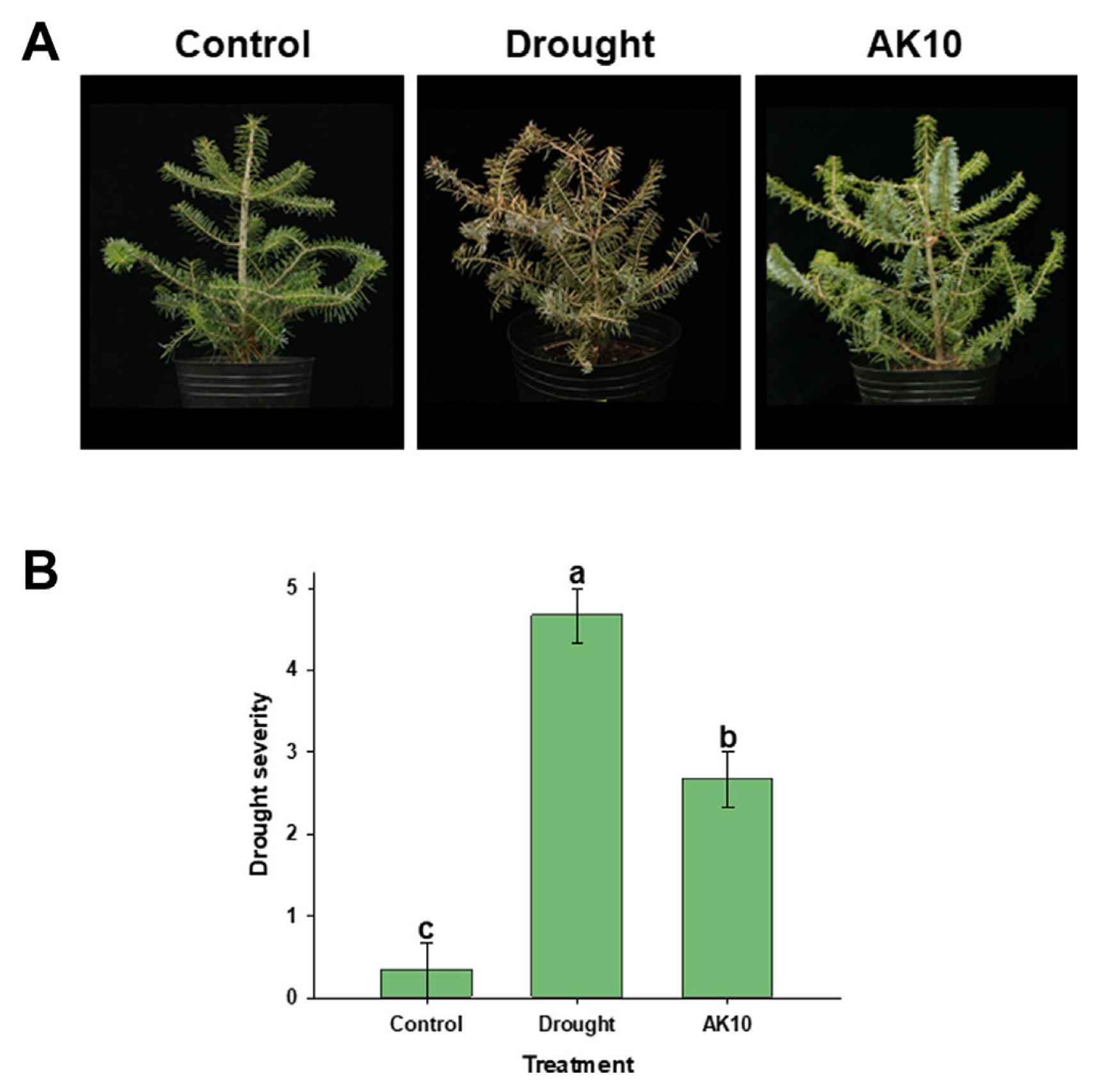
Fig. 2
Transcriptional changes in Abies koreana seedlings exposed to drought stress with Aureobasidium pullulans AK10 treatment. (A) A multidimensional scaling plot of normalized count data (log fold change) depicts the similarity among samples based on overall gene expression levels. Mean difference scatter plot (B) and Volcano plot (C), where each point symbolizes a gene in A. koreana. Differentially expressed genes (DEGs) are highlighted in red. (D) The bar graph displays the count of total DEGs, the up-regulated and downregulated genes. Criteria for DEG analysis include a >2-fold change (AK10/drought-affected seedlings) and a P-value < 0.05. FDR, false discovery rate; CPM, counts per million; FC, fold change.

