Isolation and Characterization of a Bacteriophage Preying an Antifungal Bacterium
Article information
Abstract
Several Bacillus species were isolated from rice field soils, and 16S rRNA gene sequence analysis showed that Bacillus cereus was the most abundant. A strain named BC1 showed antifungal activity against Rhizoctonia solani. Bacteriophages infecting strain BC1 were isolated from the same soil sample. The isolated phage PK16 had an icosahedral head of 100 ± 5 nm and tail of 200 ± 5 nm, indicating that it belonged to the family Myoviridae. Analysis of the complete linear dsDNA genome revealed a 158,127-bp genome with G + C content of 39.9% comprising 235 open reading frames as well as 19 tRNA genes (including 1 pseudogene). Blastp analysis showed that the proteins encoded by the PK16 genome had the closest hits to proteins of seven different bacteriophages. A neighbor-joining phylogenetic tree based on the major capsid protein showed a robust clustering of phage PK16 with phage JBP901 and BCP8-2 isolated from Korean fermented food.
The disease triangle, comprising host, pathogen, and environment, is a fundamental concept in plant disease development (Scholthof, 2007). In addition to the relationships among these three factors, each component of this triangle is related to other physicochemical and biological factors. For example, a pathogenic fungus can be affected by antifungal bacteria, which can be affected by lytic bacteriophages (Balogh et al., 2010; Song et al., 2012). However, analyses of the multifactorial biological interactions among these organisms have been rare.
Bacillus cereus is gram-positive endospore-forming bacterium belonging to the B. cereus sensu lato group, which includes B. cereus, B. thuringiensis, B. anthracis, B. mycoides, B. pseudomycoides, and B. weihenstephanensis (Tourasse et al., 2006). B. cereus is an opportunistic foodborne pathogen that causes food poisoning. No B. cereus has been reported as a plant pathogen; indeed, some strains show antifungal activity against plant-pathogenic fungi, including Fusarium oxysporum, F. solani, and Pythium ultimum (Chang et al., 2007). In addition, B. cereus strain QQ306 has been found to enhance the growth of Chinese cabbage (Chang et al., 2007). For these reasons, B. cereus is a potential biological control agent for plant disease (Emmert and Handelsman, 1999).
In contrast to B. cereus, which inhibits plant-pathogenic fungi, bacteriophages (phages) have been proposed as biological control agents to protect foods from B. cereus contamination; indeed, their use in foods is allowed by the U.S. Food and Drug Administration (FDA) (Cairns and Payne, 2008). In the present study, we characterize a B. cereus strain and phages infecting this bacterium that were isolated during a search for bacteriophages preying on the grain rot bacterium Burkholderia glumae.
B. cereus was isolated from three soil samples collected from a rice field in Geumma-myeon, Hongsung-gun, Chungchungnam-do, South Korea. Two grams of soil were mixed with fresh Luria-Bertani (LB) broth medium and incubated at 37°C for 24 h with shaking. The mixture was then centrifuged at 3,000× g for 20 min at 4°C. The super-natant was spread on LB agar plates at different dilutions and incubated at 37°C for 24 h. Three bacteria showing different morphological characteristics were isolated. Genomic DNA was isolated from these three bacteria; the 16S rRNA gene was amplified using 27F and 1492R primers (Lane, 1991) and then sequenced. Blast (https://blast.ncbi.nlm.nih.gov/Blast.cgi) analysis showed that the isolated bacteria were B. cereus, B. pseudomycoides, and Bacillus sp. B. cereus is widely distributed in various environments because it can grow at a broad range of temperatures (5–55°C) and form endospores, and it is generally resistant to penicillin-related and other antibiotics (Lee et al., 2014). It is commonly isolated from foods and food additives, and it is expected to be prevalent in rice fields. The B. cereus strain named BC1 was most abundant, and it was used for antifungal activity tests and the isolation of bacteriophages.
