The population of the world is increasing day by day and therefore, food security is being challenged in many parts of the world. Most of the suffering is due to imbalances and malnutrition (Rasul et al., 2012). The substrates for the bodyŌĆÖs regular physiological processes are provided by the consumption of a balanced diet of nutrients (Kiani et al., 2022). Legumes serve as a nutritional complement to cereals; they are a good source of protein, essential amino acids, and minerals, and therefore, can be a replacement for meat. Mungbean (Vigna radiata) is commonly known as a green gram in most countries, especially in Pakistan. It is a well-known pulse contributing to human food consumption. Mungbean is certainly a primary food and rich in source of protein and amino acids (Habibullah et al., 2007).
The growth of pulses such as mungbean is recognized to fulfill the demands for food. Several biotic challenges for instance pathogenic bacterial and fungal organisms pose a negative impact on mungbean production, therefore, limiting the maximum output. Some of the fungal diseases of mungbean include Alternaria leaf spot, Cercospora leaf spot, powdery mildew, and anthracnose which contribute to a range of losses in production. A wide variety of research based on epidemiological studies has revealed that Alternaria alternata spores are the most commonly recognized fungal spore in the environment (Woudenberg et al., 2015). Alternaria black rot, Alternaria leaf spot, and Alternaria brown spot are the most significant phenotypes of plant diseases caused by A. alternata spores. Alternaria leaf spot damage is more prominent on aerial parts and approximately 80% of losses were recorded (Singh, 1987). Due to the pathogenŌĆÖs ability to spread via seed market globalization, long-distance airborne spore dissemination, and the effects of climate change, A. alternata represent a major risk of infection to horticulture crops all over the world (Mati─ć et al., 2020). The first case of Alternaria leaf spot in Korea was observed when researchers observed many mungbean plants with dark-brown leaf spots. This study was effective in studying the presence and control of this disease in mungbean (Jung et al., 2019).
A variety of control measures including integrated pest management are being in use to control the mungbean disease (Batzer et al., 2022). Utilization of host resistance is one of the operative and economically approachable techniques of disease management in crops, especially mungbean. Screening of resistant sources is the first step towards the exploitation of host resistance (Yadav et al., 2014). Different valuable and reliable methods are in use for the screening of mungbean reactions towards diseases like Cercospora leaf spot, powdery mildew, and anthracnose. First, these diseases (the foliar diseases) are screened where the pressure of the disease is high and then it is tested by the artificial inoculation of pathogen spores (Kasettranan et al., 2010). Resistance sources are screened by the artificial inoculation of spores of pathogens. For Africa, Asia, and Australia, several disease-resistant mungbean lines have been introduced (Batzer et al., 2022). One of the most useful strategies for coping with fungal infection is the use of fungicides. Fungicides are chemicals that are employed to stop or get rid of fungal infections in plants or seeds (Price et al., 2015). To control plant infections brought on by phytopathogenic fungus, chemical-based fungicides have been utilized (Deresa and Diriba, 2023).
The majority of main diseases in growing crops meant for the market have been successfully controlled by various fungicide types (Lonergan et al., 2015). The damaging effects of overusing these chemical disease control methods on agricultural fields result in a decline in soil fertility (Carrascal-Hern├Īndez et al., 2022).To avoid chemical use, the natural defensive mechanism known as gene silencing is commonly employed which both protects against viral infection and controls endogenous gene expression (Koch et al., 2016; ┼Āe─Źi─ć and Kogel, 2021).
RNAi is known as one of the important and promising approaches for genomic analysis. Research has shown that it has a significant impact on the improvement of crops. Without altering the expression of other genes, it permits the downregulation of gene activity. This technology can be used to exploit resistance thus preventing the economic loss due to crop pathogens every year. This technology is also useful in plant stress management (Mouyna et al., 2004). Target gene suppression through RNAi-generated signals in the plant, RNA silencing plays a crucial part in the research of host-induced gene silencing and the control of phytopathogenic fungi (Carrascal-Hern├Īndez et al., 2022). The expression of specific dsRNA sequences in the host plant and the silencing of target genes are necessary for the method of host-induced gene silencing through RNAi-generated signals in the host plant (Sang and Kim, 2020). The expression of dsRNA suggests that the target pathogen will transform the host plant by creating a hairpin dsRNA that contains its gene. Small RNAi homologous to the target RNA is produced during the target pathogenŌĆÖs mRNA breakdown, protecting plants from illnesses spread by phytopathogenic fungi (Carrascal-Hern├Īndez et al., 2022; Koch et al., 2013).
Therefore keeping in view the importance of Alternaria leaf spot, a comprehensive survey was done to access the infection of A. alternata on mungbean, both morphological and molecular characterization was done and pathogenicity was confirmed as well as the current study aimed to develop an RNAi-based resistance strategy against A. alternata in model host (tobacco, Nicotiana tabacum).
Materials and Methods
Survey and sample collection in mungbean-growing areas
Different surveys of almost three growing season of different mungbean-growing areas of Faisalabad, Layyah, and Bahawalpur districts, Pakistan were done comprehensively (map attached for sampling locations) (Fig. 1A) from July to September as the disease was most prevalent in that time duration. Most of the areas have seeds sowing through the drill method. The mungbean-growing areas are plain lands as this crop needed warm climatic conditions. Thirty samples were collected from each location. After collection the infected areas of mungbean leaves were cut and leaf discs were carefully shifted to potato dextrose agar (PDA) media plates after sterilization (Devappa and Thejakumar, 2016) and incubated at 28 ┬▒ 2┬░C for 6 to 7 days (Nagrale et al., 2013).
Morphological and molecular identification
The characteristics of the colony, colony growth rate, the occurrence of sporangia, and the dimensions of conidia were observed after seven days of incubation. The extraction of genomic DNA involves a variety of techniques and methods (Zhang et al., 2010). Fungal DNA was isolated using the extraction buffer method (Masoodi et al., 2021).
Amplification of ITS and Alt a 1 gene
The ITS regions of all the isolates were amplified by the polymerase chain reaction (PCR) using primers ITS1 and ITS4 (White et al., 1990). Briefly, the primers ITS1 (5ŌĆ▓-TCCGTAGGTGAACCTGCGG-3ŌĆ▓) and ITS4 (5ŌĆ▓-TCCTCCGCTTATTGATATGC-3ŌĆ▓) were used. The PCR protocol followed Chen et al. (2000) with some modifications, the final PCR reaction mixture of 50 ╬╝l contained 15 ╬╝M each of the two primers, 50 ng of ITS DNA template, 100 ╬╝M dNTPs, 2 mM MgSO4, and 1 U Thermo Scientific Taq DNA polymerase (Waltham, MA, USA).
To further confirm that the fungal isolates belong to the Alternaria species, the major allergen gene Alt a 1 (Deards and Montague, 1991) was amplified. The primers Alt-for (5ŌĆ▓-ATGCAGTTCACCACCATCGC-3ŌĆ▓) and Alt-rev (5ŌĆ▓-ACGAGGGTGAYGTAGGCGTC-3ŌĆ▓) were designed according to Hong et al. (2005).
