Multi-Homologous Recombination-Based Gene Manipulation in the Rice Pathogen Fusarium fujikuroi
Article information
Abstract
Gene disruption by homologous recombination is widely used to investigate and analyze the function of genes in Fusarium fujikuroi, a fungus that causes bakanae disease and root rot symptoms in rice. To generate gene deletion constructs, the use of conventional cloning methods, which rely on restriction enzymes and ligases, has had limited success due to a lack of unique restriction enzyme sites. Although strategies that avoid the use of restriction enzymes have been employed to overcome this issue, these methods require complicated PCR steps or are frequently inefficient. Here, we introduce a cloning system that utilizes multi-fragment assembly by In-Fusion to generate a gene disruption construct. This method utilizes DNA fragment fusion and requires only one PCR step and one reaction for construction. Using this strategy, a gene disruption construct for Fusarium cyclin C1 (FCC1 ), which is associated with fumonisin B1 biosynthesis, was successfully created and used for fungal transformation. In vivo and in vitro experiments using confirmed fcc1 mutants suggest that fumonisin production is closely related to disease symptoms exhibited by F. fujikuroi strain B14. Taken together, this multi-fragment assembly method represents a simpler and a more convenient process for targeted gene disruption in fungi.
Introduction
Fusarium fujikuroi Nirenberg (teleomorph, Gibberella fujikuroi [Sawada] Wollenweber) is a causal agent of bakanae (foolish seedling) disease, one of the most notorious seed-borne diseases affecting rice (Oryza sativa L.) fields worldwide. Its major symptom is spindly growth of the host species, which is caused by gibberellin (GA) production by the pathogen within and around the infection site (Wiemann et al., 2013). The fungal species belongs to the mating population (MP)-C of the Gibberella fujikuroi species complex, which is composed of more than eleven MPs that are sexually infertile with one another (Bomke et al., 2008). It is known to produce fumonisin (Cruz et al., 2013; Jeon et al., 2013), moniliformin (Desjardins et al., 2000), and beauvericin (Desjardins et al., 2000). In addition to typical bakanae symptoms, a considerable number of strains or isolates trigger stunting in rice (Cruz et al., 2013; Hur et al., 2015; Jeon et al., 2013). The absolute correlation between pathogen genes located in the gibberellin biosynthetic gene cluster (GA cluster) and fungal GA biosynthesis has been determined over the last two decades (Tudzynski, 1999, 2005; Tudzynski et al., 1998, 2003). However, the contribution of GA to symptom development remains to be determined, and the functional characterizations of other genes regulating virulence or pathogenesis is only just beginning. Draft genomes of the strains IMI 58289 and B14, model strains of F. fujikuroi known to cause spindly growth and alternative stunting in rice, have been completed in consequence of the recent growth of next generation sequencing technologies for microbial genomes (Jeong et al., 2013; Wiemann et al., 2013). Comparative genome analyses and gene annotation have indicated the presence of more than several hundred genes that should be characterized during the interaction between rice and F. fujikuroi.
Similar to other biological systems, functional characterization of fungal genes has conventionally been achieved through targeted gene-specific mutation or by using mutants from pools of germplasms harboring T-DNA at random sites within the genome (Bundock and Hooykaas, 1996; Huang et al., 2011; Jeon et al., 2007; Michielse et al., 2005; Piers et al., 1996; Takahara et al., 2004). Although the latter mutant pool has made a huge contribution to the functional characterization of genes present in a limited number of model strains and genes that are conserved across strains, recent findings stress the necessity of target gene-specific gene disruption and deletion: almost all fungal genes resulting in virulence or pathogenicity are located within highly variable regions of the pathogen genome (Schirawski et al., 2010; Wiemann et al., 2013). These strain-specific virulence- and pathogenicity-regulating genes indicate the requirement for novel techniques to generate knock-out constructs efficiently. The ideal structure of a knock-out construct for the disruption of target genes in filamentous fungi is composed