Fig. 3
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of differentially expressed genes (DEGs) in Abies koreana seedlings exposed to drought stress, with or without Aureobasidium pullulans AK10 treatment. Donut charts represent the enrichment pathways of up-regulated DEGs (A) and those of downregulated DEGs (B). Segment size and color intensity are proportional to the number of genes linked to each pathway, visually representing the degree of enrichment. Essential pathways are labeled by name, and their respective P-values convey the significance of enrichment, providing a clear overview of the predominantly affected functions.

Fig. 4
Flavonoid biosynthesis molecular map illustrating the progression within the pathway (map00941) was visualized using the Cytoscape tool. The transformation of chemical compounds is illustrated alongside their associated genes, represented by rectangular nodes. Gene expression values are represented in a normalized log2 fold-change, and red color intensity corresponds to elevated gene expression when treated with AK10.
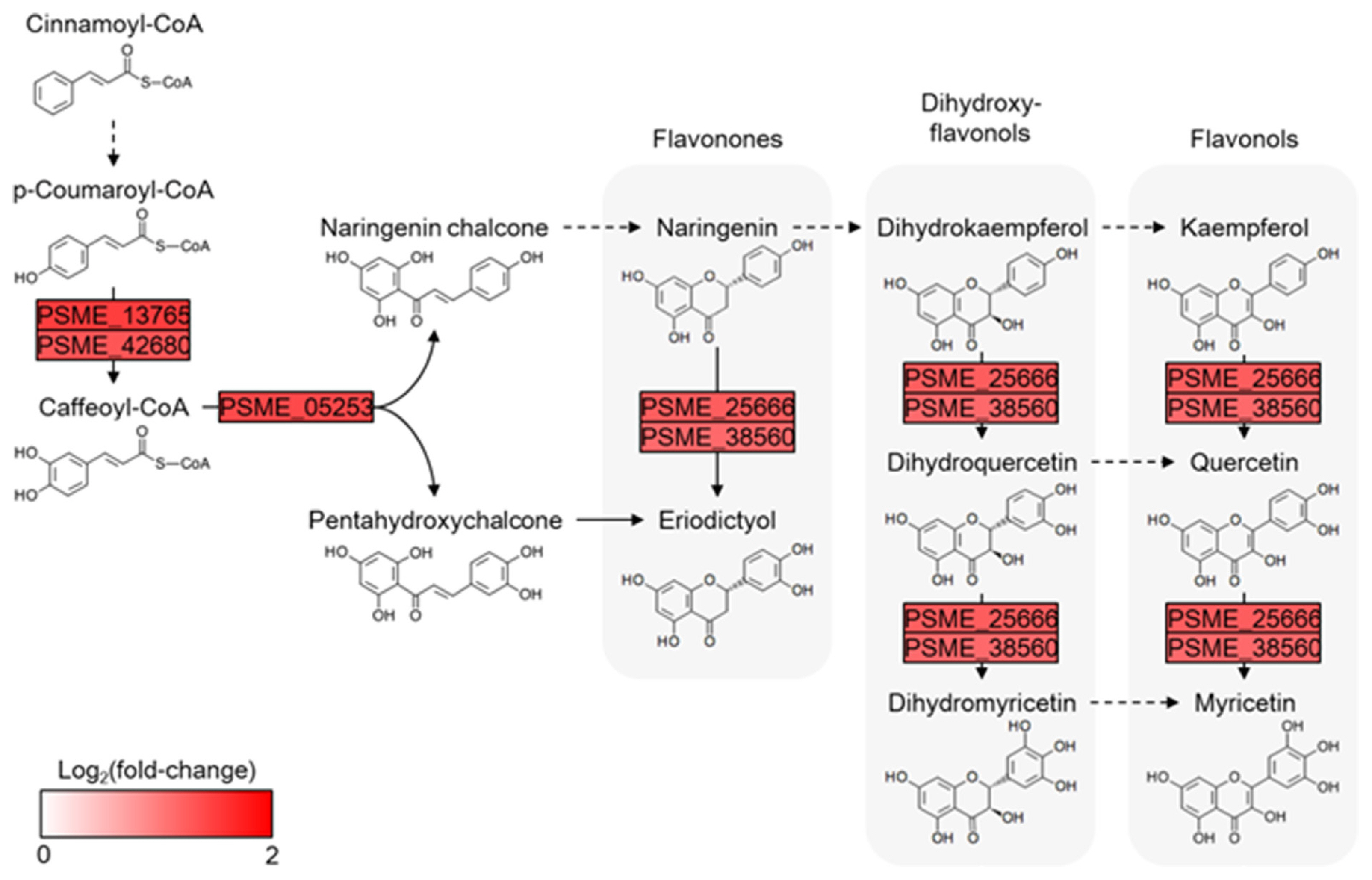
Fig. 5
Protein-protein interaction network highlighting up-regulated differentially expressed genes (DEGs) in Abies koreana seedlings exposed to drought with Aureobasidium pullulans AK10 treatment. This network, derived from the STRING database (https://string-db.org/), includes genes with interaction scores of 0.4 or above. Based on these interacting genes, enriched Gene Ontology (GO) terms for biological processes are depicted as rectangular nodes. Within the network, node size corresponds to the number of interactions for each gene, while edge thickness represents the strength of the interaction score.
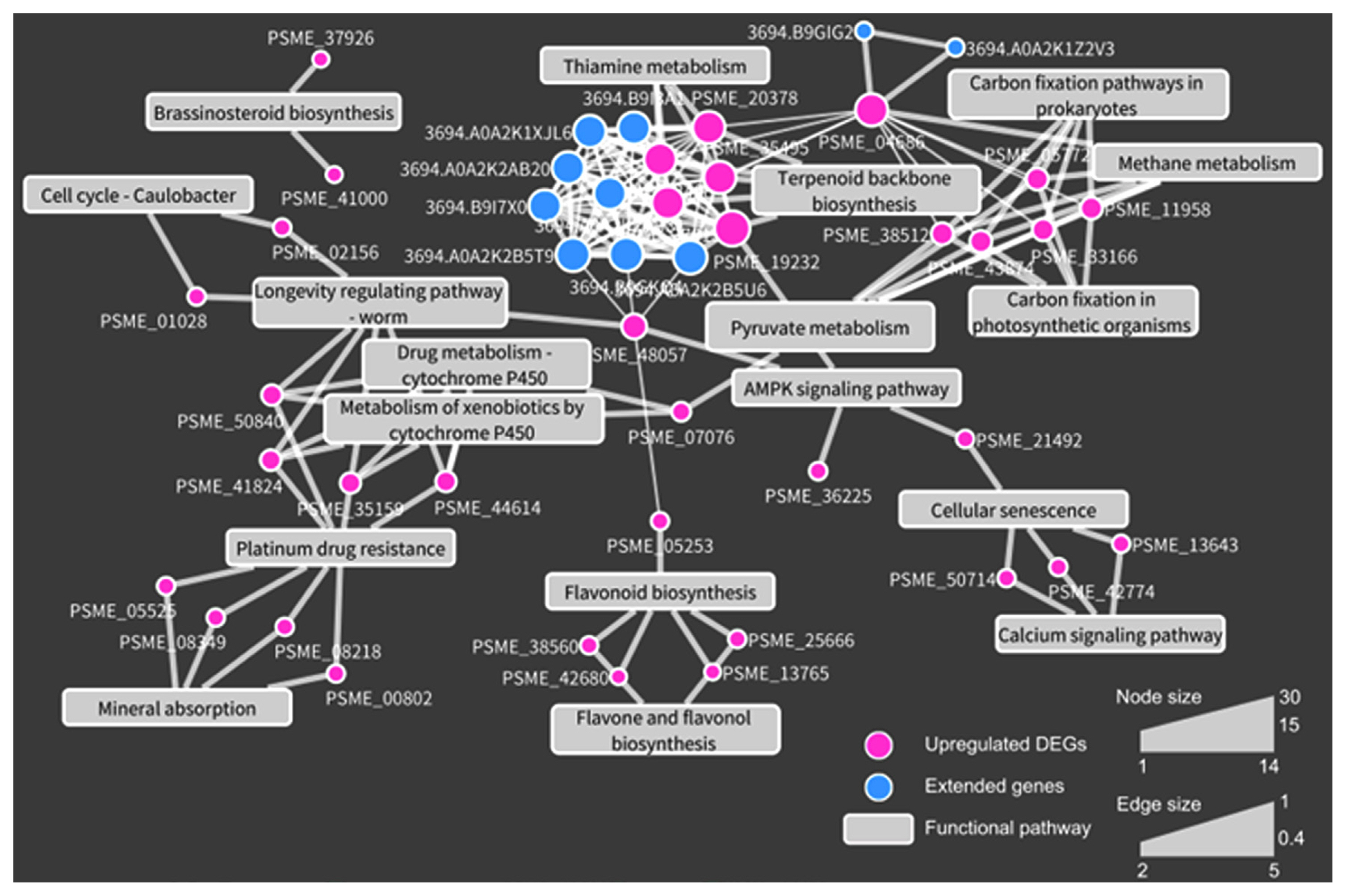
Table 1
Details on the reads processing
References
Azevedo, H., Lino-Neto, T. and Tavares, R. M. 2003. An improved method for high-quality RNA isolation from needles of adult maritime pine trees. Plant Mol. Biol. Rep. 21:333-338.


