A Rice Gene Homologous to Arabidopsis AGD2-LIKE DEFENSE1 Participates in Disease Resistance Response against Infection with Magnaporthe oryzae
Article information
Abstract
ALD1 (ABERRANT GROWTH AND DEATH2 [AGD2]-LIKE DEFENSE1) is one of the key defense regulators in Arabidopsis thaliana and Nicotiana benthamiana. In these model plants, ALD1 is responsible for triggering basal defense response and systemic resistance against bacterial infection. As well ALD1 is involved in the production of pipecolic acid and an unidentified compound(s) for systemic resistance and priming syndrome, respectively. These previous studies proposed that ALD1 is a potential candidate for developing genetically modified (GM) plants that may be resistant to pathogen infection. Here we introduce a role of ALD1-LIKE gene of Oryza sativa, named as OsALD1, during plant immunity. OsALD1 mRNA was strongly transcribed in the infected leaves of rice plants by Magnaporthe oryzae, the rice blast fungus. OsALD1 proteins predominantly localized at the chloroplast in the plant cells. GM rice plants over-expressing OsALD1 were resistant to the fungal infection. The stable expression of OsALD1 also triggered strong mRNA expression of PATHOGENESIS-RELATED PROTEIN1 genes in the leaves of rice plants during infection. Taken together, we conclude that OsALD1 plays a role in disease resistance response of rice against the infection with rice blast fungus.
Body
Living organisms need to develop their built-in immune responses against the attack of natural enemies who can colonize in host organisms (Buchmann, 2014; Jones and Dangl, 2006). Infection either pathogenic microbes or insects causes severe yield losses in crop plants (Agrios, 2005). Since plants lack specialized immune cells and circulatory systems, they have developed a unique immune system to protect themselves from pathogen infection. In general, plants have three defensive layers in their immune system. Structural barriers and stomata-associated defense, as a first layer, prevent an invasion of pathogens into the plant tissues (Arnaud and Hwang, 2015; Malinovsky et al., 2014). For an induced immunity, secondly, plants recognize microbe-associated molecular patterns by pattern recognition receptors and initiate pattern-triggered immunity (Macho and Zipfel, 2014). Additionally, disease resistance proteins of plants monitor the perturbation of plant physiology by pathogen-derived effector proteins and then activate race-specific disease resistance, mainly called effector-triggered immunity (Chisholm et al., 2006; Jones and Dangl, 2006). In addition to the local induced immunity, plants also have a capability to stimulate systemic resistance in the distal tissues of plant against subsequent pathogen infection (Fu and Dong, 2013; Ryals et al., 1996). The whole plant immunity is often developed after either localized infection with microbes or treatment of specific immune activators, which were identified for the last decade (Chanda et al., 2011; Chaturvedi et al., 2012; Dempsey and Klessig, 2012; Gao et al., 2014; Jung et al., 2009; Návarová et al., 2012).
In the local and systemic resistance responses, salicylic acid (SA) takes charge in initiation and amplification of disease resistance responses against pathogen infection (Delaney et al., 1994; Gaffney et al., 1993; Jones and Dangl, 2006; Spoel and Dong, 2012). Lots of genetic and biochemical analyses revealed that SA-dependent signaling pathway requires dozens of key regulatory genes to establish successful immune response in plants, such as ALD1 (ABERRANT GROWTH AND DEATH2 [AGD2]-LIKE DEFENSE PROTEIN1), EDS1 (ENHANCED DISEASE SUSCEPTIBILITY1), NDR1 (NONRACE SPECIFIC DISEASE RESISTANCE1), NPR1 (NONEXPRESSOR OF PATHOGENESIS-RELATED PROTEINS1), PAD4 (PHYTOALAXIN DEFICIENT4), SID2 (SALICYLIC ACID INDUCTION DEFICIENT2), and WIN3 (HopW1-INTERACTING3) (Cao et al., 1997; Century et al., 1995; Falk et al., 1999; Jirage et al., 1999; Lee et al., 2007; Nawrath and Métraux, 1999; Song et al., 2004a, 2004b). Among them, ALD1 is necessary for local and systemic resistance in Arabidopsis thaliana (Cecchini et al., 2015; Song et al., 2004b). Indeed, an ald1 mutant showed the delayed SA accumulation and PR1 (PATHOGENESIS-RELATED PROTEIN1) expression. Additionally the ald1 mutant failed to accumulate FLS2 (FLAGELLIN-SENSING2) and BAK1 (BRASSINOSTEROID INSENSITIVE1-ASSICIATED RECEPTOR KINASE1) proteins in Arabidopsis during early infection phase (Cecchini et al., 2015). As a putative aminotransferase, ALD1 protein was also involved in the biosynthesis of pipecolic acid and unidentified compound(s) important for immune response in Arabidopsis (Cecchini et al., 2015; Návarová et al., 2012). The previous studies demonstrate a crucial role of ALD1 during defense response. However, a role of ALD1’s homologous genes in monocot plants was still unclear.