of a 0.5–1 kb-long 5′ flanking region of coding DNA sequence (CDS); a gene encoding a protein conferring antibiotic-resistance, such as hygromycin B phosphotransferase (HPH) or neomycin phosphotransferase (NPTII); and a 0.5–1 kb-long 3′ flanking region of CDS. After introduction of this construct into fungal protoplasts via polyethylene glycol (PEG)/CaCl2 or Agrobacterium tumefaciens, the target CDS and both flanking regions of the fungal chromosome are replaced with the cassette HPH or NPTII by dual homologous recombination between each flanking region. Classical methods for gene knock-out vector construction are almost always dependent on DNA digestion and ligation. These techniques have long been used and remain popular; however, suitable sites for restriction enzyme digestion are frequently unavailable depending on the nucleotide sequence of the target regions, and ligation can be affected by sequence-specific variations in ligation efficiency. To overcome these problems, PCR-based multi-fragment amplification, or double-joint PCR, was designed (Kim et al., 2009; Yu et al., 2004). Although the structure of the final knock-out construct produced using this method is identical to those manipulated by classical methods, this technique is free from the limitations of finding suitable restriction sites and sequences with suitable ligation efficiency. However, a disadvantage of this method stems from the fidelity of Taq polymerase. Taq polymerase almost always integrates at an improper base(s) that is not complementary with the template sequence; the mismatch rate increases exponentially in accordance with the amplicon size and GC ratio. Genome editing techniques employing transcription activator-like effector nucleases (Aouida et al., 2014) or CRISPR/Cas9 (Laughery et al., 2015; Mans et al., 2015) represent potent, forthcoming alternatives that could replace conventional fungal gene disruption. However, the former has disadvantages in its complicated knock-out vector construction, and the latter requires genome sequencing of the candidate transformants to rule out possible off-targeting artifacts. This situation prompted us to develop an easy, versatile, and time-effective method for the production of knock-out constructs that are less affected by the presence or absence of restriction endonuclease sites, ligation efficiency, and the fidelity of Taq polymerase.
Recently, a novel gene manipulation technique enabling the simultaneous homologous recombination of several DNA fragments was developed (Raman and Martin, 2014). This technique, which is called In-Fusion HD Cloning, is sequence-independent and recombines all components in a designated order with high cloning efficiency. By invoking this technique, we demonstrate an alternative method for the efficient production of knock-out constructs suitable for the disruption or deletion of target genes in filamentous fungi, such as F. fujikuroi.
Materials and Methods
Fungal strains and growth conditions
These fungal strains were routinely cultured on oatmeal agar (OMA) at 22ºC under continuous florescent light and were preserved at −20ºC as dried mycelial agar blocks after chopping (Kim et al., 2005).
To assess the morphological characteristics of the mutants, solidified and liquid media were used to assay wild-type (WT) and mutant strains. The fungus grew on culture plates containing OMA and Fusarium complete medium (Leslie and Summerell, 2006), respectively. The culture plates were incubated for one week at 22ºC, whereas the liquid cultures were incubated for 5 days at 22ºC in a rotator shaker.
Primer design for Fusarium cyclin C1 gene (FCC1 ) and FUM1 deletions
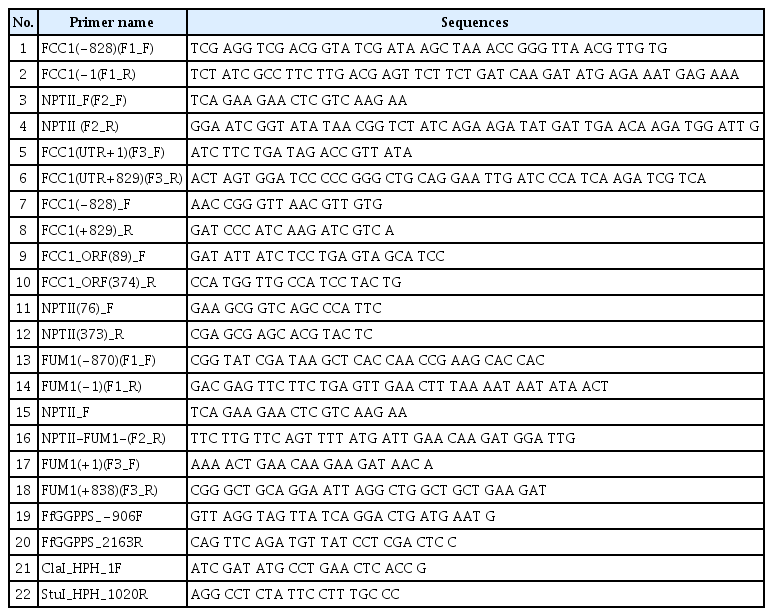
To generate target gene deletion mutants using the In-Fusion® HD Cloning system (Clontech Laboratories Inc., Mountain View, CA, USA), primers were designed using a primer design tool developed by In-Fusion® HD Cloning. Primers were also designed manually in some cases. To successfully clone any DNA fragment, the primers used must be designed to share an overlap of at least 15 homologous bases between the end of the linearized vector and/or the desired fragment. Thus, a primer will consist of a homologous sequence of 15 to 30 bases at the 5′-end and a gene-specific sequence at the 3′-end. Depending on the length and sequence of the fragments, the overlap may be longer than 15 base pairs. Primers used for target gene disruption are listed in Table 1.
