 |
 |
| Plant Pathol J > Volume 40(1); 2024 > Article |
|
Abstract
Gardenia (Gardenia jasminoides) is a popular and economically vital plant known for its ornamental and medicinal properties. Despite its widespread cultivation, there has been no documentation of plant viruses on gardenia yet. In the present study, gardenia leaves exhibiting symptoms of plant viral diseases were sampled and sequenced by both metatranscriptome and small RNA sequencing. As a consequence, bean common mosaic virus (BCMV) was identified in gardenia for the first time and named BCMV-gardenia. The full genome sequence of BCMV-gardenia is 10,054 nucleotides (nt) in length (excluding the poly (A) at the 3′ termini), encoding a large polyprotein of 3,222 amino acids. Sequence analysis showed that the N-termini of the polyprotein encoded by BCMV-gardenia is less conserved when compared to other BCMV isolates, whereas the C-termini is the most conserved. Maximum likelihood phylogenetic analysis showed that BCMV-gardenia was clustered closely with other BCMV isolates identified outside the leguminous plants. Our results indicated that the majority of BCMV-gardenia virus-derived small interfering RNAs (vsiRNAs) were 21 nt and 22 nt, with 21 nt being more abundant. The first nucleotide at the 5′ termini of vsiRNAs derived from BCMV-gardenia preferred U and A. The ratio of vsiRNAs derived from sense (51.1%) and antisense (48.9%) strands is approaching, and the distribution of vsiRNAs along the viral genome is generally even, with some hot spots forming in local regions. Our findings could provide new insights into the diversity, evolution, and host expansion of BCMV and contribute to the prevention and treatment of this virus.
Gardenia jasminoides, known as gardenia, is a perennial flowering shrub with lustrous dark-green leaves and fragrant white blooms, classified within the genus Gardenia in the Rubiaceae family (Jin et al., 2023). As a popular houseplant, gardenia has a history of cultivation in China spanning over 1,000 years, and was introduced to Europe and America in the mid-18th century (Xu et al., 2020). In addition to its ornamental value as a garden plant, gardenia holds significant medicinal value. Each part of the plant, including its fruits, leaves, flowers, and roots, possesses therapeutic properties, underscoring its role as a crucial traditional Chinese medicinal plant (Xu et al., 2023). Notably, the fruit of gardenia is extensively employed in therapeutic applications, exhibiting numerous pharmacological benefits, including its roles as an antidiabetic, choleretic, antiphlogistic, diuretic, hemostatic, antidepression, and anticancer agent (Cho, 2022; Jin et al., 2023). The economic benefits of gardenia have led to an expansion of its cultivation area. However, the growth in cultivation has been accompanied by the emergence of various diseases caused by different pathogens or pests, including fungi, nematodes, and insects (Cao et al., 2018; Fang et al., 2023; Lu et al., 2023). However, to date, there have been no documented cases of plant virus infection in gardenias.
Bean common mosaic virus (BCMV) is a significant plant virus that impacts the yield and quality of common beans (Phaseolus vulgaris L.) and other leguminous crops (Feng et al., 2019). BCMV belongs to the genus Potyvirus, the largest genus of the viruses affecting land plants. The impact of BCMV on crop yield is substantial, with an average loss of 50%, and in some cases, it can reach to 100%, depending on the cultivar and the stage of crop infection (Asensio-S.-Manzanera et al., 2006). The transmission routes of BCMV are very diverse and can be transmitted through seeds, pollen, aphids, and mechanical inoculation (Tang and Feng, 2022). In contrast to some potyviruses like soybean mosaic virus (SMV), which exhibits a narrow host range limited to a few plants (Hajimorad et al., 2018), BCMV exhibits an extensive host range, infecting over 100 plants across 44 genera. Nevertheless, the hosts of BCMV are generally thought to be restricted to leguminous plants (Sáiz et al., 1994; Wang et al., 2023). With the rapid development of high-throughput sequencing technology, more and more plants outside legume species have been reported as novel hosts of BCMV, including Cudrania tricuspidata (Seo et al., 2015), Alpinia purpurata (Larrea-Sarmiento et al., 2020), and Nandina domestica (Su et al., 2022).