Rice plant (Oryza sativa) has two homologous genes of AtAGD2 and AtALD1 (Song et al., 2004a). Based on similarities of the deduced amino acid sequences, the two genes were named as OsAGD2 (Gen-Bank accession number AY338235; Os03g0299900) and OsALD1 (Gen-Bank accession number, AY338236; Os03g0195100) (Song et al., 2004a). Especially, OsALD1 shows 58% identity and 86% similarity to AtALD1, whereas it exhibits 61% identity and 82% similarity to AtAGD2 (Song et al., 2004a). Furthermore, OsALD1 displays 62% identity and 85% similarity to OsAGD2 (Song et al., 2004a). An interesting feature of their deduced amino acid sequences is that AtAGD2 and OsAGD2, but not both AtALD1 and OsALD1, have chloroplast transit signal peptides. In the absence of typical transit peptides to target chloroplast, however, AtALD1 predominantly accumulated at chloroplast in plants (Cecchini et al., 2015; Song et al., 2004a).
Both AtALD1 and NbALD1 (Nicotiana benthamiana ALD1) (The Gene Indices [TGI databases, http://compbio.dfci.harvard.edu/tgi/], TC23014) genes were strongly expressed in the infected leaves with Pseudomonas syringae (Cecchini et al., 2015; Song et al., 2004b). Additionally, local immunization with systemic acquired resistance (SAR)-inducing P. syringae activated transcription of ALD1 in the distal systemic leaves of Arabidopsis and tobacco plants (Cecchini et al., 2015; Song et al., 2004b). To test if OsALD1 mRNA expression was also induced by pathogen infection, we inoculated rice plants (cultivar Dongjin) with the rice blast fungus, Magnaporthe oryzae KJ-105a isolate. Rice plants grew in rice nursery media (Bu-Nong, Korea) under 16-hour-day and 8-hour-night conditions at 25°C. Three-week old plants were used for the infection experiment. Fungal spore suspension (5 × 105 conidia/ml) was applied on the leaves of rice plants with a paintbrush. Inoculated plants were kept in the humid chamber under 16-hour-day and 8-hour-night conditions at 25°C to monitor disease development and to check gene expression during defense responses and pathogenesis. The infection with M. oryzae KJ-105a isolate successfully resulted in rice blast disease in cv. Dongjin plant under our experimental conditions (Fig. 1A). As well expression of M. oryzae β-Tubulin2 (MoTUB), a fungal gene, was dramatically increased in the infected leaves (Fig. 1B). These results indicate that the M. oryzae KJ-105a isolate successfully colonized in the leaves of cv. Dongjin plant after inoculation. Next, to examine if the infection with M. oryzae induced transcriptional change of defense/pathogenesis-related genes in the leaves of cv. Dongjin plant, total RNAs were extracted from the leaves of rice plants after mock-inoculation and pathogen infection by using Trizol® reagent (Thermo Fisher Scientific Inc., San Jose, CA, USA). Experimental procedure and data analysis for quantitative real-time RT-PCR were described in the previous studies (Jung et al., 2009; Livak and Schmittgen, 2001; Pfaffl, 2001). Nucleotide sequences of primers used in this study are presented in Table 1. A basic PR1, OsPR1b gene (Os01g0382000) was strongly transcribed in the leaves after M. oryzae infection (Fig. 1C), suggesting that the pathosystem between cv. Dongjin plant and M. oryzae KJ-105a isolate was sufficient to analyze gene expression and disease resistance response in rice plant against the fungal infection. Transcription of OsALD1 gene was also increased in the infected leaves, as compared with that in mock-treated plants (Fig. 1D). Additionally, it is likely that high humidity can slightly affect transcription of OsALD1 in rice plant, since even mock-inoculation partly induced OsALD1 mRNA expression. The strong expression of OsALD1 was continued till 4 days after infection. The result shows a possibility that OsALD1 takes part in defense response or pathogenesis in rice plant.