Construction of FCC1 and FUM1 deletion strains
Construction of targeted gene deletion of FCC1 and FUM1 was performed following Changlin’s method, with modifications (Fu et al., 2014). Here, we used a pBlue-script sk(−) vector that had been linearized by digestion with EcoRI and HindIII to obtain the targeted DNA fragment containing the 5′ flanking region of the target gene, selection marker and the 3′ flanking region of the target gene. The NPTII gene, which was used as a selectable marker for the transformation of F. fujikuroi by conferring geneticin resistance, was amplified from the pII99 vector. After PCR amplification using specific primer pairs developed by In-Fusion® HD Cloning, the reaction products were analyzed by electrophoresis on a 1% agarose gel to remove non-specific DNA bands and to avoid false-positive clones. A total of 4 purified DNA fragments, including a linearized vector, a 5′ target gene flanking region, NPTII and a 3′ target gene flanking region were combined in a tube containing 1 μl of 5× In-Fusion reaction mix. For multiple fragment assembly, 100 ng/μl of vector and 250 ng/μl of each insert were used. The reaction tube was incubated for 1 hour at 50ºC and transformed directly.
Construction of GGPPS1 deletion strains
To generate a GGPPS1 deletion mutant, a 2.3 kb fragment of genomic DNA containing a 5′ and 3′ sequence flanking GGPPS1 in the F. fujikuroi strain was amplified by PCR and cloned into the pGEM-T easy vector (Promega, Madison, WI, USA). Next, a 1,257 bp ClaI-StuI fragment of the GGPPS1 gene in the pGEM-T easy vector was replaced with a 1,020 bp ClaI-StuI fragment amplified from pAN7-1 containing the E. coli HPH gene as a selection marker. This 2.6 kb fragment, which has a 5′ sequence flanking GGPPS1, was amplified by PCR from the pGEM-T easy vector. Purified DNA was used to transform the B14 F. fujikuroi strain. The primers used for GGPPS1 gene disruption are listed in Table 1.
Fungal genomic DNA isolation
For genomic DNA isolation, strains were grown in 10 ml of Fusarium complete medium at 25ºC. After harvesting and lyophilizing the mycelial mass, gDNA was extracted following cetyltrimethyl- ammonium bromide (CTAB)-based methods modified in our laboratory. The lyophilized mycelium was filled with 600 μl of CTAB buffer and beads. These were mixed thoroughly on a Geno/Grinder (SPEX CertiPrep., Metuchen, NJ, USA) for 5 minutes. The samples were incubated for 30 minutes at 65ºC; then, chloroform was added and shaken for 1–2 minutes. Tubes were centrifuged for 10 minutes at 12,000 rpm, and the supernatant was collected in a new tube containing ice-cold isopropanol. After centrifugation for 10 minutes, the pellet was washed with ice cold 70% (v/v) ethanol, dried and dissolved in 50 μl of TE buffer (4.5 ml, 10 mM Tris, 1 mM ethylenediamine tetraacetic acid, pH set to 8.0 with HCl). Extracted genomic DNA was verified by agarose gel and stored at −20ºC until use.
Fungal transformation
Fungal gene deletion was performed by homologous recombination as described previously (Kim et al., 2005). Briefly, two primer pairs were used to amplify the 5′- and 3′-flanking regions of the target gene from each construct by PCR. The purified PCR products were used for fungal transformation. To harvest the protoplasts of the B14 strain for transformation, young mycelia were incubated at 25ºC for 3 hours in 20% sucrose containing 25 mg/ml Driselase and 10 mg/ml lysing enzyme (L1412; Sigma-Aldrich, St. Louis, MO, USA). Samples were centrifuged, and the resulting protoplast pellets were suspended in STC buffer (20% sucrose, 50 mM Tris-HCl pH 8.0 and 50 mM CaCl2) and then mixed with a PTC solution (40% PEG 3350 in STC buffer) containing DNA fragments to obtain transformants. The transformed protoplasts were plated in regeneration agar medium (0.3% yeast extract, 0.3% casamino acid, 1.0% glucose, 20% sucrose, and 0.8% agar) with 50 μg/ml geneticin and incubated at 25ºC. Hygromycin and geneticin-resistant transformants appeared after 5 to 7 days, and the fungi were grown in potato dextrose agar (PDA; Difco, Sparks, MD, USA) for 3 days for selection. To confirm successful gene deletion, PCR was performed using specific primer pairs (Table 1).