Phylogenetic analysis
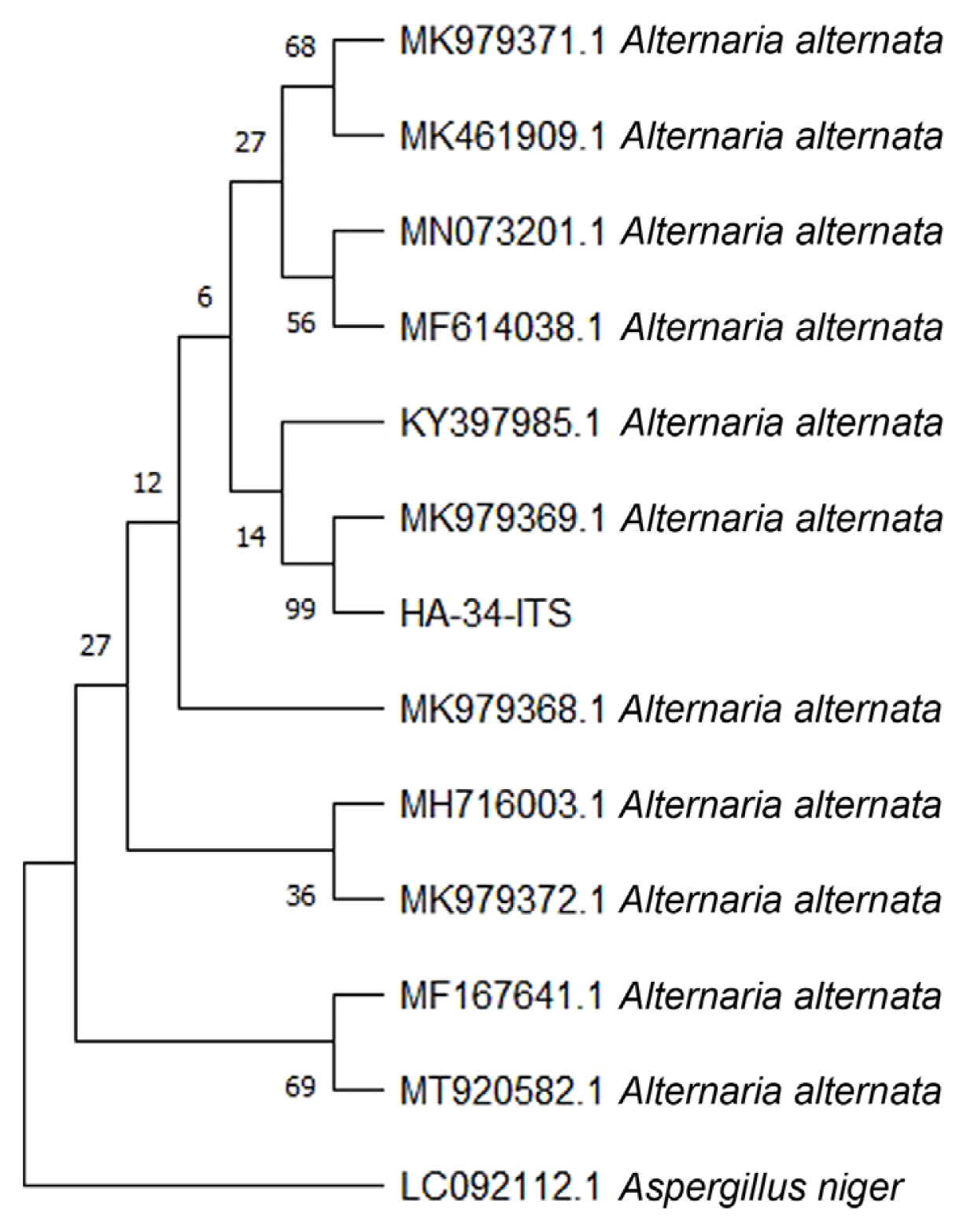
Isolated ITS PCR products were sequenced from Macrogen Korea, and their sequences were specifically analyzed by BLAST to identify them. The sequences from the GenBank database that indicated a substantial similarity with both isolates during BLAST were downloaded. Phylogenetic analysis was carried out MEGA X (https://www.megasoftware.net/) was used to conduct the phylogenetic analysis. The Maximum Composite Likelihood approach was used to build a neighbor-joining tree and bootstrap confidence values were obtained after 1,000 replications.
Diversity of mungbean germplasm for resistance against A. alternata
Fifty genotypes of mungbean (14104, 14108, 14113, 14114, 14115, 14117, 14123, 14128, 14129, 14133, 14134, 14141, 14150, 14165, 14180, 14198, 14203, 14205, 14208, 14215, 14243, 14250, 14256, 14261, 14265, 14266, 14279, 14289, 14291, 14292, 14294, 14295, 14299, 14300, 14304, 14305, 14306, 14310, 14311, 14314, 14349, 14368, 14369, 14378, 14438, 14443, 14473, 14478, 14480, and 14540) were evaluated for resistance to A. alternata under field conditions. The data on various pathological traits; disease incidence, disease intensity, leaf area, lesion area and infection percentage was observed.
Construction of PVX-based VIGS vector
Specific PCR amplification and cloning in PVX
Oligonucleotide primers were designed with the addition of restriction sites. SalI restriction site G/TCGAC was added at 5ŌĆ▓ end of Alt forward primer Alt-SalI-For (5ŌĆ▓-GTCGACATGCAGTTCACCACCATCGC-3ŌĆ▓) and ClaI AT/CGAT was added at 5ŌĆ▓ end of Alt Reverse primer Alt-ClaI-Rev (5ŌĆ▓-ATCGATACGAGGGTGAYGTAGGCGTC-3ŌĆ▓) as our desired cloning vector PVX (VIGS pgR107) contains ClaI-SmalI-SalI.
The disease-causing genes of the virus have been deleted and the modified cDNA of the virus genome is ligated into a binary vector (pGreen 0000). A PCR fragment amplified by Alt-SalI-For and Alt-ClaI-Rev primers was cloned at respective sites in the vector pgR107for the construction of the PXV-based RNAi construct used for VIGS in tobacco plants. Double restriction digestion initially with ClaI 1├Ś Tango buffer followed by incubation at 37┬░C for 1 h. A final 2├Ś concentration of Tango buffer along with a 2-fold excess of SalI was added after completion of initial digestion.
Construction of Agrobacterium tumefaciens electro-competent cells
Luria-Berani (LB) broth medium was cultured followed by agitation for 48 h at 28┬░C with the appropriate antibiotics, i.e., rifampicin and tetracycline (Wise et al., 2006). The culture was further transferred to fresh LB media and incubated at 28┬░C until the cell attained an OD600 value of 0.5 to 1.0 (1010 cells/ml). Cells were pelleted and re-suspended thrice and were finally suspended in 1.5 ml of cold filter-sterilized glycerol, aliquots were made and kept at ŌłÆ80┬░C.
Electroporation-based clone transformation in Agrobacterium tumefaciens
Agrobacterium-competent cells were electroporated using the method reported by (Sambrook et al., 1989). The electro-cell manipulator 600 (BTX, San Diego, CA, USA) was used to carry out the electroporation.