In this study, we conducted metatranscriptome and small RNA sequencing on gardenia leaves exhibiting typical plant virus symptoms to study the potential plant virus infecting gardenia. The virome analysis results indicated that a BCMV isolate (with about 92% nucleotide similarity to the closest sequence) was the sole plant virus in the sample. The full genome sequence of BCMV identified in gardenia was recovered by a combination of reverse transcription polymerase chain reaction (RT-PCR) and rapid amplification of cDNA ends (RACE). Moreover, we also characterized the antiviral small-interfering RNA response to BCMV in gardenia. Our study signifies the first documented case of BCMV infecting gardenia and represents the first research to study the plant virus pathogen in this plant species.
Leaves and twigs of gardenia showing mosaic, mottle, or chlorotic symptoms were collected from a small garden at Ningbo University in Ningbo City, Zhejiang Province, China. After collection, samples were rapidly frozen in liquid nitrogen to preserve RNA integrity. Following grinding, approximately 50 mg of the ground powder underwent total RNA extraction with the TRIzol reagent (Invitrogen, Carlsbad, CA, USA). After the assessment of RNA quality and quantity, RNAprep Pure Plant Kit (Polysaccharides & Polyphenolics-rich) (TianGene, Beijing, China) was employed for total RNA extraction, and the samples were subsequently sent to Novogene (Beijing, China) for both metatranscriptome (RNA sequencing [RNA-Seq]) and small RNAs sequencing.
The sequencing library preparation and sequencing were carried out by Novogene. In brief, for RNA-Seq, the RNA sequencing library was prepared by deleting ribosomal RNAs from the total RNAs using the Ribo-Zero rRNA Removal Kit (Plant) (Illumina, San Diego, CA, USA), following the manufacturer’s instructions. Subsequently, the Illumina Novaseq 6000 system was employed for sequencing in a paired-end mode with a read length of 150 bp on both ends (150 bp × 2). For small RNA sequencing, the small RNA sequencing library was prepared with TrueSeq Small RNA Library Prep Kit (Illumina) and the sequencing was carried out on an Illumina HiSeq 4000 system with a read length of 50 bp.
Raw RNA-Seq dataset was processed to remove adapter regions and low-quality reads using fastp (Chen et al., 2018). Reads shorter than 36 nucleotides were removed. De novo assembly of the remaining reads were conducted using Trinity assembler (version 2.8.5) (Hass et al., 2013). To identify and annotate the virus-associated contigs, the assembled transcriptome contigs were subjected to BLASTx/BLASTn analysis against the NCBI (https://www.ncbi.nlm.nih.gov) NR and NT databases (accessed on May 2023) (Altschul et al., 1990). Additionally, a prebuilt RdRP Hidden Markov Model database was searched using the hmmsearch tool embedded in HMMER (http://hmmer.org/) to detect highly divergent viruses which may exist.
Total RNA was extracted from leaf samples of eight plants, including five symptomatic plants and three asymptomatic plants, using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. To confirm the infection of potyvirus, the potyvirus degenerate primers PotyF and PotyR (Marie-Jeanne et al., 2000) were used in RT-PCR. Subsequently, to recover the full length of BCMV, the whole genome regions were amplified by overlapping RT-PCR and RACE. Each amplicon was inserted into the pClone007 Blunt Simple Vector (Tsingke, Beijing, China) and sequenced by Sanger sequencing. The primers used in this study are listed in Supplementary Table 1.
To confirm the evolutionary status of BCMV-gardenia in the genus Potyvirus, genome sequences of representative members within the genus Potyvirus were retrieved from NCBI GenBank database. The polyprotein encoding regions of these viruses were extracted and aligned using MAFFT (Katoh and Standley, 2013). A phylogenetic tree deduced based on the maximum likelihood method was constructed using IQ-TREE (v1.6.6) (Nguyen et al., 2015). The nucleotide substitution model GTR + F + I + G4, selected by ModelFinder (Kalyaanamoorthy et al., 2017), was applied. Confidence in the topology was assessed using 1,000 bootstrap replicates.