The antifungal activity of the isolated B. cereus BC1 strain was tested against Rhizoctonia solani, F. solani, and Magnaporthe oryzae. Burkolderia pyrrocinia CH-67, which is known to have antifungal activity (Song et al., 2012), and Escherichia coli DH5α were used as positive and negative controls, respectively. The bacteria R. solani and F. solani were inoculated onto a potato dextrose agar plate at the same time and incubated at 37°C. Owing to differences in growth rates, M. oryzae was cultured for 6 days at 24°C before being inoculated onto the plates. The isolated B. cereus strain showed no inhibition against M. oryzae and F. solani, but showed detectable inhibition against R. solani (Fig. 1). B. cereus was reported to have antifungal activity against several plant-pathogenic fungi (Chang et al., 2007), perhaps due to hydrolytic enzymes, including chitinase, chitosanase and protease, that are induced when the bacterium is cultured in a medium containing shrimp and crab shell powder. In addition to hydrolytic enzymes, several antifungal agents have been reported from B. cereus (Fujiu et al., 1994; He et al., 1994). The cell walls of the plant-pathogenic fungi used in the present experiment consisted of chitin. Thus, a similar inhibitory effect is expected against these fungi if the main antifungal activity is mediated by hydrolytic enzymes. The difference in the growth inhibition of different fungi by B. cereus strain BC1 suggests that the antifungal activity may due to unidentified agents rather than hydrolytic enzymes.

Antifungal activity of Bacillus cereus isolated from a rice field. B. cereus BC1, Burkolderia pyrrocinia CH-67 (positive control), and Escherichia coli DH5α (negative control) were simultaneously inoculated with Rhizoctonia solani on potato dextrose agar plates. The plate is a representative from three replications and 4 days incubation at 37°C.
Bacteriophages infecting the isolated B. cereus BC1 strain were isolated from the same soil samples. For enrichment of phages, 2 g of soil sample was homogenized in 2 ml of overnight culture of B. cereus and 1 ml of fresh LB broth, and then incubated at 37°C for 48 h. After centrifugation at 3,000× g for 20 min at 4°C, the supernatant was filtered using a 0.2-μm filter (GVS Filter Technology, Seoul, Korea). Diluted filtrates were mixed with exponentially growing host cells and molten 0.7% LB soft agar, and then plated on LB medium. After incubation at 37°C for 15 h, plaques were picked and kept in 1 M SM buffer (100 mM NaCl, 8 mM MgSO4·7H2O, and 50 mM Tris-Cl [pH 7.5]). Plaque isolation was repeated three times to purify single phages from two original plaques of different size. One of the phages producing smaller plaques was named PK16 and used for further study.
The host range of the phage PK16 was tested against six B. cereus strains obtained from Korean Collection of Type Cultures (KCTC), Biological Resource Center (BRC), Korean Research Institute of Bioscience and Biotechnology (KRIBB), and three bacterial strains isolated in the present study. Phage PK16 infected all three of the latter bacterial strains and three of six B. cereus strains from KCTC (Supplementary Table 1). Interestingly, phage PK16 could infect B. pseudomycoides in addition to B. cereus. Despite the large number of phages that infect the Bacillus group, to date, there has been no report of phages infecting B. pseudomycoides. One explanation may be that this species has not been found within the host range. Information on phage PK16 will be useful for future research on phages infecting B. pseudomycoides.
The one step growth of the phage was investigated by inoculating exponentially growing host with the phage at the multiplicity of infection of 0.01. After 5 min incubation, the bacteria was centrifuged at 3,000× g for 20 min to remove unbound phage in the supernatant and the pelleted cells were resuspended with fresh medium and incubated at 37°C with shaking. Five hundred microliter sample was taken every 10 min, spun down with centrifugation (3,000× g, 4°C, 5 min) and virus titer in the supernatant was determined by plaque assay. The one-step growth curve of phage PK16 showed an incubation period of about 30 min and a complete replication cycle of 30 min, which is similar to that of other B. cereusinfecting phages such as JBP901 (Shin et al., 2011).