Agrobacterium-mediated infiltration of host plants
Following the procedure outlined by Yang et al. (2000), Agrobacterium cultures were introduced into the Nicotiana tabacum and mungbean plants. A. tumefaciens strain GV3101 was electroporated with the infectivity constructs. The bacteria were suspended in 10 mm MgCl2 after being pelleted down at 4,000 rpm for 15 min. One hundred and fifty g/ml of acetosyringone was administered to the bacterial suspension. After 3 h of incubation, the cells were then infiltrated into the fresh, properly matured leaves of N. tabacum that were 3 to 4 weeks old.
Statistical analysis
The experiments were repeated and the average of data collected was subjected to cluster analysis and principal component analysis (PCA) by using statistical software SPSS version 12 and version 5.0 of Statisca (Sneath and Sokal, 1973). K-means clustering was used to perform cluster analysis while the elucidation distances was done by using WardŌĆÖs method.
Semi-quantitative reverse transcriptase polymerase chain reaction
Based on the manufacturerŌĆÖs recommendations, total RNA was extracted from 10 samples (S1 to S2) and 50 more samples (PH1 to PH 50) using TRIzol (Invitrogen, Carlsbad, CA, USA). Superscript III reverse transcriptase (Invitrogen) and random primers were used for cDNA synthesis in accordance with the manufacturerŌĆÖs instructions (Ali et al., 2014). ╬▓-Actin gene of Alterneria alternata was used as an internal control (Li et al., 2021).