To study the evolutionary relationship among different isolates of BCMV identified in various hosts, the BCMV isolate sequences were retrieved from NCBI GenBank database and similar evolutionary analysis was carried out with the best-fitted model selected by ModelFinder.
The adapter sequence was removed using the cutadapt tool (v1.16) (Martin, 2011). Reads that were 18 nt to 30 nt and contained no ambiguous nucleotides (‘N’) were retained as clean reads. These clean reads were then clustered into unique reads using an in-house Python script. The remaining small RNA clean reads were subsequently aligned to the BCMV-gardenia virus reference genome (accession no. OR832357) using the bowtie tool (Langmead et al., 2009), allowing for up to one mismatch. The profile of virus-derived small interfering RNA (vsiRNAs), including length distribution, 5′ terminal nucleotide preference, genome distribution, and polarity, was explored through the analysis of small RNAs that could be mapped to the virus reference genome.
To elucidate the potential pathogens of gardenia exhibiting disease symptoms, leaves and twigs showing symptoms such as mosaic, chlorosis were collected from a small garden in Ningbo University, Ningbo City, Zhejiang Province, China (Fig. 1). About 5-7 different leaves were selected randomly to extract the total RNA for both RNA-Seq and small RNA sequencing. After RNA-Seq, a total of 43,103,034 reads were produced. Following quality control, more than 41,319,142 reads (95.86%) were retained for sequence assembly. For small RNA sequencing dataset, after strict quality control, only 14,807,281 of the 30,234,844 raw reads remained for subsequent analysis (Table 1).
Then the clean reads in RNA-Seq library were assembled to long contigs and annotated to NCBI NR/NT reference database. A large contig with a length of 10,043 nt was annotated to a BCMV isolate genome sequence (KF114860) with significant similarity (91.66% nucleotide similarity in BLASTn). A large polyprotein sequence predicted from that viral contig, 3,222 aa in length, could be annotated to polyprotein of BCMV (AYE54419) in NCBI-BLASTp with 96.61% similarity. This large contig also could be annotated to contain a potyviridae domain with e-value of 5.99e-230 in hmmsearch analysis. Moreover, both BLASTx search-based and hmmsearch-based methods showed that there were no other known or unknown viruses in the RNA-Seq library. According to species demarcation criteria of the family Potyviridae proposed by International Committee on Taxonomy of Viruses (<76% nucleotide identity and <82% amino acid identity in complete ORF sequence), the viral contig we identified was considered as a strain of BCMV and named BCMV-gardenia.
To confirm the presence of the potyvirus, the potyvirus degenerate primers PotyF and PotyR (Marie-Jeanne et al., 2000) were synthesized and used in RT-PCR. After PCR amplification, the bright bands with expected length (~300 bp) were only detected in samples from five symptomatic leaves and not in three asymptomatic leaves (Fig. 2). The sequences of amplicons showed approximately 95.41% nucleotide sequence identity to BCMV isolate Ir-MSC1 (MF498888). Subsequently, the complete nucleotide sequences of this BCMV candidate virus were amplified by overlapping RT-PCR and RACE. The gel electrophoresis image depicted in Supplementary Fig. 1 displayed that all these sets of specific primers produced specific bands with corresponding length (Supplememtary Table 1).