Southern blot
Three micrograms of genomic DNA from WT and mutant fungi was digested at 37ºC with HindIII (New England Biolabs, Ipswich, MA, USA), which did cut in the mutant but not in the WT probe region. After resolving the digested products on a gel, the DNA was transferred to a nylon membrane (Hybond-N+; Amersham, Piscataway, NJ, USA) by capillary blotting. Hybridization was carried out according to the instructions provided with the DIG High Prime DNA Labeling and Detection Starter Kit II (Roche Applied Science, Penzberg, Germany). The digoxigenin (DIG)-dUTP labeled 5′ flanking region of FCC1 was used as a probe.
Virulence assays
Healthy cv. Dongjin rice seeds were surface sterilized as previously described (Sahoo et al., 2011) and germinated on half-strength Murashige and Skoog (MS) medium under a 16-hour-light/8-hour-dark cycle at 26ºC. The fungal mycelial blocks (0.5 cm in diameter) of F. fujikuroi strains grown for 7 days were transferred onto the center of vermiculite soil in the test tube (inner diameter × length = 1.6 × 14.7 cm) and recovered with a 2 cm layer of vermiculite soil. Then, 5-day-old rice seedlings were placed on the upper layer of the soil to prevent direct contact between the inocula and plants. The test tubes were covered with a cap to maintain the humidity for 7 days. Growing roots contacted the inocula within 24–36 hours after inoculation. Five days after inoculation (dai), the seedling height was measured. Seedlings were photographed at 7 dai.
Results
Constructs used for target gene disruption
During the interaction of rice and the F. fujikuroi strain B14, B14-infected rice plants showed severe root rot symptoms due to fumonisin production (Hwang et al., 2013). In particular, FCC1 (Fusarium cyclin C1) plays an important role in fumonisin B1 (FB1) biosynthesis (Shim and Woloshuk, 2001). To determine FCC1 function in the B14 strain, we used a modified gene-disruption method via In-Fusion cloning, which can be used to assemble multiple fragments. This strategy for targeted gene disruption requires a total of three PCR reactions and one linearized vector. Fig. 1A outlines our procedure for gene disruption. First, an amplification containing the 5′- and 3′-flanking regions of the FCC1 locus was performed with specific primer sets—1/2 and 5/6, respectively—from B14 genomic DNA. In parallel, 795 bp of NPTII, a geneticin-resistance gene used as a selectable marker, was amplified from the pII99 vector by PCR using the primer pair 3/4. The observed size of the PCR products resolved by agarose gel electrophoresis corresponded to a predicted size of approximately 800 bp (Fig. 1B). pBluescript sk(−) vector linearized with the restriction enzymes EcoRI and HindIII was used as the carrier. The purified amplicons and linearized vector were input directly into the In- Fusion cloning system and linked using 15–30 bp overlapping sequences between adjacent fragments. These multiple fragments were mixed in the single tube and incubated at 50ºC for 1 hour. The DNA fragments containing the 5′- and 3′-flanking regions of FCC1 and the NPTII gene were amplified from a construct of confirmed sequence and transformed into B14 fungus to delete the FCC1 gene. Subsequent PCR resulted in a PCR product of approximately 2.4 kb, the expected size of the FCC1 disruption (Fig. 1C). These results indicated that our homologous recombination-based cloning system generated a construct for the disruption of the FCC1 gene.

Fusarium cyclin C1 (FCC1) disruption in the Fusarium fujikuroi strain B14 using the In-Fusion cloning system. (A) Strategies for FCC1 disruption by homologous recombination. (B) Amplification of the three primary products: the 5′ flanking region of FCC1, neomycin phosphotransferase (NPTII), and the 3′ flanking region of FCC1. (C) Verification of the constructs prior to target gene disruption. (D) PCR screening results from the selected transformants using specific primers. P, positive control. (E) Southern blot analysis of wild-type and fcc1 mutants.