Results
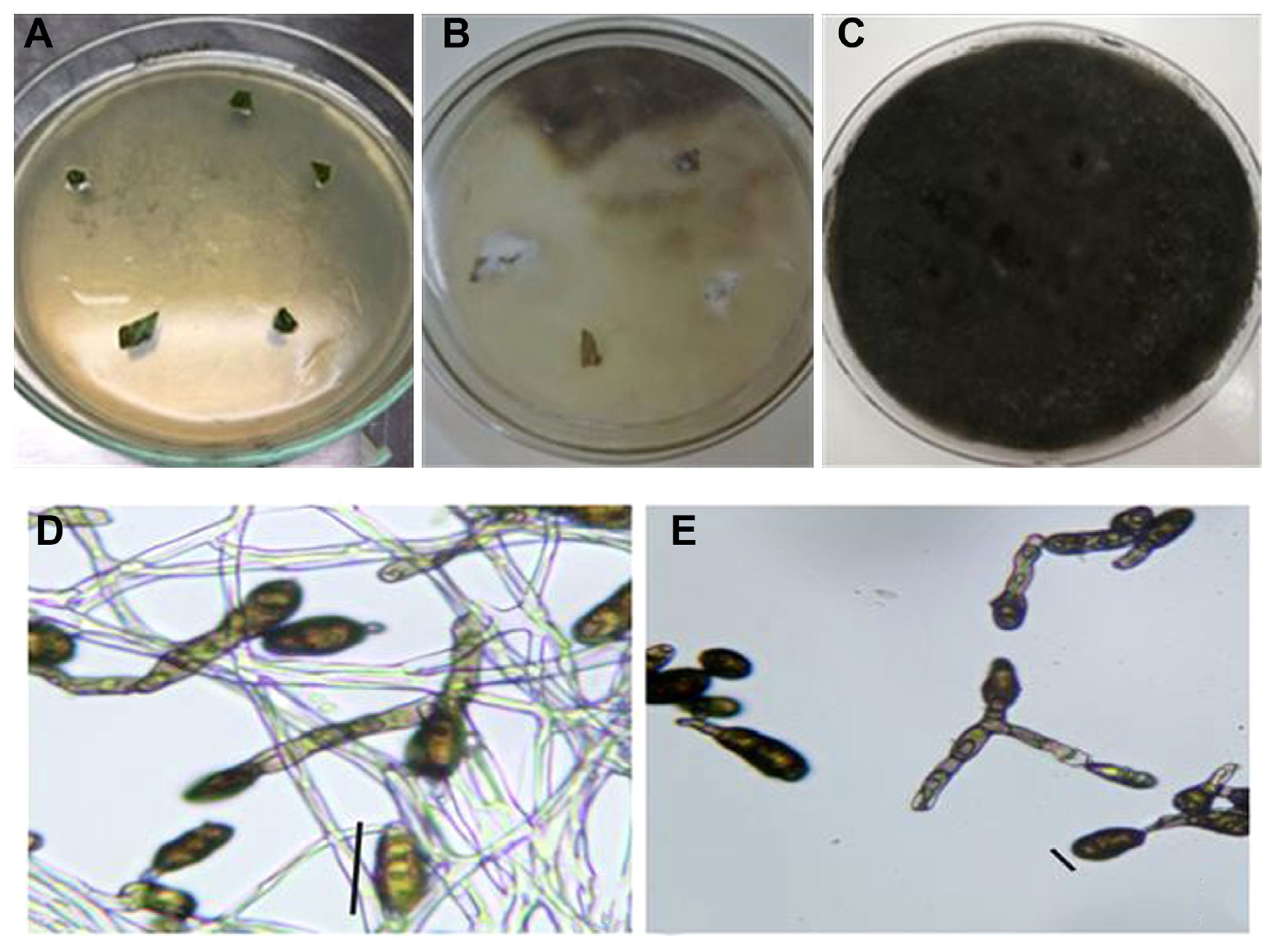
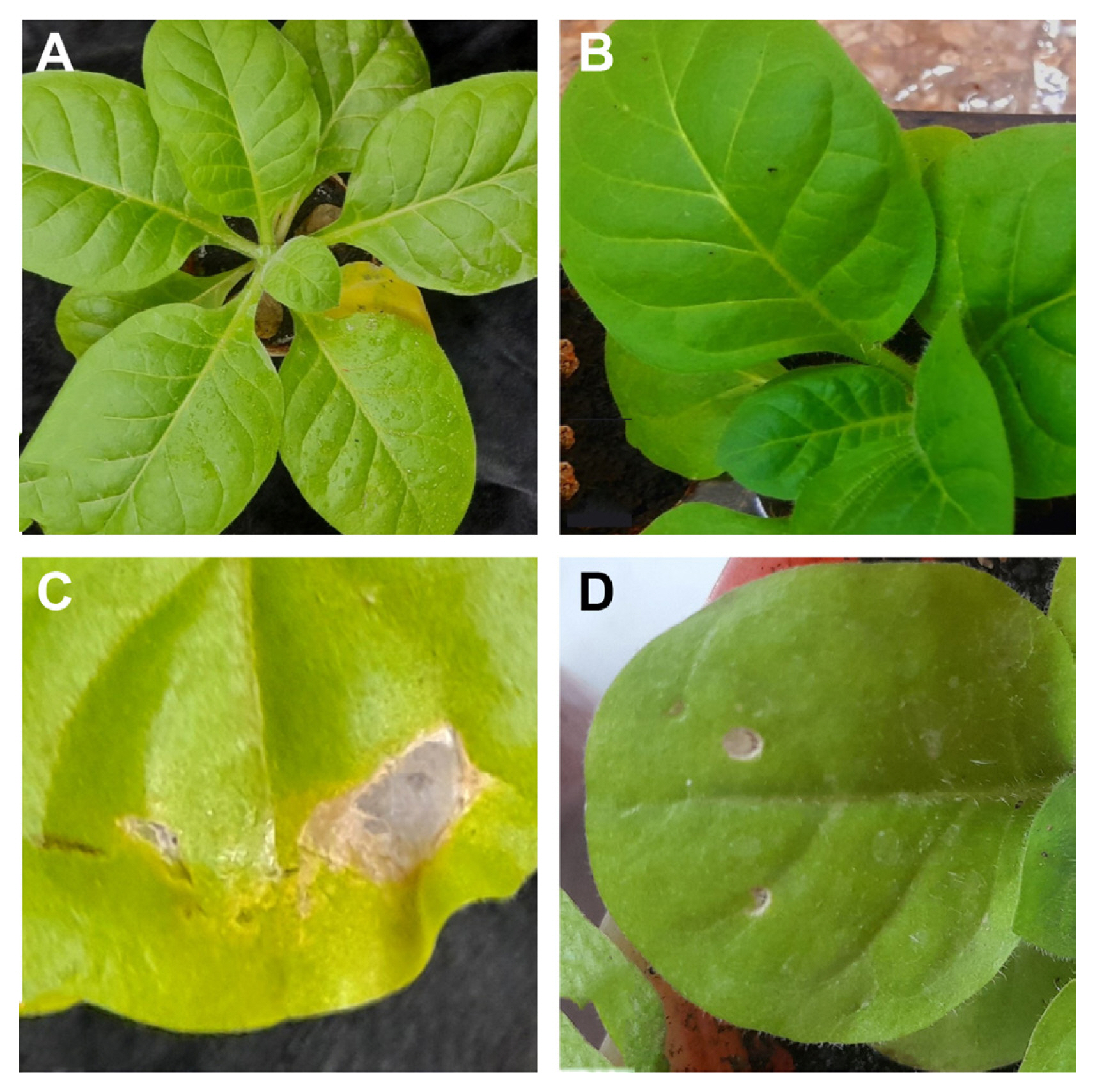
Collection and fungus culturing through diseased leaves of mungbean. A thorough study of the various mungbean-growing regions was conducted in Punjab, Pakistan (Fig. 1A). Small, circular, and atypically shaped necrotic spots were seen (Fig. 1C). Dark to gray-brown color spots were present (Fig. 1B). The spots typically had a yellow halo that surrounded them and had brown rings surrounding their pale centers.
Morphological identification
In order to verify and identify the exact fungal pathogen the infected leaf areas sterilized with 15% household sodium hypochlorite were cultured on PDA plates (Fig. 2A). Dull white woolly mycelia growth was observed on leaf discs (Fig. 2B). Mycelia quickly matured, changed into gray and eventually turned greenish-brown to black (Fig. 2C). The initial stages of the mycelium were translucent, later it changed to a greenish-brown color. It was randomly branched, multicelled, and septate (Fig. 2D). Conidiophores were both short and long, arranged in single and small groups. On conidiophores, conidia were arranged in chains of five or more. Smooth, elongated conidia ranged in color from light olivaceous to dark brown with overall length of 19 ╬╝m and typically 5 ╬╝m wide (Fig. 2E). Based on its morphological traits, the fungus was suspected to be a species from the genus Alternaria.