After Sanger sequencing of the amplification product from the overlapping RT-PCR and RACE, the full-length genome sequence of BCMV-gardenia was found to be 10,054 nt in length, excluding the poly(A) at the 3′ termini, with a 5′ untranslated region (UTR) of 134 nt and a 3′ UTR of 286 nt (GenBank accession no. OR832357) (Fig. 3A). Consistent with other potyviruses, nine highly conserved proteolytic cleavage sites were predicted, and ten putative mature proteins could be identified, including P1 (443 aa, 49.96 kDa), HC-Pro (457 aa, 52.06 kDa), P3 (347 aa, 39.57 kDa), 6K1 (52 aa, 5.93 kDa), CI (634 aa, 71.25 kDa), 6K2 (53 aa, 6.15 kDa), VPg (190 aa, 21.90 kDa), NIa-Pro (243 aa, 27.56 kDa), NIb (516 aa, 59.62 kDa), and CP (287 aa, 32.20 kDa) from N-termini to C-termini. The putative P3N-PIPO, encoding a 74 aa protein with 8.77 kDa, was also identified at nucleotide position from nucleotide position 3,292 nt to 3,514 nt (Fig. 3A and C). To compare both the similarities and variation in the conserved protein cleavage sites among different BCMV isolates, as well as bean common mosaic necrosis virus (BCMNV) and SMV, all belonging to the genus Potyvirus, the conserved protein cleavage sites were predicted and indicated in the Fig. 3A. From the results, it was evident that despite differences in the polyprotein sequence between BCMV-gardenia and BCMV with accession number NC003397 (identified in black-eye cowpea), their conserved protein cleavage sites exhibit complete identity. For BCMV isolates, BCMNV, and SMV, differences were only observed in the protein cleavage sites between P1 and HC-Pro proteins (Fig. 3A). We also compared the amino acid and nucleotide similarity of each protein between BCMV-gardenia and another BCMV isolate (NC003397). We found that these two BCMV isolates showed higher similarity at the C-termini of the polyprotein, such as CP protein region, while the sequence similarity is lowest at the N-termini, especially in the P1 protein region (Fig. 3C). When clean reads were mapped back to the BCMV-gardenia reference genome, we found that every region of the viral genome was covered, with the highest depth at both ends, especially the 3′ end of the viral genome (Fig. 3B).
To confirm the evolutionary status of BCMV-gardenia in the genus Potyvirus, the phylogenetic tree of BCMV-gardenia and other representative member of the genus Potyvirus, was constructed by a maximum likelihood algorithm using IQ-TREE program. The multiple sequence alignment was performed by MAFFT. As shown in Supplementary Fig. 2, BCMV-gardenia was highly associated and clustered with BCMV isolate (NC003397), reinforcing the conclusion that virus identified in this research is an isolate of BCMV. To study the evolutionary relationships among different BCMV isolates, a midpoint-rooted phylogenetic tree was constructed using the various isolate of BCMV identified in different hosts (Fig. 4). Except for the BCMV-gardenia, the genome of other BCMV isolates were retrieved from NCBI GenBank database. The final dataset used to construct evolutionary tree comprised 101 sequences, the vast majority of which represented the BCMV genomes identified in leguminous plant hosts. However, there were three exceptions: BCMV-gardenia (identified in gardenia), KM076650 (identified in Cudrania tricuspidata), and MZ670770 (identified in Nandina domestica). From the evolutionary tree, we found that BCMV isolate identified in the same host species tend to cluster into a phylogenetic group. Remarkably, the three BCMV isolates identified outside leguminous hosts, were adjacent to each other on the whole evolutionary tree (Fig. 4). Notably, among the three BCMV isolate identified outside the leguminous hosts, KM076650 (identified in Cudrania tricuspidata) and MZ670770 (identified in Nandina domestica) formed a monophyletic group and BCMV-gardenia (identified in gardenia) seemed to have a closer evolutionary relationship with other BCMV identified in leguminous plants (Fig. 4).
RNA silencing serves as a highly effective defense mechanism against viral infections employed by plants. This defense strategy relies on the production of the vsiRNAs. Therefore, the characterization of the vsiRNAs profile contributes to our understanding of the virus-host interaction. We found that the vsiRNAs derived from BCMV-gardenia are mainly 21nt and 22nt, with 21nt being the most abundant (Fig. 5A). We investigated the 5′-terminal nucleotide preference of vsiRNAs derived from BCMV-gardenia. Our analysis revealed a predominant U/A preference for vsiRNAs with lengths of 21 nt and 22 nt (Fig. 5A) or across all vsiRNAs (Fig. 5B). Regarding the genomic regions from which vsiRNAs were generated, our observations indicated a relatively even distribution across the entire viral genome. However, specific local regions emerged as hot spots for vsiRNA production, with 21 nt and 22 nt vsiRNAs being the predominant types in each region (Fig. 5C). We further explored the strand polarity and specific origin sites of vsiRNAs along BCMV-gardenia in detail. The results indicated that 51.1% of vsiRNAs originated from the sense strand, while 48.9% originated from the antisense strand (Fig. 5D).