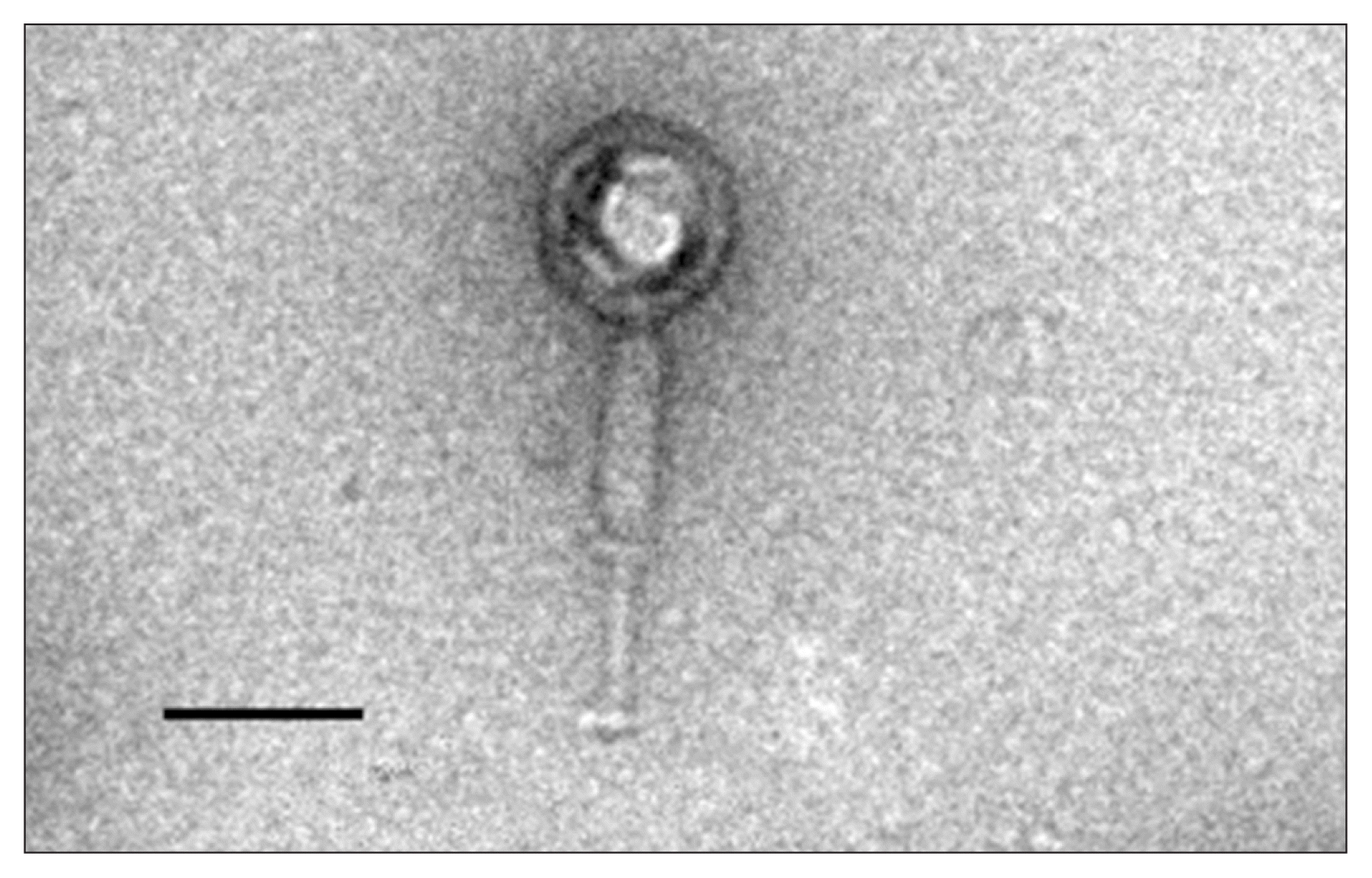
Phage PK16 was purified using procedures described by Zink and Loessner (1992) with modifications. The purified virus was fixed with 0.2% formaldehyde, stained with 1% uranyl acetate, and observed using a bio-transmission electron microscope (Tecnai G2 Spirit Twin; FEI, Hillsboro, OR, USA) at the Korean Basic Science Institute, Ochang. The average diameter of the isometric head was about 100 ± 5 nm, and the tail was about 200 ± 5 nm in length × 23 ± 5 nm (Fig. 2). All of the phages infecting B. cereus are known to have a dsDNA genome and to belong either to the order Caudovirales or to the family Tectiviridae. Phages belonging to the order Caudovirales are divided into three families: Myoviridae (long contractile tails), Siphoviridae (long non-contractile tails), and Podoviridae (short non-contractile tails) based on the morphology of their tails; phages of Myoviridae are the most abundant phages of the B. cereus group. Many myoviruses infecting the B. cereus group are reported to have characteristics of the recently proposed subfamily Spounavirinae, which have heads about 84–94 nm in diameter and striated tails of 140–219 nm in length (Gillis and Mahillon, 2014). Phage PK17 was morphologically similar to other myoviruses of the B. cereus group, and its genomic DNA was analyzed for further study.