Diversity of mungbean germplasm for resistance against A. alternata
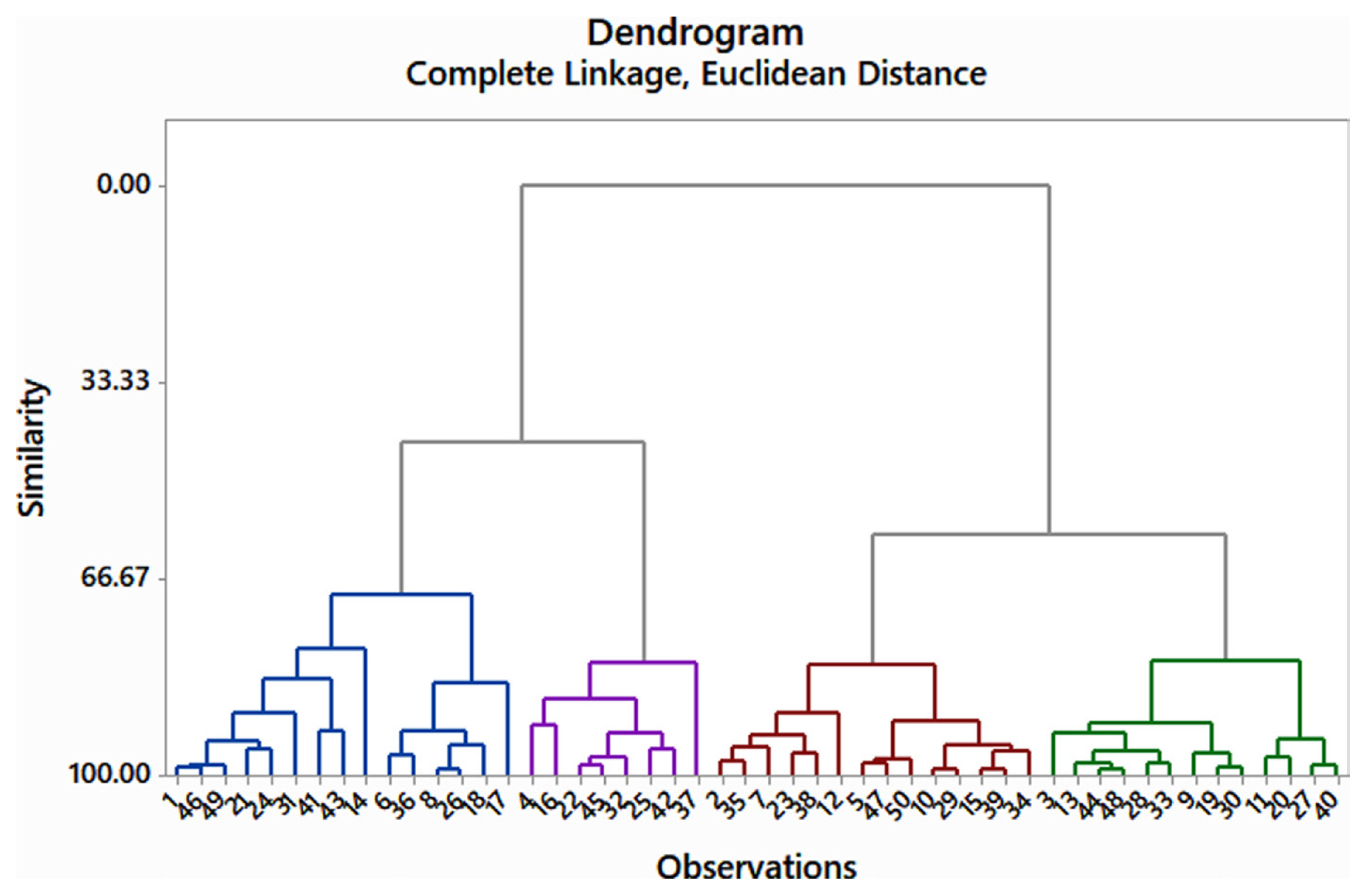
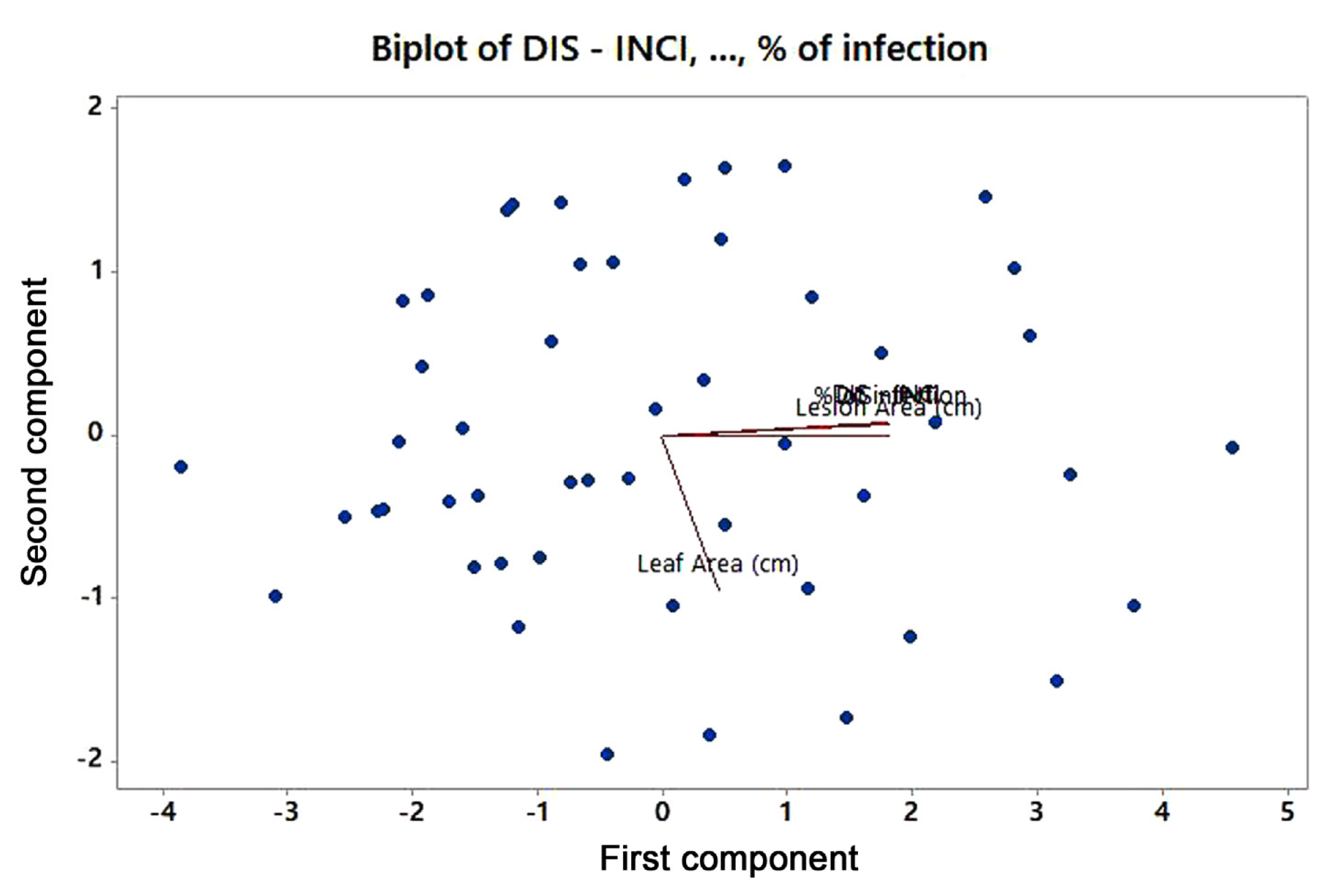
Fifty genotypes of mungbean were evaluated for resistance to A. alternata under field conditions (Table 1). The data on various pathological traits were subjected to cluster analysis. The results demonstrated that 50 genotypes of mungbean were grouped into four clusters based on resistance parameters under the influence of disease. Cluster analysis showed that the genotypes in cluster 4 have maximum disease intensity, incidence, and infection percentage (Table 2). However, on the other hand, minimum disease parameters were observed in members grouped into cluster 3. The most diverse members were of cluster 4 and cluster 3 derivatives followed by members of cluster 2 and cluster 4. The most similar clusters were cluster 2 and cluster 3. Similar results were shown by the distance metric between the clusters where clusters 3 and 4 had maximum value (61.5108). The members of different clusters in the dendrogram are displayed in (Fig. 3). The PCA biplot demonstrated that all the disease-related parameters were strongly correlated with each other while they showed a weak correlation with leaf area (Fig. 4).