BCMV, a member within the genus Potyvirus, is one of the most widespread and destructive pathogens primarily affecting bean crops, including common beans, soybeans, and other legume plants. Some potyvirus, such as SMV, which is a virus present worldwide but has a very narrow host range in nature (Hajimorad et al., 2018). As a contrast, BCMV has a much wider host range, including over 100 plant species across 44 genera (Tang and Feng, 2022). With the extensive investigation of plant viruses and the widespread use of high-throughput sequencing technology, more and more hosts outside the leguminous plants have been found to be natural hosts of BCMV. According to the published literature, there are currently at least four plant species outside the family Leguminosae found to be the natural host of BCMV, including Cudrania tricuspidata (Seo et al., 2015), Alpinia purpurata (Larrea-Sarmiento et al., 2020), Nandina domestica (Su et al., 2022), and Populus alba (Wang et al., 2023). Among them, only BCMV identified in Cudrania tricuspidata and Nandina domestica are reported with complete genome sequence. Interestingly, the BCMV isolates identified in Cudrania tricuspidata and Nandina domestica and the BCMV isolate identified in gardenia in this study were positioned closely together in the phylogenetic tree constructed with BCMV isolates identified in other legumes. Moreover, the BCMV isolate found in gardenia plants shows a stronger evolutionary connection to the BCMV isolates found in other leguminous plants. This suggests that gardenia plants could potentially serve as a reservoir for BCMV, facilitating its transmission to leguminous plants and resulting in harmful consequences.
In this study, it was observed that BCMV could be detected by RT-PCR in all leaves showing disease symptoms, but no RT-PCR bands could be obtained in all normal leaves, which suggests a connection between BCMV and these plant symptoms. However, whether these symptoms such as mosaic and chlorosis are caused by BCMV requires strict inoculation experiments.
The complete genome of BCMV identified in gardenia was determined by a combination of RT-PCR and RACE. The genome sequence of BCMV-gardenia has been submitted to the NCBI nucleotide database under the accession number OR832357. The availability of this complete sequence will enhance our comprehension of genome variation, evolution, and diversity among different BCMV isolates. Our molecular characterization showed that compared with other BCMV isolates, BCMV-gardenia showed higher conservation in the C-termini of the polyprotein (such as the CP protein region), but exhibited higher variability in the N-termini of the polyprotein (such as the P1 protein region), which may be the results of BCMV adapting to different hosts.
The transmission routes of BCMV are highly diverse, including transmission through seeds, pollen, aphids, and mechanical inoculation. When we inspected the polyprotein of BCMV-gardenia, we found the presence of some known sequence motifs required for systemic movement and aphid transmission, including “KLSC” (polyprotein amino acid position 496-499), “CCC” (position 734-736), “PTK” (position 752-754) motifs in HC-Pro, as well as “DAG” motif (position 1815-1817) near N-terminus of the coat protein (Atreya et al., 1991; Fresnillo et al., 2022; Maia et al., 1996). Since there were no leguminous crops around the gardenia we sampled in a small garden in Ningbo University, we speculated that BCMV infection of gardenia might be caused by the feeding activities of aphids flying from other places.