Viral nucleic acid was extracted from purified viruses and confirmed to be double-stranded DNA based on digestion with DNase, RNase, and selected restriction enzymes. Thus far, there have been no reports of single-stranded DNA or RNA phages infecting B. cereus, and all of the dsDNA B. cereus phages belong to the order Caudovirales or the family Tectiviridae (King et al., 2012). Among the three families of tailed phages in the order Caudovirales, Myoviridae, Siphoviridae, and Podoviridae, which have long contractile tails, and Siphoviridae, which have long non-contractile tails, are the most abundant among the B. cereus phages (Gillis and Mahillon, 2014).
The entire genome sequence of phage PK16 was analyzed at the National Instrumentation Center for Environmental Management (NICEM), Seoul National University, using a hierarchical genome-assembly process (HGAP) and a single, long-insert shotgun DNA library in conjunction with single-molecule, real-time (SMRT) DNA sequencing. Prediction of open reading frames (ORFs) was carried out using the RAST server (http://rast.nmpdr.org/), and Blastp analysis of amino acid sequences was performed using the NCBI database (https://blast.ncbi.nlm.nih.gov/Blast.cgi). tRNA prediction was conducted using the tRNAscan-SE program (http://lowelab.ucsc.edu/tRNAscan-SE). A total of 1,355,071,057 bp were read from 81,484 reads containing 24,082 reads of over 50 nucleotides. The genome of phage PK16 is 158,127 bp, with G + C content of 39.9% (GenBank accession No. KX495186), which is within the range of genome sizes of B. cereus phages in the family Myoviridae (which range from 94 to 219 kb) (Lee et al., 2014). A total of 235 putative protein coding sequences, of which 156 encode hypothetical proteins, and 19 tRNA genes (including 1 pseudogene), were identified. Identifiable proteins encoded by the PK16 genome included structural genes (major capsid, tail sheath and tubes, base plate), as well as proteins involved in the infection process (two tail lysins), cell lysis (autolysin and holin), and DNA replication and modification (DNA polymerases, helicases, a primase, a recombinase, methylase) (Supplementary Fig. 1). Although over 30 genome sequences of bacteriophages infecting the B. cereus group have been reported, only 34.9–93.5% of the annotated ORFs were assigned hypothetical proteins (as in the case of phage PK16), owing most likely to the lack of sufficient gene annotation data for these phage genomes (Lee et al., 2014).
A protein of interest among those encoded by phages of B. cereus is the peptidoglycan hydrolyzing enzyme autolysin. After the 2001 anthrax attacks in the USA, several autolysins of B. cereus phages have been studied intensively as a control agent of B. anthracis, and have shown lytic effect on both vegetative cells and germinating spores (Schuch et al., 2002). Endolysins have also shown a strong antimicrobial effect on food-borne pathogenic strains of B. cereus. In particular, autolysin LysBPS13, encoded by phage BPS13, showed antimicrobial activity against Bacillus species and some gram-negative bacteria even after incubation at 100°C for 30 min in the presence of glycerol, suggesting possible applications in the food industry (Park et al., 2012). Phage PK16 encodes a putative autolysin in ORF 12 that shows 98% amino acid sequence identity to that of Bacillus phage BM15, and requires further characterization in order to determine its activity and potential utility.