An OsALD1 gene, whose products accumulated at chloroplast, was strongly expressed in the infected leaves of rice plants with rice blast fungus. (A) Symptom development of rice blast disease in Oryza sativa cv. Dongjin infected by a Magnaporthe oryzae KJ-105a isolate. (B) Fungal growth was verified by quantifying expression of M. oryzae β-Tubulin2 (MoTUB). The relative expression level was calculated by a 2−ΔΔCT method (Livak and Schmittgen, 2001). A rice ubiquitin gene was used as an internal reference gene. (C, D) Infection with the rice blast fungus triggered expression of O. sativa PATHOGENESIS-RELATED PROTEIN1b (OsPR1b) (C) and OsALD1 (D) genes in the infected leaf tissues. Relative expression ratios were computed by a standard curve-based method (Pfaffl, 2001). mRNA levels of each sample were normalized by that of cv. Dongjin plants before mock-inoculation. Data represent the average with standard deviation (n = 3). Either fungal spore suspension (gray bars) (5 × 105 conidia/ml) or water (white bars) was inoculated with a paintbrush on the leaves (A–D). These experiments were repeated twice with the same results. (E) OsALD1 proteins localized at chloroplast in the leaves of Nicotiana benthamiana. OsALD1:GFP construct, whose expression was conditionally controlled by dexamethasone (DEX)-inducible promoter, was introduced in the leaves of N. benthamiana in accordance with an Agrobacterium-mediated transient expression protocol. Green fluorescence protein (GFP) was visualized 1 day after DEX (30 μM) treatment under a confocal microscopy (× 100). DAI, days after inoculation.
AtALD1 proteins localized to chloroplasts in plant cells, even though AtALD1 did not have typical transit signal peptides for chloroplast localization (Cecchni et al., 2015; Song et al., 2004a). As like AtALD1, an OsALD1 protein also does not possess a chloroplast transit signal. In order to visualize where in the cell OsALD1 acts, we expressed fusion proteins of OsALD1 with green fluorescence protein (GFP) under the control of dexamethasone (DEX)-inducible promoter in the leaves of N. benthamiana. Full-length cDNA fragments of OsALD1 gene were amplified with gene-specific primers (Table 1), and then sub-cloned into Gateway® pENTR™/D-TOPO vector (Thermo Fisher Scientific Inc.), which forming an entry clone, named a pENTR:OsALD1. After LR reaction via Gateway technology (Thermo Fisher Scientific Inc.), the full length OsALD1 cDNA was cloned into a pBAV150 plant binary vector (Vinatzer et al., 2006). The resulting construct (pBAV150:OsALD1) was introduced into Agrobacterium tumefaciens GV3101 via electroporation (BioRad, CA, USA). A. tumefaciens GV3101 harboring pBAV150:OsALD1 was sequentially incubated into Agrobacterium induction media (0.1 × Murashige and Skoog [MS] salt, 0.1% B5 vitamins, 1% [w/v] glucose, 2% [w/v] sucrose, and 100 μM acetosyringone in 20 mM 3-(N-morpholino) propanesulfonic acid [MOPS], pH 5.4) and infiltration media (0.1 × MS salt, and 200 μM acetosyringone in 20 mM MOPS, pH 5.4) (Rathjen et al., 1999). The transient expression assay was performed in the leaves of 3-week old N. benthamiana plants as described previously (Jung et al., 2005; Rathjen et al., 1999). In order to induce expression of OsALD1:GFP fusion proteins, 30 μM DEX solution was applied onto the leaves of plants 1 day after Agro-infiltration (optical density measured at a wavelength of 600 nm [OD600] = 0.1). GFP localization and red auto-fluorescence were visualized 1–2 days after treatment using a confocal microscopy. In agreement with a chloroplast localization of AtALD1, green fluorescence from OsALD1:GFP proteins predominantly co-detected with the red fluorescence of chloroplast in the leaves of N. benthamiana (Fig. 1E).