In addition, our research also examined the features of small interfering RNAs derived from the BCMV-gardenia, which were generated by the host’s RNA interference pathway against viral infections. In this study, we found that vsiRNAs derived from BCMV-gardenia were mainly 21nt and 22nt, with 21nt being the most abundant. Various Dicers/DCLs are involved in the processing of vsiRNAs of distinct sizes. DCL4 is primarily accountable for generating 21-nt vsiRNAs, whereas DCL2 is responsible for producing 22-nt vsiRNAs. The latter plays a crucial role in counteracting RNA viruses when the activity of DCL4 is impaired or suppressed (Diaz-Pendon et al., 2007; Ding, 2003). Hence, it can be inferred that DCL2 and DCL4 play a key role against BCMV infection in gardenia. Our study revealed a preferential occurrence of “U” or “A” residues at the 5′-termini of BCMV-gardenia vsiRNAs. Previous researches have suggested that the initial nucleotides at the 5′ end of small RNAs (sRNAs) regulate the sorting of sRNAs to particular AGO complexes in plants (Mi et al., 2008). The vsiRNAs of BCMV-gardenia exhibited a preference to “A” and “U”, suggesting that AGO1, AGO2, or AGO4 may be involved in recruiting and sorting these vsiRNAs. Analysis of the polarity and genome distribution of BCMV-gardenia vsiRNAs showed that vsiRNAs were relatively evenly distributed on the viral genome and showed close sense and antisense strand preference, which suggests that the vsiRNAs are mainly produced from the dsRNA replication intermediates of BCMV-gardenia.
In summary, our work evidenced the existence of BCMV infection in gardenia for the first time. Our virus detection experiments emphasized the association between BCMV infection and disease symptoms in gardenia. The full-length genome of BCMV-gardenia was recovered by a combination of RT-PCR, RACE, and Sanger sequencing. Collectively, these data provide valuable information for understanding to diversity, evolution of BCMV, contributing to the virus control as well as disease management.
Acknowledgments
This work was funded by the National Natural Science Foundation of China (U20A2036) and Ningbo Yongjiang Grant (2022A-220-G).
Electronic Supplementary Material
Supplementary materials are available at The Plant Pathology Journal website (http://www.ppjonline.org/).
Fig. 1
Comparison between healthy gardenia and gardenia exhibiting disease symptoms. (A) Overview of gardenia twigs with leaves showing mosaic, chlorosis, yellowing symptoms. (B) Leaves showing disease symptoms. (C) Heathy leaves.
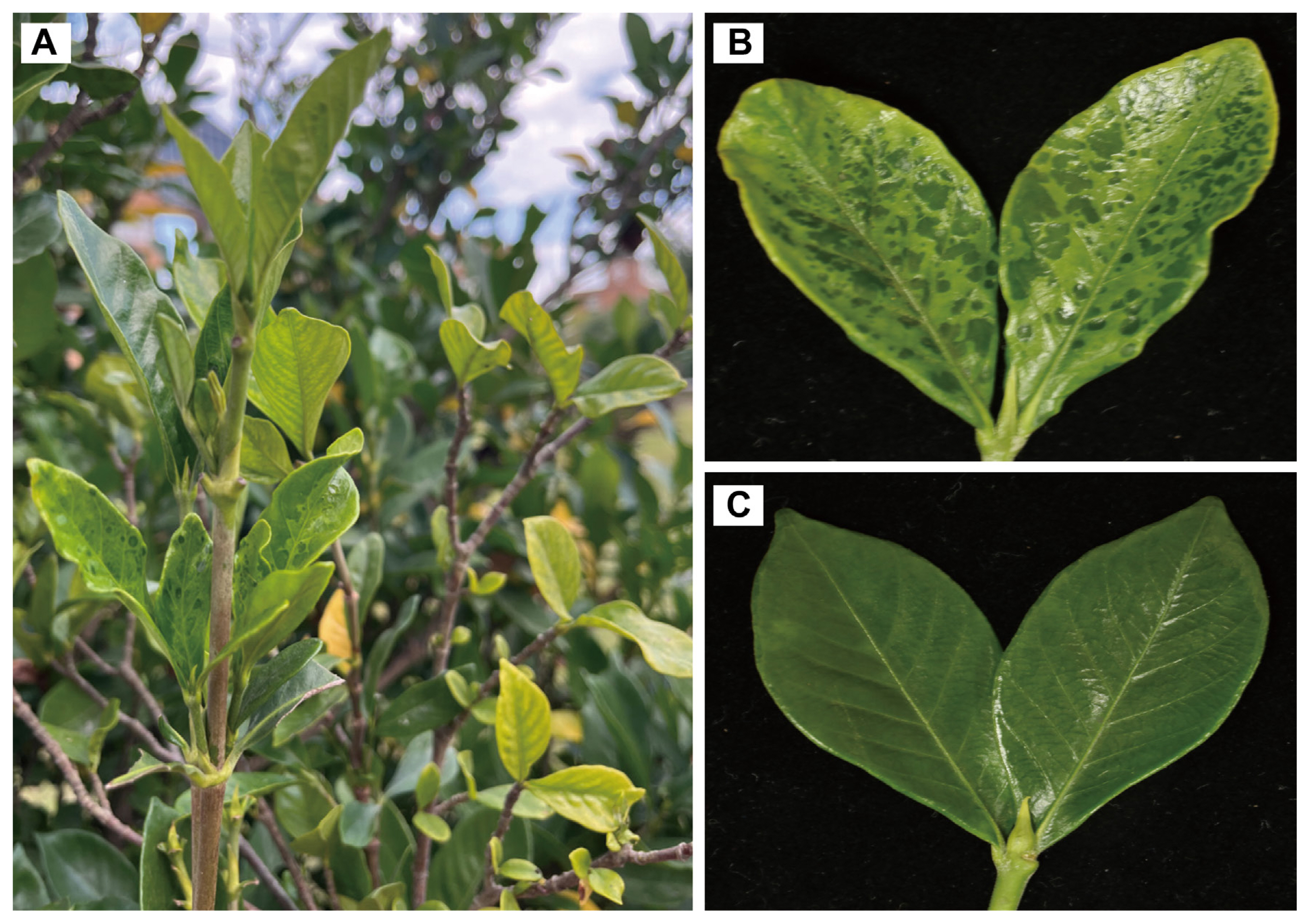
Fig. 2
Confirmation of the presence of bean common mosaic virus (BCMV)-gardenia by reverse transcription polymerase chain reaction (RT-PCR) using PotyR/PotyF primer pair. M, marker, Trans2K Plus DNA Marker; lanes S1-S5, RT-PCR in gardenia leaves showing disease symptoms; lanes S6-S8, RT-PCR in heathy gardenia leaves.
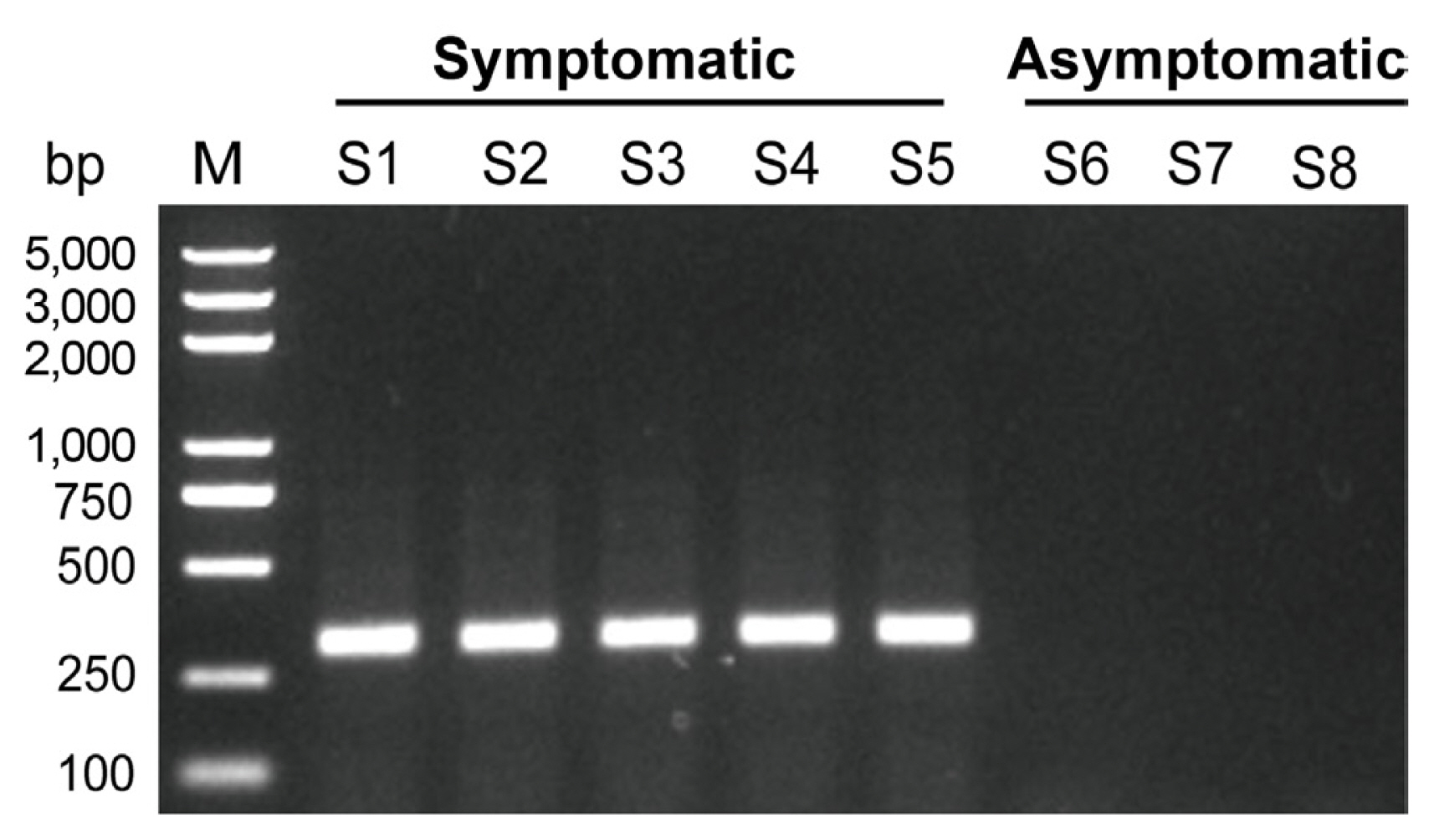
Fig. 3
Characterization of bean common mosaic virus gardenia isolate (BCMV-gardenia). (A) Genome organization of BCMV-gardenia. Open reading frames are denoted by the color boxes. The predicted cleavage sites of the BCMV-gardenia are compared to those of BCMV identified in black-eye cowpea (NC003397), bean common mosaic necrosis virus (BCMNV; NC004047), and soybean mosaic virus (SMV; NC002634) polyproteins. (B) Transcriptome clean reads coverage along the BCMV-gardenia genome. (C) Characteristics of the ten mature proteins encoded in the genome of BCMV-gardenia. The percentage of sequence identity are calculated through global alignments of the nucleotide (nt) and amino acid (aa) sequence between BCMV-gardenia and another BCMV isolate (NC003397, identified in black-eye cowpea).
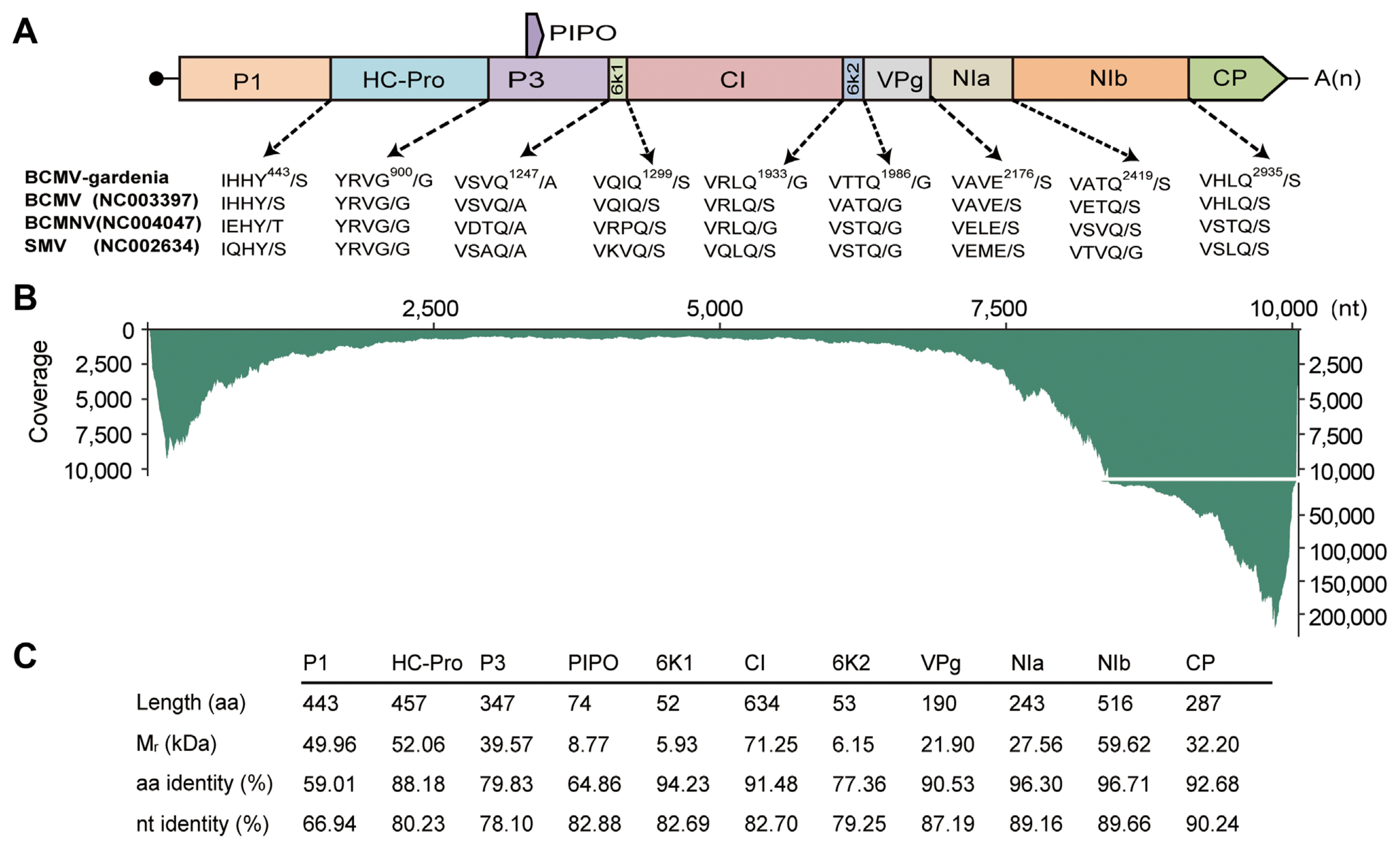
Fig. 4
Evolutionary relationships of bean common mosaic virus (BCMV) isolates. Maximum likelihood phylogenetic tree based on the polyprotein coding region of BCMV-gardenia and other BCMV isolates. One thousand bootstrap replicates were employed and bootstrap support value was calculated and given at each node. The substitution model selected by ModelFinder is GTR + F + I + G4. The bar represents the number of substitutions per site (0.04).
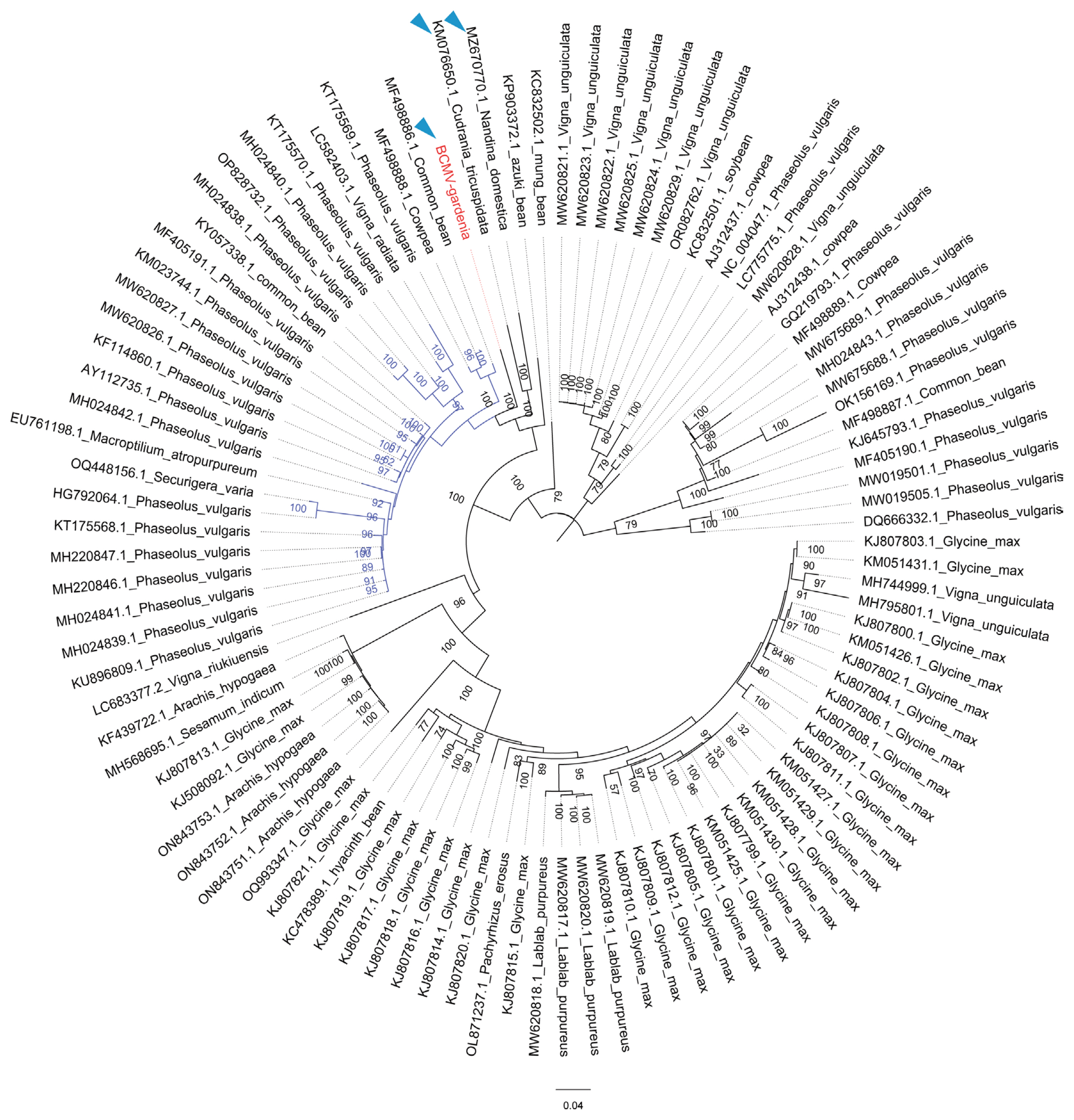
Fig. 5
Profile of bean common mosaic virus (BCMV)-gardenia derived small interfering RNAs (vsiRNAs). (A) Length distribution of BCMV-gardenia vsiRNAs. (B) 5′ Terminal nucleotide preference of BCMV-gardenia vsiRNAs. (C) Distribution of BCMV-gardenia vsiRNAs alongside the viral genome. (D) BCMV-gardenia vsiRNAs polarity.