Baozhu, L., Ruonan, F., Yanting, F., Runan, L., Hui, Z., Tingting, C., Jiong, L., Han, L., Xiang, Z. and Chun-peng, S. 2022. The flavonoid biosynthesis regulator PFG3 confers drought stress tolerance in plants by promoting flavonoid accumulation. Environ. Exp. Bot. 196:104792.

Benjamini, Y. and Hochberg, Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57:289-300.


Chi, Z., Wang, F., Chi, Z., Yue, L., Liu, G. and Zhang, T. 2009. Bioproducts from Aureobasidium pullulans, a biotechnologically important yeast. Appl. Microbiol. Biotechnol. 82:793-804.



Di Francesco, A., Di Foggia, M., Corbetta, M., Baldo, D., Ratti, C. and Baraldi, E. 2021. Biocontrol activity and plant growth promotion exerted by Aureobasidium pullulans strains. J. Plant Growth Regul. 40:1233-1244.


Hardie, D. G. 2011. AMP-activated protein kinase: an energy sensor that regulates all aspects of cell function. Genes Dev. 25:1895-1908.



Kanehisa, M., Sato, Y. and Morishima, K. 2016. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 428:726-731.


Kim, D. W., Jeong, D. Y. and Park, H. C. 2020. Identification of molecular markers for population diagnosis of Korean fir (Abies koreana) vulnerable to climate change. Proc. Natl. Inst. Ecol 1:68-73.
Kim, H. J., Le, Q. K., Lee, M. H., Kim, T. S., Lee, H.-K., Kim, Y. H., Bae, K. and Lee, I.-S. 2001. A cytotoxic secocycloartenoid from Abies koreana. Arch. Pharm. Res. 24:527-531.



Kim, J.-K., Koh, J.-G., Yim, H.-T. and Kim, D.-S. 2017. Changes of spatial distribution of Korean fir forest in Mt. Hallasan for the past 10 years (2006, 2015). Korean J. Environ. Ecol 31:549-556.

Kim, K., Bu, Y., Jeong, S., Lim, J., Kwon, Y., Cha, D. S., Kim, J., Jeon, S., Eun, J. and Jeon, H. 2006. Memory-enhancing effect of a supercritical carbon dioxide fluid extract of the needles of Abies koreana on scopolamine-induced amnesia in mice. Biosci. Biotechnol. Biochem. 70:1821-1826.


Kim, Y.-S., Chang, C.-S., Kim, C.-S. and Gardner, M. 2011.
Abies koreana. The IUCN Red List of Threatened Species 2011:e.T31244A9618913. International Union for Conservation of Nature and Natural Resources, Gland, Switzerland. pp. 11.
LaouĂŠ, J., Fernandez, C. and OrmeĂąo, E. 2022. Plant flavonoids in mediterranean species: a focus on flavonols as protective metabolites under climate stress. Plants 11:172.



Li, H. and Durbin, R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754-1760.




Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., Marth, G., Abecasis, G. and Durbin, R. 2009. The Sequence Alignment/Map (SAM) format and SAMtools. Bioinformatics 25:2078-2079.


Liao, Y., Smyth, G. K. and Shi, W. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30:923-930.



Mandlik, A., Livny, J., Robins, W. P., Ritchie, J. M., Mekalanos, J. J. and Waldor, M. K. 2011. RNA-seq-based monitoring of infection-linked changes in Vibrio cholerae gene expression. Cell Host Microbe 10:165-174.