The presence of 18 functional tRNA genes in the PK16 genome is intriguing, and suggests that these genes may assist in translation of the phage mRNAs. Bailly-Bechet et al. (2007) showed that tRNAs were selected to facilitate phage integration into prokaryotic chromosomes and that there was a significant association between tRNA distribution and codon usage. In a comparative codon usage analysis of the B. cereus host and its phage BCP78, Lee et al. (2012) also revealed different codon preferences for asparagine, phenylalanine, and serine. Conversely, analysis of translational codon usage bias in B. cereus phage TsarBomba, which has 20 tRNA-coding genes, indicated that these tRNAs do not alter the host tRNA pool (Erill and Caruso, 2015). Comparison of the codon usage of phage PK16 and B. cereus NC7401 strain (GenBank accession No. AP007209.1) showed that 9 of 17 codons (excluding the AUG start codon) that had corresponding tRNA genes in the phage genome showed higher usage in the phage genome than did the bacteria. In particular, a significant preference of the codons corresponding to phage-encoded tRNA was observed in proline, glycine, and threonine (data not shown). Although more codon usage analysis in B. cereus strains that are susceptible to and resistant against phage PK16 is necessary, it seems that the contribution of tRNA genes encoded by phages infecting B. cereus differs among host-phage combinations, which may explain the range of single phages that infect different strains of the same B. cereus species.
Blastp analysis showed that the proteins encoded by the PK16 genome had the closest hits to proteins of seven bacteriophages (Supplementary Table 1). Bacillus phage JBP901 was most similar to PK16, and 105 ORFs of phage JBP901 showed the greatest similarity to corresponding ORFs. The relative locations of ORFs showing amino acid similarity are shown in Fig. 3 (Sullivan et al., 2011). Despite the high degree of similarity, some ORFs present in PK16 are not present in phage JBP901, and vice versa. A phylogenetic tree based on the amino acid sequence of the major capsid protein, constructed using the neighbor-joining method, showed a clustering of phage PK16 with B. cereus phages JBP901 and BCP8-2 (Supplementary Fig. 2). Phages JBP901 and BCP8-2 were both isolated from Korean fermented food and exhibited a lysis profile specific to the B. cereus group, with a broad spectrum of hosts among the B. cereus isolates (Shin et al., 2011). These two phages have been tentatively classified as members of the SPO1-like group of the family Myoviridae, but genome analysis revealed that JBP901 lacks deoxyuridylate hydroxymethyl transferase, a signature gene of SPO1 phages (Asare et al., 2015). Phage PK16 also lacks this gene, and it seems that these three phages form a distinguishable group within the sub-family Spounavirinae of the family Myoviridae, as suggested by Asare et al. (2015).

Comparative genomic analysis of phage PK16 and related phages preying on the Bacillus cereus group. Phages that share over 10 best-hit proteins with PK16 were selected and compared using the Easyfig 9 program (Sullivan et al., 2011).
Because phages JBP901 and BCP8-2 were found in Korean fermented food, these phages have been considered candidates for removal of the opportunistic foodborne pathogenic B. cereus group (Bandara et al., 2012). However, our data showed that the host bacterium has antifungal activity against the plant-pathogenic fungus R. solani. This reveals the complicated nature of interactions among microorganisms in nature, and further analysis on the multifactorial relationship among the host plant, pathogenic fungus, antifungal bacteria and bacteriolytic phages may provide insight into the coevolution of these organisms. Phage PK16 is a lytic phage and no gene related to antifungal activity was identified in its genome. Therefore, is seems that the phage itself is not involved in the antifungal activity of the bacterium. However, prevalence of the Bacillus infecting phage in diverse environment can affect the population of the B. cereus sensu lato group and dynamic relationship in nature.
Supplementary materials
Acknowledgments
This work was supported by a Research Grant of Pukyong National University (2015 year). The authors appreciate Professor Min Hee Lee of the Department of Earth & Environmental Sciences, Pukyong National University for providing soil samples.