References
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. and Lipman, D. J. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410.


Asensio-S-Manzanera, M. C., Asensio, C. and Singh, S. P. 2006. Gamete selection for resistance to common and halo bacterial blights in dry bean intergene pool populations. Crop Sci. 46:131-135.


Atreya, P. L., Atreya, C. D. and Pirone, T. P. 1991. Amino acid substitutions in the coat protein result in loss of insect transmissibility of a plant virus. Proc. Natl. Acad. Sci. U. S. A 88:7887-7891.



Cao, Y., Zhi, J., Zhang, R., Li, C., Liu, Y., Lv, Z. and Gao, Y. 2018. Different population performances of Frankliniella occidentalis and Thrips hawaiiensis on flowers of two horticultural plants. J. Pest Sci. 91:79-91.


Chen, S., Zhou, Y., Chen, Y. and Gu, J. 2018. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34:i884-i890.




Cho, Y. S. 2022. Genipin, an inhibitor of UCP2 as a promising new anticancer agent: a review of the literature. Int. J. Mol. Sci. 23:5637.



Diaz-Pendon, J. A., Li, F., Li, W.-X. and Ding, S.-W. 2007. Suppression of antiviral silencing by cucumber mosaic virus 2b protein in Arabidopsis is associated with drastically reduced accumulation of three classes of viral small interfering RNAs. Plant Cell 19:2053-2063.




Ding, S.-W. 2023. Transgene silencing, RNA interference, and the antiviral defense mechanism directed by small interfering RNAs. Phytopathology 113:616-625.