Mannaa, M., Han, G., Jung, H., Park, J., Kim, J.-C., Park, A. R. and Seo, Y.-S. 2023.
Aureobasidium pullulans treatment mitigates drought stress in Abies koreana via rhizosphere microbiome modulation. Plants 12:3653.



Mortazavi, A., Williams, B. A., McCue, K., Schaeffer, L. and Wold, B. 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5:621-628.



Park, J., Jeon, H. W., Jung, H., Lee, H.-H., Kim, J., Park, A. R., Kim, N., Han, G., Kim, J.-C. and Seo, Y.-S. 2020. Comparative transcriptome analysis of pine trees treated with resistance-inducing substances against the nematode Bursaphelenchus xylophilus. Genes 11:1000.



Peleg, Z. and Blumwald, E. 2011. Hormone balance and abiotic stress tolerance in crop plants. Curr. Opin. Plant Biol. 14:290-295.


Qureshi, R. and Sacan, A. 2013. Weighted set enrichment of gene expression data. BMC Syst. Biol. 7(Suppl 4):S10.



Rodriguez, M. C. S., Petersen, M. and Mundy, J. 2010. Mitogen-activated protein kinase signaling in plants. Annu. Rev. Plant Biol. 61:621-649.


Shao, H.-B., Song, W.-Y. and Chu, L.-Y. 2008. Advances of calcium signals involved in plant anti-drought. Comptes Rendus Biol. 331:587-596.

Shen, J.-L., Li, C.-L., Wang, M., He, L.-L., Lin, M.-Y., Chen, D.-H. and Zhang, W. 2017. Mitochondrial pyruvate carrier 1 mediates abscisic acid-regulated stomatal closure and the drought response by affecting cellular pyruvate content in Arabidopsis thaliana. BMC Plant Biol. 17:217.




Shukla, N., Yadav, R., Kaur, P., Rasmussen, S., Goel, S., Agarwal, M., Jagannath, A., Gupta, R. and Kumar, A. 2018. Transcriptome analysis of root-knot nematode (Meloidogyne incognita)-infected tomato (Solanum lycopersicum) roots reveals complex gene expression profiles and metabolic networks of both host and nematode during susceptible and resistance responses. Mol. Plant Pathol. 19:615-633.




Smith, A. M., Zeeman, S. C. and Smith, S. M. 2005. Starch degradation. Annu. Rev. Plant Biol. 56:73-98.


Szklarczyk, D., Gable, A. L., Lyon, D., Junge, A., Wyder, S., Huerta-Cepas, J., Simonovic, M., Doncheva, N. T., Morris, J. H., Bork, P., Jensen, L. J. and con Mering, C. 2019. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47:D607-D613.



Tiedge, K., Li, X., Merrill, A. T., Davisson, D., Chen, Y., Yu, P., Tantillo, D. J., Last, R. L. and Zerbe, P. 2022. Comparative transcriptomics and metabolomics reveal specialized metabolite drought stress responses in switchgrass (Panicum virgatum). New Phytol. 236:1393-1408.




Wang, L., Feng, Z., Wang, X., Wang, X. and Zhang, X. 2010. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26:136-138.



Watkins, J. M., Chapman, J. M. and Muday, G. K. 2017. Abscisic acid-induced reactive oxygen species are modulated by flavonols to control stomata aperture. Plant Physiol. 175:1807-1825.



Yang, L-L, Yang, L., Yang, X., Zhang, T., Lan, Y.-M., Zhao, Y., Han, M. and Yang, L.-M. 2020. Drought stress induces biosynthesis of flavonoids in leaves and saikosaponins in roots of Bupleurum chinense DC. Phytochemistry 177:112434.


Yang, Y. H., Dudoit, S., Luu, P., Lin, D. M., Peng, V., Ngai, J. and Speed, T. P. 2002. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 30:e15.



Ye, H., Liu, S., Tang, B., Chen, J., Xie, Z., Nolan, T. M., Jiang, H., Guo, H., Lin, H.-Y., Li, L., Wang, Y., Tong, H., Zhag, M., Chu, C., Li, Z., Aluru, M., Aluru, S., Schnable, P. S. and Yin, Y. 2017. RD26 mediates crosstalk between drought and brassinosteroid signalling pathways. Nat. Commun 8:14573.




- TOOLS
-
METRICS

-
- 0 Crossref
- 0 Scopus
- 647 View
- 98 Download
- ORCID iDs
-
Young-Su Seo

https://orcid.org/0000-0001-9191-1405 - Related articles



 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Supplement
Supplement Print
Print