Gene replacement with the FCC1 gene disruption construct
To disrupt the FCC1 gene, B14 fungi were transformed with the following disruption cassette: an amplified DNA fragment of approximately 2.4 kb consisting of 5′- and 3′-FCC1 flanking regions and the NPTII gene. In this manner, the FCC1 gene was disrupted in B14 by homologous recombination, replacing FCC1 with the NPTII gene. This strategy is based on selection for geneticin resistance stemming from the precise replacement of FCC1 with the FCC1 gene disruption construct containing an NPTII cassette. The NPTII selectable marker gene allows the selection of correct transformants using geneticin. Geneticin-resistant transformants were verified by PCR with specific primers (Fig. 1D). Four transformants were confirmed to yield the expected PCR results. These transformants also exhibited the expected restriction enzyme digestion patterns via Southern blot analysis (Fig. 1E).
Phenotypic characterization of fcc1-disrupted B14 strains
The transformants obtained by successful homologous recombination were designated fcc1 mutants. In terms of the growth rate, the culture broth of the selected FCC1-disrupted B14 strains exhibited significantly more red pigmentation at all time-points compared with the WT strain, which showed significantly less coloration. The fcc1 strains exhibited slow radial growth and decreased conidia production when grown on PDA, suggesting that FCC1 may be involved in the growth of mycelium and sporulation in this fungus. These mutants also exhibited increased pigmentation compared with the WT strain (Fig. 2A). To determine whether the fcc1 mutation influences fumonisin production, fumonisin content was measured in the WT and fcc1 mutants in vitro. First, fumonisin production by the fcc1 mutants and WT strain was tested after growth in PDB for 5 days. As expected, fumonisin-producing ability was repressed by FCC1 disruption. The fcc1 mutants produced a lower or undetectable amount of fumonisin compared with the WT strain (Fig. 2B, left panel). Next, we tested whether fumonisin production during interaction with F. fujikuroi and rice was affected in fcc1 mutant-infected rice plants. Significantly reduced fumonisin production was observed in rice infected by fcc1 mutants compared with WT-infected rice (Fig. 2B, right panel). These findings clearly show that the deletion of FCC1 has an influence on fumonisin production, indicating that FCC1 is involved in fumonisin synthesis during infection.