Molecular identification
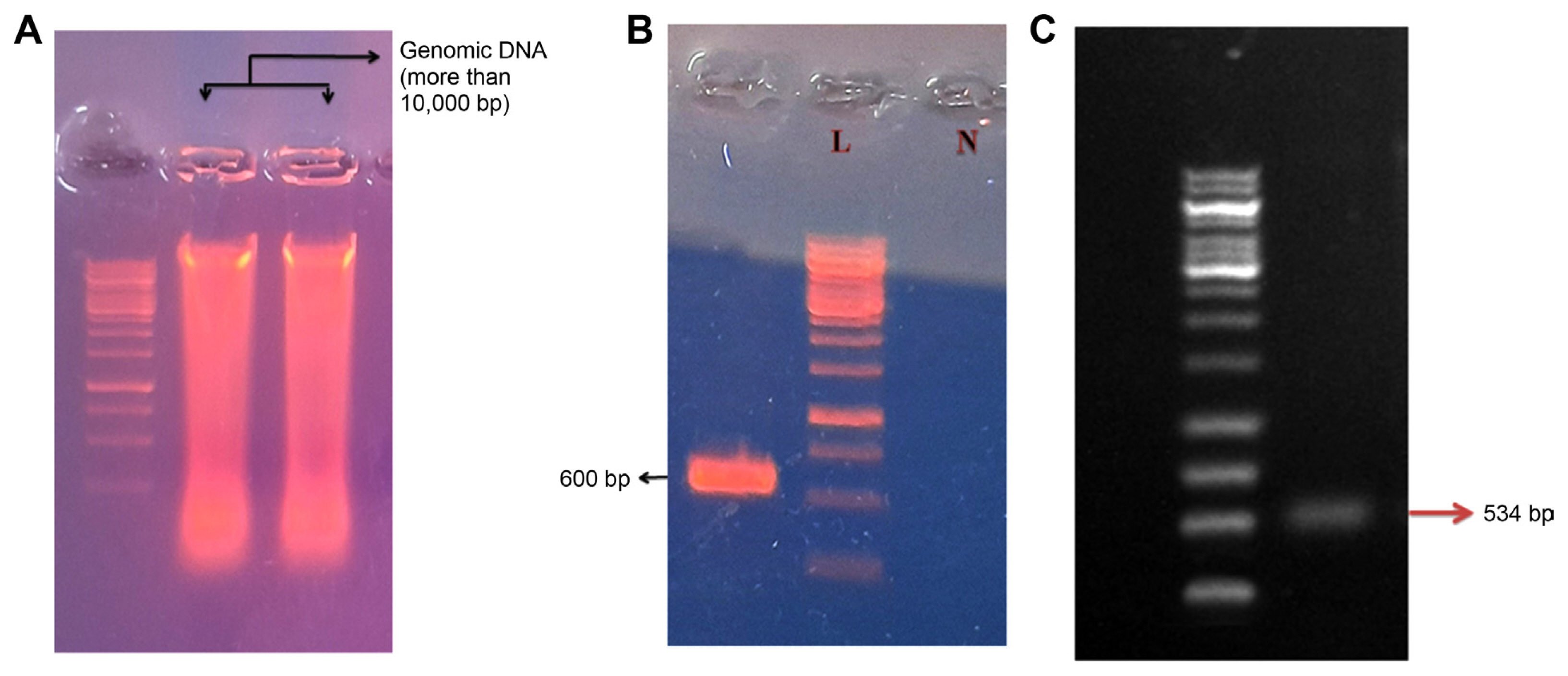
The genomic DNA of the fungus was isolated using the extraction buffer technique and further examined using agarose gel (Fig. 5A). To further confirm and characterize the fungal isolate the ITS region was amplified. A primer pair of ITS1 and ITS4 was employed for the PCR amplification. Approximately a 600 bp DNA fragment was amplified from the DNA of fungus samples and resolved on 1% agarose gel (Fig. 5B). Alt a 1 gene amplification was carried out by using Alt-for and Alt-rev primers. The PCR products were approximately 534 bp (Fig. 5C) typical of the size of Alt a 1 gene. Selected PCR products were purified and sequenced. The nucleotide sequence from ITS region of the isolate was submitted to NCBI with GenBank accession no. OR349656.
Sequencing and phylogenetics
The sequences were analyzed using BLASTn and the results revealed 100% similarity to A. alternata isolates. The nucleotide sequences of these isolates were downloaded from the GenBank and analyzed with HA-34-ITS, the nucleotide sequence of a PCR product selected from this study. A phylogenetic relationship established through a neighbour-joining statistical approach supported the results that the isolate HA-34-ITS is an isolate of A. alternata (Fig. 6). The morphological information with both visual observations of fungus growth and microscopic analysis revealed that the fungus pathogen infecting mungbean in the field is Alternaria sp. Amplification of Alt a 1 gene specific to the Alternaria spp. further supported the findings. Furthermore, the sequence analysis of the ITS region showed that leaf spot disease of mungbean is caused by the A. alternata.
Construction of PVX-based VIGS vector
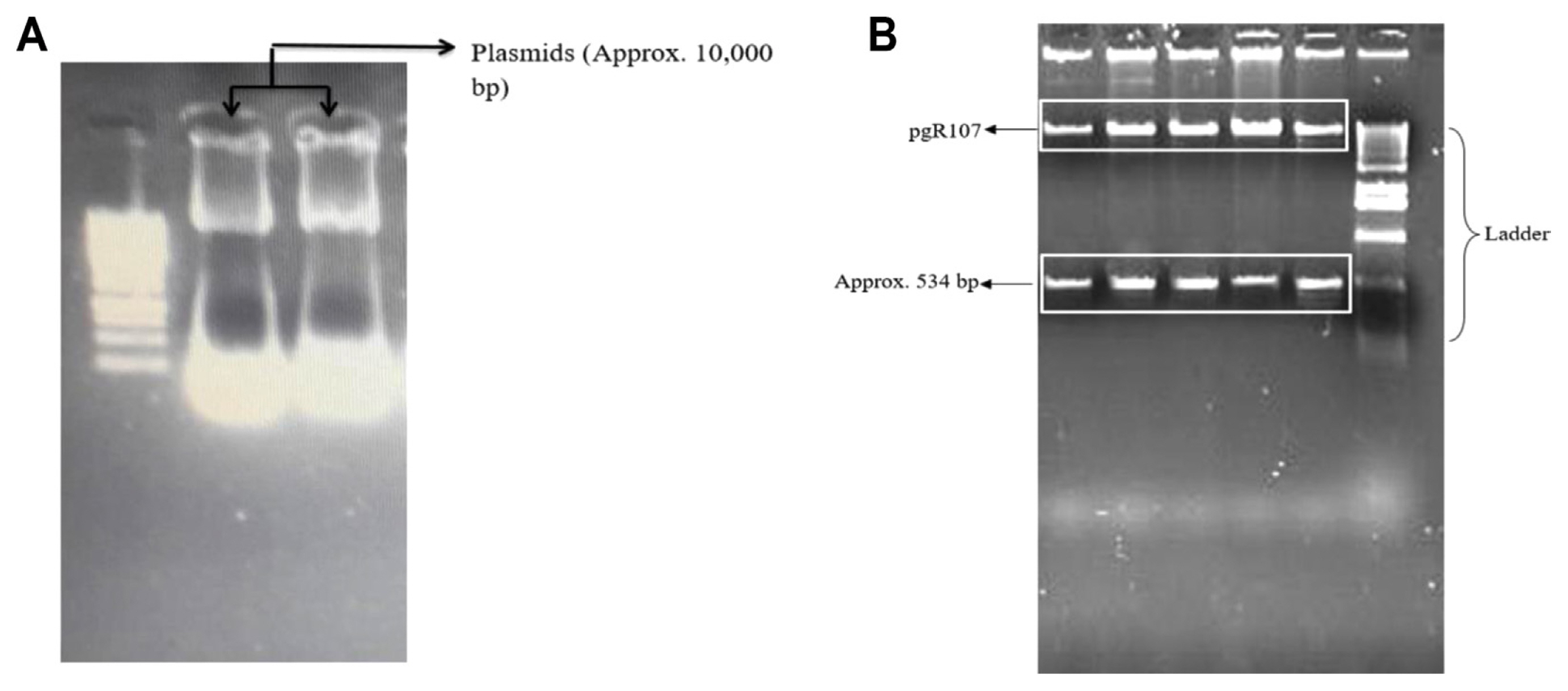
To use RNAi technique for gene silencing, PVX-based VIGS (pgR107) vector was selected. ClaI and SalI were used to ligate the PCR product in pgR107 vector. An amplified fragment of 534 bp was observed. DNA fragment specific to Alt a 1 gene was amplified using Alt-SalI-For and Alt-ClaI-Rev primers and ligated into PVX-based VIGS vector pgR 107 at SalI and ClaI site. The ligated plasmids were transformed into competent E. coli cells (DH5╬▒) and clones were selected on kanamycin. Plasmids were isolated from selected colonies (Fig. 7A) and restriction digestion was performed using SalI and ClaI enzymes. Restriction digestion generated two fragments each corresponding to vector pgR107 (about 10,000 bp) and Alt a 1 (534 bp). A clone designated as HA-107-3 (lane 3, Fig. 7B) was selected for transformation in Agrobacterium and VIGS analysis.