To test whether or not OsALD1 gene was involved in disease resistance response of rice plant, we developed transgenic rice plants over-expressing OsALD1. As a result of LR reaction between the entry clone (pENTR: OsALD1) and the destination vector (pB7WG2D) (Karimi et al., 2002), the full-length OsALD1 cDNA was introduced into the plant expression vector. Transgenic rice plants were generated by Agrobacterium-mediated transformation with rice calli obtained from cv. Dongjin plant, as previously described (Shin et al., 2012). Each individual transformant was verified by PCR with primers for Bialophos resistance gene- and OsALD1 gene-specific primers (Table 1) (data not shown). Regenerated T0 plants were grown for massive reproduction under a green house. We checked OsALD1 mRNA expression in the leaves of independent T1 transgenic lines without pathogen infection. Two independent transgenic plants over-expressing OsALD1 (OsALD1-ox, line # 1 and #5) under the control of Cauliflower mosaic virus 35S promoter exhibited the strong increase of OsALD1 mRNA expression in the rice leaves without pathogen infection (Fig. 2A). On the other hand, several transgenic plants failed to vigorously express OsALD1, compared with non-transgenic plants. Thus we cultivated and maintained these two transgenic plants for further analysis.
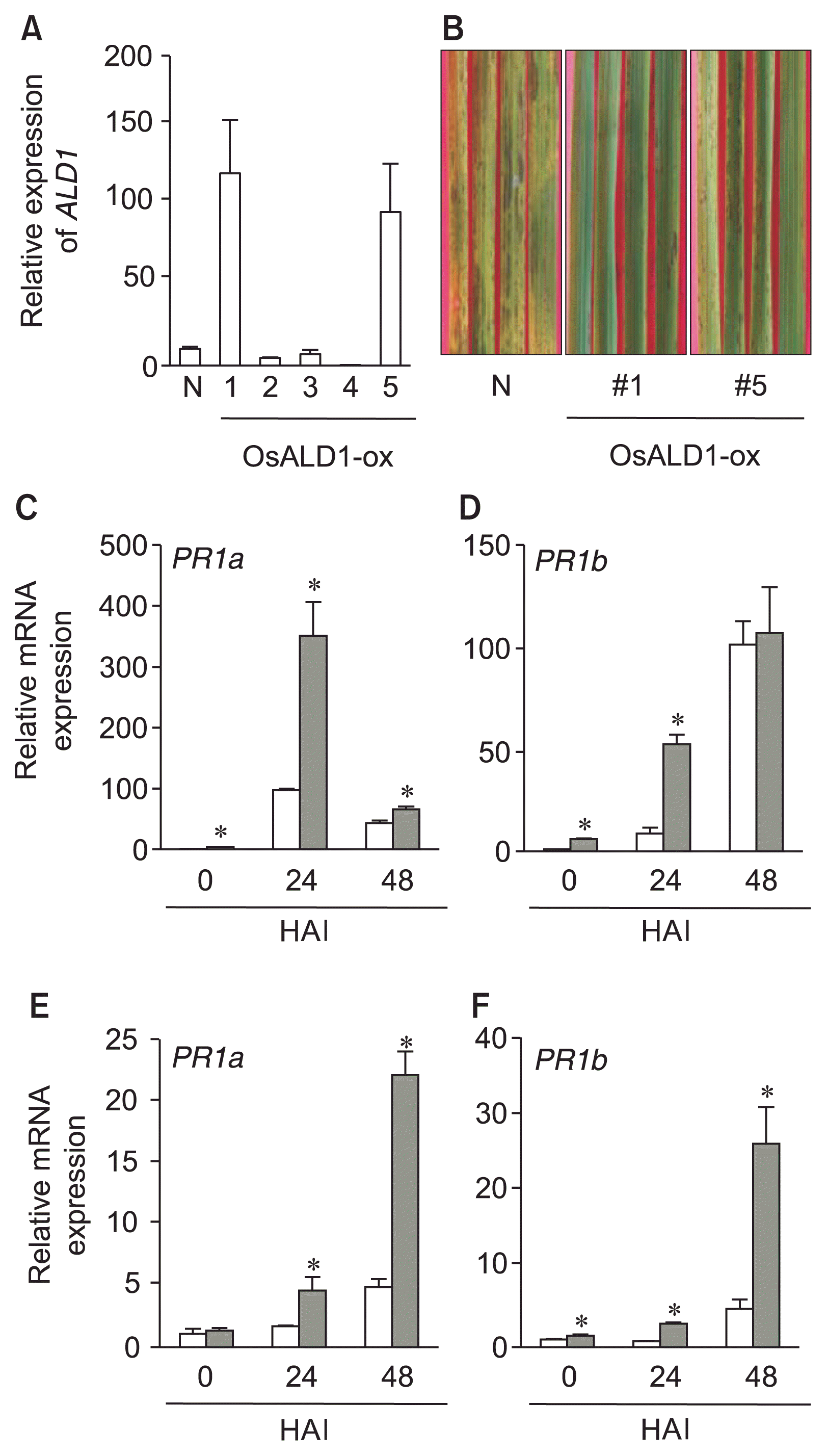
OsALD1 conferred disease resistance response on rice plant against the infection with rice blast fungus. (A) The level of OsALD1 transcript in the leaves of non-modified (N) and individual transgenic T1 plants. Relative OsALD1 mRNA level in over-expressing plants was compared with that of the non-infected plants (OsALD1/actin). The standard curve-based method was used to analyze mRNA expression ratios. Data represent the average with standard deviation (n = 3). (B) Disease symptoms of non-modified (N), and OsALD1 over-expressing plants (line #1 and #5) after the fungal infection (5 × 105 conidia/ml). The photos were taken on day 4 after inoculation. The experiments were repeated three times with similar results. (C–F) mRNA expression of PATHOGENESIS-RELATED PROTEIN1a (PR1a) (C, E) and PR1b (D, F) genes in the leaves of non-transgenic (white bars) and OsALD1-ox plants (line #1 [C, D] and #5 [E, F]) (gray bars) after the infection by M. oryzae (5 × 105 conidia/ml). Expression levels of PR1a and PR1b were calculated by the comparative CT method (2−ΔΔCT) (Livak and Schmittgen, 2001). The asterisks indicate statistically differences between wild type and transgenic plants at each time point after infection (*P < 0.05, two-tailed student t-test). The experiments were repeated twice with similar results. HAI, hours after inoculation.