Phenotypic characterization and fumonisin analysis in the fcc1 mutant strains compared with the wild-type (WT). (A) Phenotypical analysis of the WT and the fcc1 mutants in PDB and on PDA for 5 and 7 days. (B) The effect on fumonisin production by Fusarium cyclin C1 (FCC1) disruption in vitro and in vivo. Error bars are represent as ± standard deviation. Statistical significance was analyzed by Student’s t-test. Asterisks indicate significant differences (**P < 0.01).
Fungal virulence assay on rice
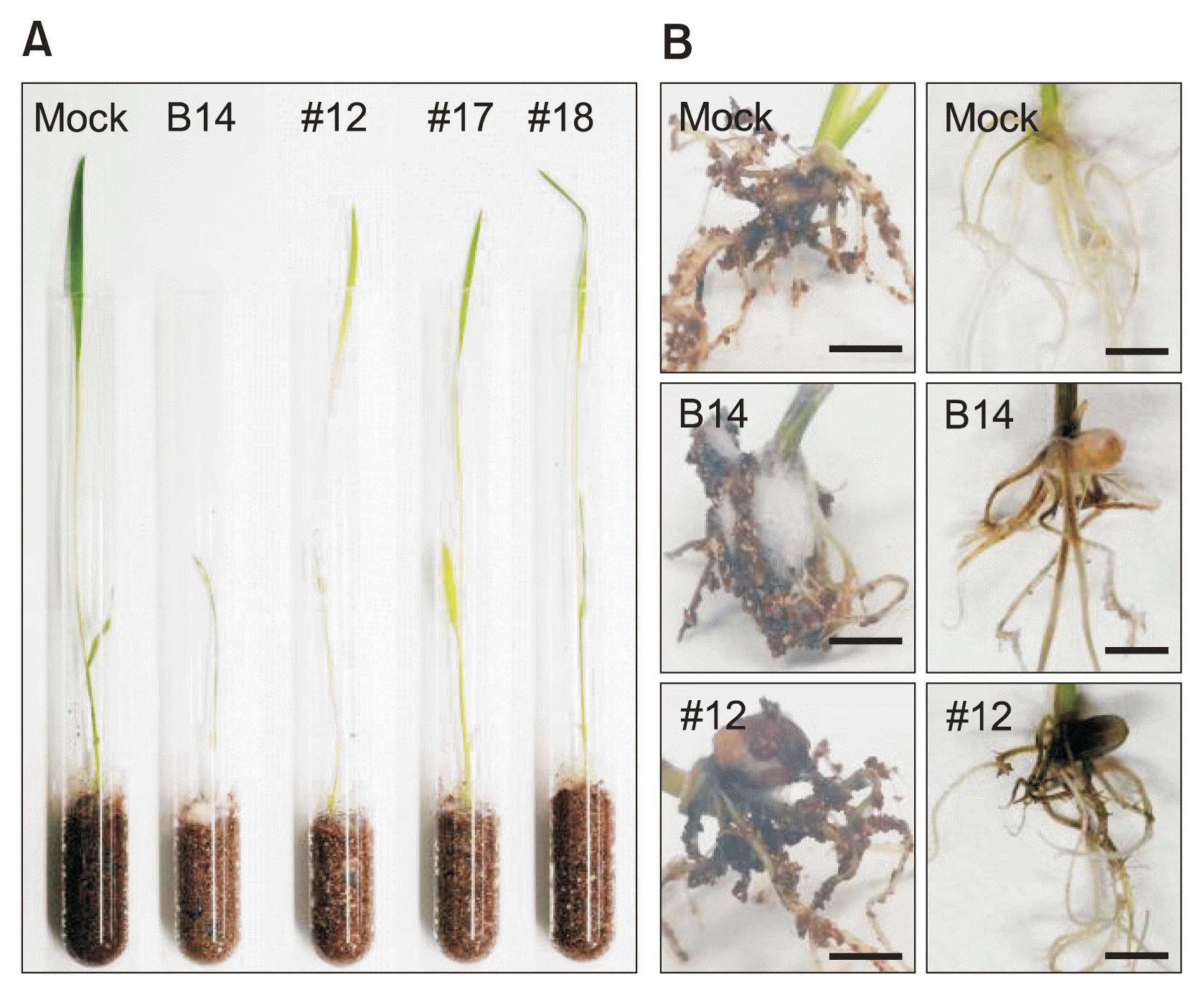
Next, we performed a virulence assay in fcc1 mutant-inoculated rice seedlings. Seedlings inoculated with the WT strain showed typical B14-mediated disease symptoms, such as root rot and growth restriction. In contrast, seedlings inoculated with fumonisin-deficient fcc1 mutant strains exhibited a significant reduction in B14-mediated disease symptoms (Fig. 3). Intriguingly, root rot progression was delayed or decreased by fcc1 mutant infection. This result indicated that the amelioration of disease symptoms by fcc1 mutants may be related to the ability of these plants to produce fumonisin.
A comparative study of the efficiency of cloning systems
Compared with restriction enzyme- and ligase-dependent cloning strategies for fungal gene deletion, multiple fragment assembly using the In-Fusion cloning system is simple. To generate a construct for GGPPS1 disruption in F. fujikuroi, a 2.3 kb fragment of genomic DNA containing a 5′ flanking GGPPS1 region (0.8 kb), GGPPS1 (1.2 kb), and a 3′ flanking GGPPS1 region (0.8 kb) from F. fujikuroi is required. Either naturally occurring restriction enzyme sites or engineered sites introduced during PCR amplification are essential for the insertion of the HPH gene for targeted gene replacement (Fig. 4A). However, construction of the FUM1 gene disruption cassette, which can delete the FUM1 gene (8.1 kb in total), required only a single PCR amplification and one reaction to assemble multiple fragments (Fig. 4B). Screening by PCR using NPTII gene specific primer pairs showed that this method reached over 90% efficiency. The final cloning efficiency by enzyme digestion and sequence analysis dropped to approximately 60–70% due to some mutations such as deletions or mismatches. However, these clones contained a correctly assembled insert of the 5′ flanking sequence, NPTII, and the 3′ flanking sequence. These clones were base perfect, suggesting that multi-fragment assembly by the In-Fusion cloning system can be easily applied for fungal gene disruption due to its high efficiency (Fig. 4C).