Fang, L., Xie, Y., Wu, J., Wang, L. and Wang, H. 2023. First report of Diaporthe gardeniae causing branch blight of Gardenia jasminoides Ellis in Zhejiang Province, China. Plant Dis. 107:1218.
Feng, X., Orellana, G. E., Green, J. C., Melzer, M. J., Hu, J. S. and Karasev, A. V. 2019. A new strain of bean common mosaic virus from lima bean (Phaseolus lunatus): biological and molecular characterization. Plant Dis. 103:1220-1227.


Fresnillo, P., Jover-Gil, S., Samach, A. and Candela, H. 2022. Complete genome sequence of an isolate of Passiflora chlorosis virus from passion fruit (Passiflora edulis Sims). Plants 11:1838.



Hajimorad, M. R., Domier, L. L., Tolin, S. A., Whitham, S. A. and Saghai Maroof, M. A. 2018. Soybean mosaic virus: a successful potyvirus with a wide distribution but restricted natural host range. Mol. Plant Pathol. 19:1563-1579.




Haas, B. J., Papanicolaou, A., Yassour, M., Grabherr, M., Blood, P. D., Bowden, J., Couger, M. B., Eccles, D., Li, B., Lieber, M., MacManes, M. D., Ott, M., Orvis, J., Pochet, N., Strozzi, F., Weeks, N., Westerman, R., William, T., Dewey, C. N., Henschel, R., LeDuc, R. D., Friedman, N. and Regev, A. 2013.
De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc 8:1494-1512.




Jin, C., Zongo, AW-S, Du, H., Lu, Y., Yu, N., Nie, X., Ma, A., Ye, Q., Xiao, H. and Meng, X. 2023. Gardenia (Gardenia jasminoides Ellis) fruit: a critical review of its functional nutrients, processing methods, health-promoting effects, comprehensive application and future tendencies. Crit. Rev. Food Sci. Nutr. Advanced online publication.https://doi.org/10.1080/10408398.2023.2270530.

Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A. and Jermiin, L. S. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14:587-589.




Katoh, K. and Standley, D. M. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30:772-780.



Langmead, B., Trapnell, C., Pop, M. and Salzberg, S. L. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25.



Larrea-Sarmiento, A., Wang, X., Borth, W. B., Barone, R. P., Olmedo-Velarde, A., Melzer, M. J., Sugano, J. S. K., Galanti, R., Suzuki, J. Y., Wall, M. M. and Hu, J. S. 2020. First report of bean common mosaic virus infecting flowering ginger (Alpinia purpurata) in Hawaii. Plant Dis. 104:603.

Lu, X. H., Solangi, G. S., Huang, J. L., Liu, Z. M. and Qin, L.-P. 2023. First report of root-knot nematode Meloidogyne enterolobii on Antirrhinum majus in China. Plant Dis. 107:2555.

Maia, I. G., Haenni, A. and Bernardi, F. 1996. Potyviral HC-Pro: a multifunctional protein. J. Gen. Virol 77:1335-1341.


Marie-Jeanne, V., Ioos, R., Peyre, J., Alliot, B. and Signoret, P. 2000. Differentiation of Poaceae potyviruses by reverse transcription-polymerase chain reaction and restriction analysis. J. Phytopathol 148:141-151.

Martin, M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17:10-12.

Mi, S., Cai, T., Hu, Y., Chen, Y., Hodges, E., Ni, F., Wu, L., Li, S., Zhou, H., Long, C., Chen, S., Hannon, G. J. and Qi, Y. 2008. Sorting of small RNAs into Arabidopsis Argonaute complexes is directed by the 5′ terminal nucleotide. Cell 133:116-127.



Nguyen, L.-T., Schmidt, H. A., von Haeseler, A. and Minh, B. Q. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32:268-274.



Sáiz, M., Dopazo, J., Castro, S. and Romero, J. 1994. Evolutionary relationships among bean common mosaic virus strains and closely related potyviruses. Virus Res. 31:39-48.


Seo, J.-K., Kang, M., Shin, O. J., Kwak, H.-R., Kim, M.-K., Choi, H.-S. and Ko, S.-J. 2015. First report of bean common mosaic virus in Cudrania tricuspidata in Korea. Plant Dis. 99:292.

Su, X., Zhou, X., Li, Y., Ma, L., Cheng, X. and Guo, K. 2022. First report of bean common mosaic virus infecting heavenly bamboo (Nandina domestica) in China. Plant Dis. 106:1079.


Tang, M. and Feng, X. 2022. Bean common mosaic disease: etiology, resistance resource, and future prospects. Agronomy 13:58.

Wang, L., Zhang, W., Shen, W., Li, M., Fu, Y., Li, Z., Li, J., Liu, H., Su, X., Zhang, B. and Zhao, J. 2023. Integrated transcriptome and microRNA sequencing analyses reveal gene responses in poplar leaves infected by the novel pathogen bean common mosaic virus (BCMV). Front. Plant Sci. 14:1163232.



Xu, X., Chen, B., Zhang, J., Lan, S. and Wu, S. 2023. Whole-genome resequencing analysis of the medicinal plant Gardenia jasminoides. PeerJ. 11:e16056.




Xu, Z., Pu, X., Gao, R., Demurtas, O. C., Fleck, S. J., Richter, M., He, C., Ji, A., Sun, W., Kong, J., Hu, K., Ren, F., Song, J. J., Wang, Z., Gao, T., Xiong, C., Yu, H., Xin, T., Albert, V. A., Giuliano, G., Chen, S. and Song, J. 2020. Tandem gene duplications drive divergent evolution of caffeine and crocin biosynthetic pathways in plants. BMC Biol. 18:63.




- TOOLS
-
METRICS

-
- 0 Crossref
- 0 Scopus
- 688 View
- 93 Download
- ORCID iDs
-
Yi-Yuan Li

https://orcid.org/0000-0003-1426-0033 - Related articles



 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Supplement1
Supplement1 Print
Print