In order to examine if a stable expression of OsALD1 conferred disease resistance on rice plant against the infection with rice blast fungus, we inoculated the detached leaves of non-transgenic rice plants (cv. Dongjin) and OsALD1-ox transgenic plants (T2 generation) with M. oryzae KJ-105a isolate (5 × 105 conidia/ml). The infected leaves were incubated on 25°C for 4 days under high humidity conditions (16-hour day/8-hour night). Over-expression of OsALD1 led to the decrease of symptom development by the infection of rice blast fungus, whereas non-transgenic plants showed severe disease symptom after infection (Fig. 2B). We independently repeated the experiments three times, and got the same results. This strongly proposes that OsALD1 plays a role in disease resistance response in rice plant. In Arabidopsis, PR1 expression was suppressed in the ald1 mutant during Pseudomonas infection (Cecchini et al., 2015; Song et al., 2004b). On the other hand, over-expression of AtALD1 in Arabidopsis plant strongly induced PR1 expression in early infection phase, as compared with wild-type plant (Cecchini et al., 2015). These previous studies presented a possibility that ALD1 regulates PR1 expression in the infected leaves by pathogen. To test this, we checked the mRNA levels of two different PR1 genes, acidic PR1 (Os-PR1a) (Os07g0129200) and basic PR1 (OsPR1b), in the leaves of non-transgenic and OsALD1-ox plants (line #1 and #5) during infection. Transcript levels of PR1 genes were higher in the OsALD1-ox plants than those in the non-transgenic plants in the presence and absence of M. oryzae infection (Fig. 2C–F). In general, SA-dependent defense signaling regulates expression of PR1 genes in plants (Loake and Grant, 2007). In rice plant, however, SA directs basal and constitutive defense responses rather than inducible immunity after pathogen infection (Silverman et al., 1995). Based on these previous studies and our results, we propose that ALD1 may act as a key regulator to control transcription of PR1a and PR1b genes in rice plant after pathogen infection.
In conclusion, a main strategy to enhance disease resistance in rice plant against rice blast fungus is to use disease resistance (R) genes (Liu et al., 2010; Miah et al., 2013). However the race-specific resistance was fragile, because of arms race between plants and pathogenic microbes (Jones and Dangl, 2006). Thus it is also necessary to gather defense regulatory genes to develop more durable disease resistance in plants (Fukuoka et al., 2015; Helliwell and Yang, 2013; Liu et al., 2014). At this point of view, ALD1 is an excellent candidate to increase basal disease resistance in rice plant, since ALD1 is involved in the synthesis of signal compounds and expression of defense-related genes without any morphological and developmental defects. As well, it seems that OsALD1 overexpressing plant may be a good material to study defense signaling in rice plant against M. oryzae infection.
Acknowledgments
This research was supported by Wu Jang-Choon Project from the Rural Development Administration (RDA) grant (PJ007850052014) and Basic Science Research Program from the National Research Foundation (NRF) of Korea (2010-0006441) to Ho Won Jung.
Notes
Articles can be freely viewed online at www.ppjonline.org.