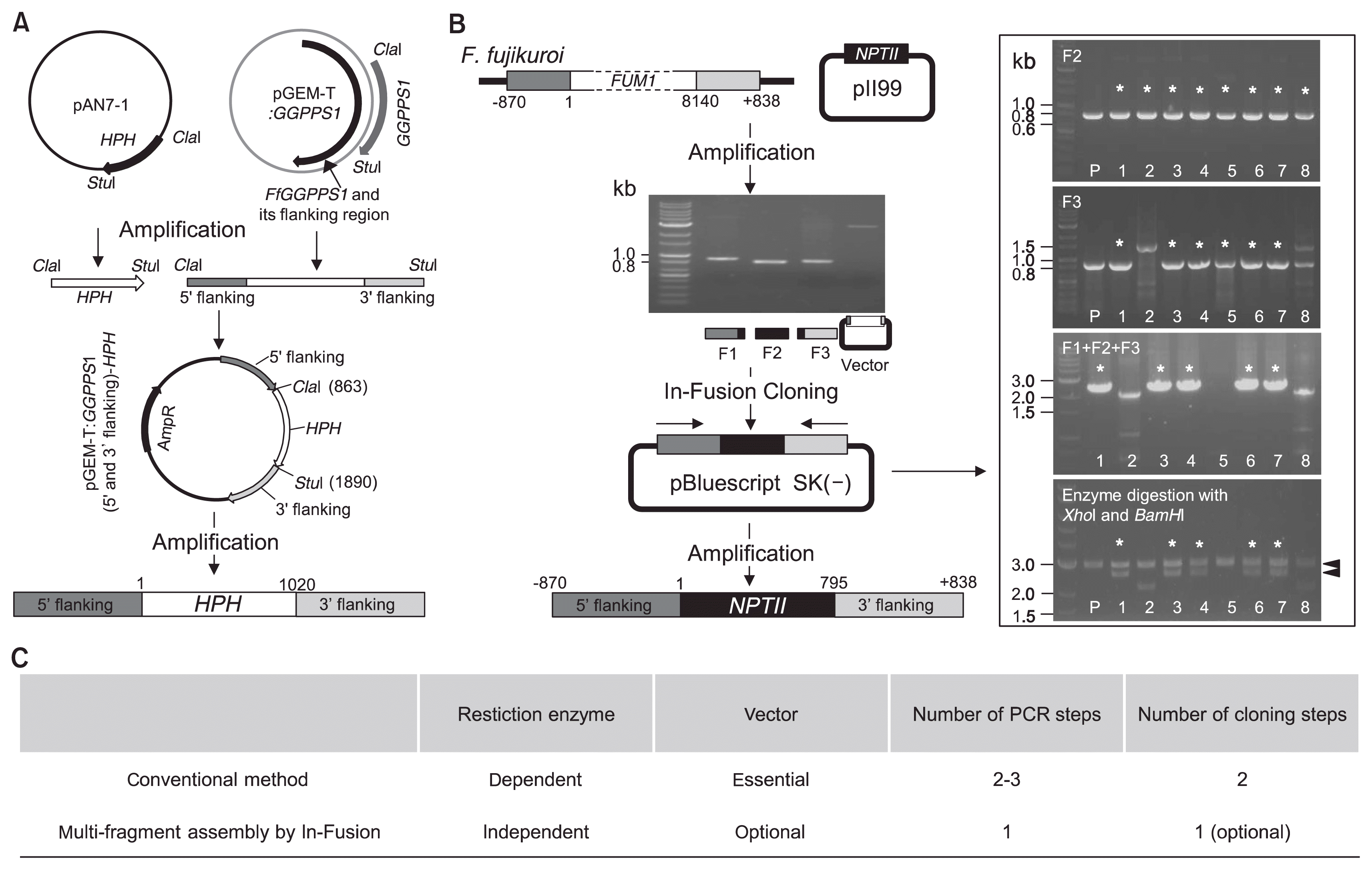
Schematic representation of the construction of a gene replacement cassette. (A) Construction of a disruption and replacement cassette using restriction enzyme- and ligase-dependent cloning. HPH, hygromycin B phosphotransferase. (B) Construct used for FUM1 (8.1 kb) disruption in Fusarium fujikuroi created using the In-Fusion cloning system. NPTII, neomycin phosphotransferase. *The marks indicate clones exhibiting expected results. (C) Comparison of the construction systems used in these experiments for targeted gene deletion in fungi.