Transformation of Agrobacterium
For inoculation on the plant host and the VIGS analysis, the clone HA-107-3 was transformed into A. tumefaciens strain LBA 4404 and was grown on LB plates containing kanamycin and rifampicin as a selection marker. Bacterial colonies containing HA-107-3 were confirmed through colony PCR using Alt a 1 gene primers. Colony PCR produced a 534 bp fragment representative of the size of the Alt a 1 gene. The individual colonies containing HA-107-3 plasmid were grown in LB broth containing antibiotics.
Infiltration of VIGS construct on tobacco (Nicotianatabacum)
Agrobacterium culture containing PVX VIGS construct HA-107-3 was infiltrated on 20-day-old 60 tobacco plants. After 3 days, the inoculum of Alternaria fungus was sprayed on 50 plants (labeled from PVH1 to PVH50) keeping the remaining 10 plants as control (designated as treated control i.e., TC1, TC2, TC3, TC4, TC5, TC6, TC7, TC8, TC9, and TC10). Ten more plants were used as untreated control (neither infiltrated with PVX VIGS nor the fungus inoculum i.e., UC1, UC2, UC3, UC4, UC5, UC6, UC7, UC8, UC9, and UC10). Ten plants were used as inoculated control (only fungus inoculum was applied i.e., S1, S2, S3, S4, S5, S6, S7, S8, S9, and S10). The plants were covered with polythene bags and kept under controlled conditions. The plants were monitored on alternate days. The symptoms of fungus infection started appearing on the inoculated control plants after 5 days. All the 10 plants displayed visible symptoms of fungal infection. Moving toward the 50 plants (PH1 to PH50) infiltrated with PVX VIGS and fungus inoculum, the symptoms were delayed with mild infection in some plants.
Multiple observations were derived from the infectivity analysis (Table 3). Control plants (TC1 to TC10) displayed no symptoms, TC1 is shown in (Fig. 8A). All fungal inoculated plants (S1 to S10) showed clear symptoms of fungal infection, S1 is shown in (Fig. 8C). Plants infiltrated only with PVX VIGS (UC1 to UC10) displayed no symptoms. In 50 plants infiltrated with PVX VIGS and fungus inoculum (PH1 to PH50), the symptoms were delayed up to 15 days and very mild infection was observed compare to the control plants. Out of 50 inoculated plants, 12 plants (PH1, PH6, PH7, PH9, PH11, PH14, PH15, PH30, PH33, PH41, PH45, and PH50) showed no infection and 9 plants (PH2, PH4, PH5, PH8, PH19, PH21, PH31, PH35, and PH42) showed delayed on set of disease, PH1and PH2 is shown in Fig. 8B and D, respectively. Remaining 29 plants (PH3, PH10, PH12, PH13, PH16, PH17, PH18, PH20, PH22, PH23, PH24, PH25, PH26, PH27, PH28, PH29, PH32, PH34, PH36, PH37, PH38, PH39, PH40, PH43, PH44, PH46, PH47, PH48, and PH49) displayed the symptoms of fungal infection. These results showed that the symptom development was associated with the expression of Alt a 1 gene that is precisely responsible for virulence and pathogenicity of Alterneria alternata, hence giving evidence that the target gene (Alt a 1) was silenced. Support for this evidence comes from the observation that fungal infection was reduced to a considerable extent only in plants that were infiltrated with PVX VIGS.
Expression analysis using semi-quantitative reverse transcriptase polymerase chain reaction
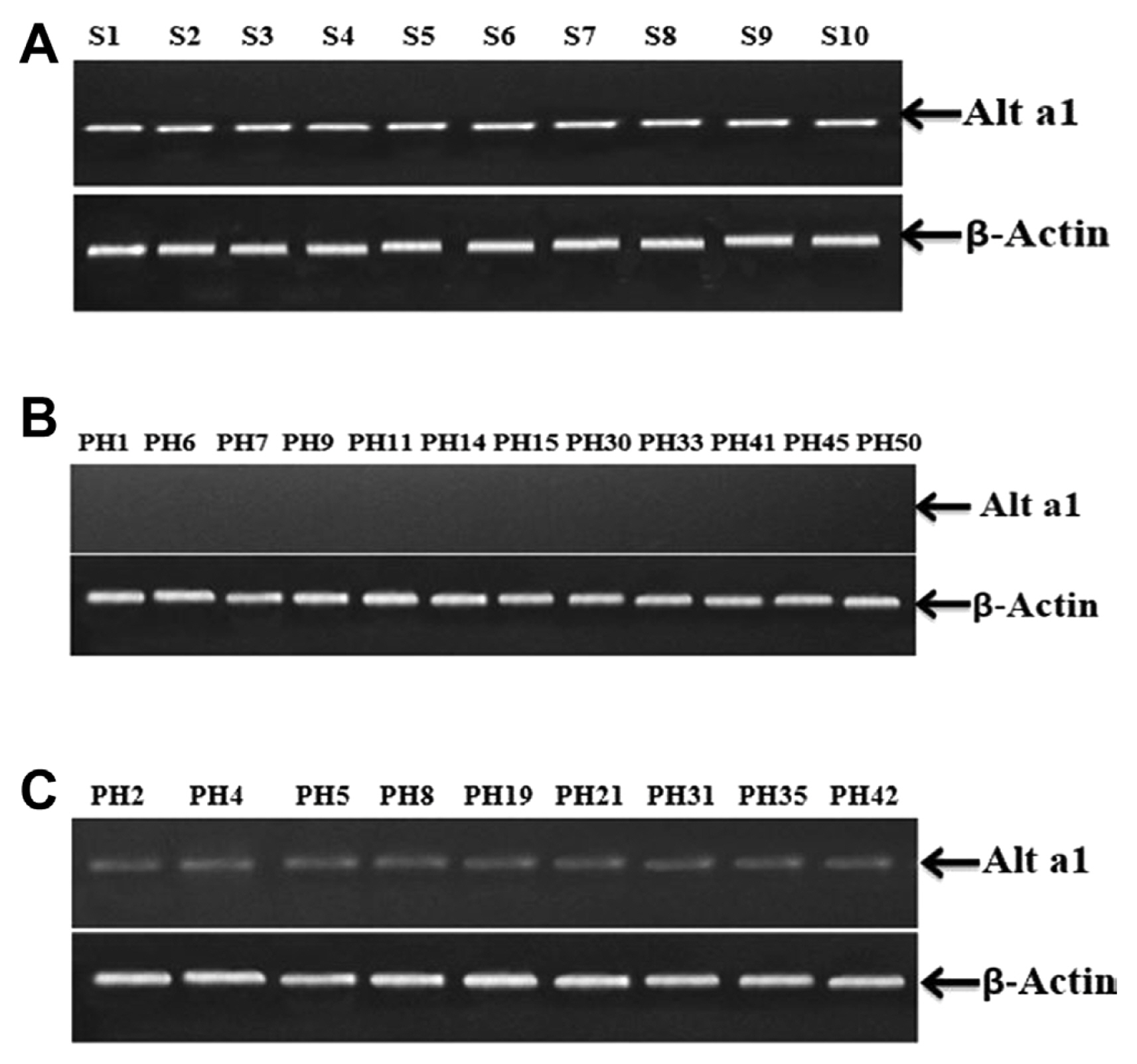
Semi-quantitative reverse transcriptase polymerase chain reaction was performed to further confirm the efficacy of PVX VIGS. Samples from the inoculated control on which only fungus inoculum was applied (S1, S2, S3, S4, S5, S6, S7, S8, S9, and S10) displayed high expression of Alt a 1 gene (Fig. 9A). Results from the analysis of 50 plants infiltrated with PVX VIGS were according to the phenotypic impression. Twelve plant samples (PH1, PH6, PH7, PH9, PH11, PH14, PH15, PH30, PH33, PH41, PH45, and PH50) displayed almost negligible Alt a 1 expression (Fig. 9B). While 9 plant samples (PH2, PH4, PH5, PH8, PH19, PH21, PH31, PH35, and PH42) visibly shown relatively low Alt a 1 expression (Fig. 9C). Furthermore, there was no alteration in the expression of actin throughout the analysis.
Discussion
Mungbean is an economically important crop with numerous practical applications (Brar et al., 2022). Every year, economic loss occurs as a result of fungal and viral infections that lead to reduced mungbean production as a whole (Schreinemachers et al., 2015; Tak and Kumar, 2020). Fungal diseases pose the biggest threat to crops grown for human consumption (Bankole and Adebanjo, 2003; Fisher et al., 2012). The fungus A. alternata, which causes Alternaria leaf spot, can lower plant grains yield and quality (Bas─▒m et al., 2017; Nauanova, 2020). Each year, this disease reduces economic output. It is a foliar mungbean disease that can affect the crop at various growth stages.
A comprehensive survey of different mungbean-growing areas of Punjab, Pakistan was done. Leaf spot disease was noticed in various mungbean varieties. The symptoms on numerous different plants were marked with dark-brown leaf spots. There were abnormally shaped tiny, round, and irregular necrotic patches. There were concentric bands of dark to gray-brown color dots. The spots often had brown rings around their light-colored cores and a yellow halo around them. A study carried out in Korea noticed the same brown spots on the leaves of mungbean that were brown to black (Jung et al., 2019). Similar brown spots were also noticed in kidney bean plants in the fields in China (Jin et al., 2022). Previously from Pakistan, there is a lack of information available about the pathogen causing leaf spot diseases in mungbean. However, in the present study, the causal organism responsible for leaf spot disease is identified as A. alternata. The leaf spots recorded in this study were similar to the previous reports of A. alternata infection on mungbean from Korea (Jung et al., 2019) and kidney beans in China (Jin et al., 2022).
To verify and identify the accurate fungal species the infected leaf areas were cultured on PDA plates. The initial stages of the mycelium were translucent, later it changed to a greenish-brown color. It was randomly branched, multicelled, and septate. Conidiophores were both short and long, arranged in single and small groups, and typically 2 to 6 nm in length. On conidiophores, conidia were arranged in chains of five or more. Smooth, elongated conidia ranged in color from light olivaceous to dark brown. The morphological characters observed were identical to the description of A. alternata explained by Woudenberg et al. (2015) and Jin et al. (2022). Microscopic observations carried out by Aung et al. (2020) and Nagrale et al. (2013) further confirmed that the characteristics of the isolates mentioned above were similar to Alternaria sp.
Mungbean genotypes were assessed for their resistance against A. alternata. The results demonstrated that transgressive segregates could be developed between the members of cluster 4 and the members of either cluster 2 or cluster 3 to find some genotypes with enhanced resistance against A. alternata. Cluster analysis and distances between them determined diversity among germplasm (Adhikari et al., 1994, 1999; Mukherjee et al., 1999). The PC biplot showed positively closed correlation. The biplot display provides differential interactions among various traits (Gabriel, 1971; Onasanya et al., 2004). To ensure correct identification of the isolates, molecular analysis of the genomic DNA was performed (Cenis, 1992; Saleem and El-Shahir, 2022; Watanabe et al., 2010). For amplification and molecular characterization of fungal pathogens, the ITS sequences were used according to Toledo et al. (2013). A 600 bp amplified fragment was seen on the gel. The same PCR amplification analysis with the ITS region was used in the study conducted by Aloi et al. (2021) and Tarini et al. (2010). To further confirm that the fungal isolates belong to the Alternaria species, the major allergen gene Alt a 1 was amplified. PCR products were approximately 534 bp long. Results were similar to the studies carried out by (Aung et al., 2020; Gabriel et al., 2017; Hong et al., 2005; Nayyar et al., 2017; Sk├│ra et al., 2015). RNAi technique was used for gene silencing. Agrobacterium-mediated VIGS system could potentially be effective in silencing genes (Zhou et al., 2012). Alt a 1 gene was silenced by following VIGS using PVX in tobacco plants. PVX-based VIGS (pgR107) vector was selected. This PVX-based RNAi construct when agro-infiltrated in the tobacco plant was able to induce resistance against fungus pathogens. Results showed that 12 out of the 50 plants remained symptomless, whereas 9 plants exhibited a delayed onset of fungal infection. These findings revealed confirmation that the target gene (Alt a 1) was silenced as the symptom development is associated with the expression of the Alt a 1 gene, which is specifically responsible for virulence and pathogenicity of A. alternata.
Furthermore, to confirm the phenotypic expression, semi-quantitative reverse transcriptase PCR was performed to investigate the expression of Alt a 1 gene at molecular level. S1, S2, S3, S4, S5, S6, S7, S8, S9, S10 (from inoculated control plants that were sprayed with fungus inoculum only) displayed the presence and adequate expression of Alt a 1 gene. While out of 50 plants infiltrated with (PVX(VIGS) + Inoculum), 12 plant samples (PH1, PH6, PH7, PH9, PH11, PH14, PH15, PH30, PH33, PH41, PH45, and PH50) displayed almost negligible Alt a 1 expression that is also confirmed by phenotypic expression of plant samples that was showing no infection symptoms. While 9 plant samples (PH2, PH4, PH5, PH8, PH19, PH21, PH31, PH35, and PH42) visibly showed relatively low Alt a 1 expression, these 9 plants showed delayed and minor infection symptoms apparently. ╬▓-Actin was used as control and showed no difference in the expression. Both the phenotypic results and molecular evidence hence confirm that PVX(VIGS) played a promising role in RNAi-mediated gene silencing. Conclusively, these findings suggest evidence that VIGS was effective in controlling the Alterneria alternata infection in tobacco.
The stable expression of this RNAi construct in host can provide more effective and durable resistance against A. alternata. Furthermore, as the construct is designed to silence the Alt a 1 gene, a major allergen-producing gene the construct will be helpful in minimizing or restricting the expression of allergen in plants. Therefore, could act as a model system in plant protection.




 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Print
Print