Discussion
In F. verticillioides, the FCC1 is involved in fumonisin production by inhibiting the transcription of FUM5, which encodes a polyketide synthase necessary for FB1 biosynthesis (Shim and Woloshuk, 2001). This gene exhibits high homology across different Fusarium species, and FCC1 of the IMI58289 and B14 strains shares 100% sequence identity. In this study, we introduced a simple method to generate constructs for FCC1 gene disruption in the F. fujikuroi strain B14 in order to identify this gene’s function. Although restriction enzyme- and ligase-dependent cloning strategies for constructing gene targeting vectors are the most commonly used methods, they are at times inconvenient due to the need for unique restriction enzyme sites. In addition, it can be difficult to clone large fragments; sometimes, fungal genomic DNA has over 10 kb, including the 5′- and 3′-flanking regions necessary for homologous recombination. As mentioned, the conventional method requires complicated steps and takes several days to several weeks, depending on the success of the restriction enzyme digests necessary to insert a selectable marker cassette. Recently, strategies for vector construction that do not require multiple cloning steps have been developed to overcome these issues (Frandsen, 2011). Yu et al. (2004) reported a double-joint PCR method that does not require restriction enzyme sites for cloning steps. Similarly, construction by fusion PCR has been widely used for gene deletion in fungi (Kuwayama et al., 2002). However, these methods typically require multiple PCR steps to generate the final amplicon and can be complicated by low efficiency caused by the presence of repetitive DNA sequences. To get through these difficulties, many strategies have been developed for directional cloning and/or multi-gene/fragment assembly by homologous recombination. In Saccharomyces cerevisiae, cloning method by homologous recombination with high efficiency is also employed for single and multiple fragment assembly (Joska et al., 2014).
In this report, we introduced a simple method for the generation of a targeted gene disruption construct in F. fujikuroi. This method, which is termed multi-fragment assembly by In-Fusion cloning, requires only one standard PCR reaction and one fragment assembly reaction (Zhu et al., 2007). To generate a construct for FCC1 deletion, we first performed PCR using specific primers containing 15–30 bp overlapping sequences and then incubated 3 fragments and the linearized vector digested by a restriction enzyme at 50ºC for 1 hour. To obtain high levels of the final DNA construct containing 5′ and 3′ FCC1 flanking regions and the NPTII gene, we selected a strategy that is compatible with any linearized vector; multi-fragment assembly offers single-step construction regardless of the presence of a vector. This one-step method is simple, includes primer design, and is faster than restriction enzyme- and ligase-dependent cloning systems and/ or any other time-consuming methods used to generate constructs for fungal gene deletion. Using this strategy, we successfully created constructs for FCC1 deletion in a short period of time. We used these constructs to determine the relationship between fumonisin production and B14-mediated disease symptoms. Correct deletion of FCC1, which regulates fumonisin synthesis, in B14 resulted in compromised fumonisin production, leading to weakened disease symptoms during infection. This result demonstrates that fumonisin promotes non-typical bakanae symptoms in rice seedlings. In addition, our results showed that fcc1 mutants produce small amounts of fumonisins in planta during their interaction compare with when fcc1 mutants were grown in media, suggesting that F. fujikuroi produces numerous secondary metabolites such as fumonisins that may be not essential for life in vitro but are indispensable for survival under natural conditions, including during fungal-plant interaction.
In conclusion, we herein report the function of FCC1 via gene disruption in the F. fujikuroi strain B14. We have obtained several mutants that exhibited reduced fumonisin production or different growth patterns in vitro. Furthermore, we demonstrated the excellence of multi-fragment assembly. This method is a highly efficient process that is not dependent on restriction enzyme sites and/or ligation. Multi-fragment assembly by the In-Fusion cloning system can be easily controlled and modified for gene deletion in fungi, regardless of which restriction enzyme sites are needed, the size of the gene and its 5′- and 3′-flanking regions, or how many fragments are necessary for gene disruption. We expect that multi-fragment assembly will provide a wide range of possibilities for functional analysis through gene deletion in fungi.
Acknowledgments
The B14 F. fujikuroi strains used in this study were provided by Prof. Sung-Hwan Yun of Soonchunhyang University, Korea.
This research was supported by grants from the Research Program for National Institute of Agricultural Sciences (Project No. PJ01006401) and the Next-Generation BioGreen 21 (Project No. PJ01164301), Rural Development Administration, Korea.
Notes
Articles can be freely viewed online at www.ppjonline.org.