Lucerne transient streak virus; a Recently Detected Virus Infecting Alfafa (Medicago sativa) in Central Saudi Arabia
Article information
Abstract
A survey was conducted to determine the status of Lucerne transient streak virus (LTSV) in three high-yielding alfalfa regions in central Saudi Arabia (Riyadh, Qassim, and Hail) during 2014. Three hundred and eight symptomatic alfalfa, and seven Sonchus oleraceus samples were collected. DAS-ELISA indicated that 59 of these samples were positive to LTSV. Two isolates of LTSV from each region were selected for molecular studies. RT-PCR confirmed the presence of LTSV in the selected samples using a specific primer pair. Percentage identity and homology tree comparisons revealed that all Saudi isolates were more closely related to each other but also closely related to the Canadian isolate-JQ782213 (97.1–97.6%) and the New Zealand isolate-U31286 (95.8–97.1%). Comparing Saudi isolates of LTSV with ten other sobemoviruses based on the coat protein gene sequences confirmed the distant relationship between them. Eleven out of fourteen plant species used in host range study were positive to LTSV. This is the first time to document that Trifolium alexandrinum, Nicotiana occidentalis, Chenopodium glaucum, and Lathyrus sativus are new host plant species for LTSV and that N. occidentalis being a good propagative host for it.
Introduction
Lucerne transient streak virus (LTSV) belongs to the sobemo group of plant viruses (Rogalska, 2012). It is an isometric virus with 27–28 nm diameter (Jones et al., 1983). LTSV isolates are distinct in host ranges and symptomologies (Paliwal, 1983; Rogalska, 2012). LTSV was reported from alfalfa in Australia, New Zealand, and Canada (Blackstock, 1978; Forster and Jones, 1979; Paliwal, 1983). Al-Shahwan et al. (2013), in 2013 reported the presence of LTSV for the first time in Saudi Arabia. According to Ministry of Agriculture in 2014 alfalfa comprises 67% of the total production of fodder crops in Saudi Arabia and 74% of alfalfa production comes from the three regions; Riyadh, Qassim, and Hail regions in the Central Saudi Arabia (Ministry of Agriculture, Kingdom of Saudi Arabia, 2014). In a recent study, LTSV found to be the third most important virus infecting alfalfa in Saudi Arabia (Al-Shahwan et al., 2013). Its absence from Europe and Africa, and its recent detection in Asia (Al-Shahwan et al., 2013) makes it more worthwhile to be studied. The main objective of this study was to survey alfalfa fields in these three high yielding alfalfa-producing regions in the central Saudi Arabia for determining the status of this virus and to characterize representative isolates biologically, serologically and molecularly from these regions and their phylogenetic relationships with other isolates deposited in the GenBank.
Materials and Methods
Field survey and analysis
Alfalfa fields in Riyadh, Qassim, and Hail regions were surveyed during 2014 for detection of LTSV. Three hundred and fifteen symptomatic samples were collected from alfalfa and Sonchus oleraceus growing in or adjacent to alfalfa fields (190 from Riyadh, 63 from Qassim, and 62 from Hail) to identify LTSV in these samples. Different symptoms like mosaic, mottling, stunting, interveinal chlorosis and small leaves were mostly observed on the collected plant samples. These samples were kept in plastic bags to avoid contamination and dehydration. Samples were either processed immediately or stored at −80°C for further analysis.
DAS-ELISA was used for detection of LTSV from surveyed alfalfa and S. oleraceus symptomatic samples. Kits were purchased from a commercial producer, AC Diagnostics Inc. (Fayetteville, AR, USA), and company protocol was followed for detection. Samples were ground in extraction buffer and then tested by DAS-ELISA in duplicates. Absorbance values were measured at 405 nm on ELx808™ absorbance microplate reader. Positive and negative controls provided by the AC Diagnostics Inc. were also used. Absorbances of at least twice the mean value of negative controls were considered positive.
RT-PCR analysis
Two LTSV isolates from each region were selected for nucleic acid extraction on the basis of their highest reactions to DAS-ELISA. Isolates selected for molecular studies from Riyadh region were 56-R, and 83-R, those from Qassim region were 22-Q, and 25-Q, and those from Hail region were 4-H, and 23-H. Total RNA extractions from these selected isolates were carried out using Total RNA isolation kit (Jena Bioscience, Jena, Germany) according to the manufacturer’s instructions. Eluted RNA was stored at −80°C for further use.
A primer pair was designed to amplify a fragment of 1,033 bp in the coat protein region of LTSV genome by the Primer-BLAST tool of National Center for Biotechnology Information (NCBI) using the LTSV isolates already submitted to NCBI database (Ling et al., 2013). The forward primer designed for RT-PCR was 5′-GGATCGTTGCTATCTCCAAT-3′ while the reverse primer was 5′-CGAAAACCATCATACGGCTA-3′. RT-PCR was carried out using SCRIPT One-Step RT-PCR Kit (Jena Bioscience) according to manufacturer’s protocol. Advanced-Primus 96 machine (PEQLAB Ltd., Fareham, UK) was used for RT-PCR test. Each reaction contained 2 μl of sample RNA, 1 μl of each primer, 1 μl of SCRIPT RT-PCR enzyme mix, 7.5 μl of RNase-free water, and 12.5 μl of SCRIPT RT-PCR Reaction Mix in 25 μl of final volume. The reverse transcription started with a reverse transcription cycle of 45 min at 50°C. The amplification program started with an initial denaturation step at 95°C for 5 min, followed by 35 cycles of 95°C for 10 s, 50°C for 20 s, 72°C for 1 min, and a final extension at 72°C for 5 min. Amplified RT-PCR products were then visualized at 1% agarose gel as the target DNA size was about 1 kb (Voytas, 2000). The gel was stained with ethidium bromide. RT-PCR products were stored at −20°C for further use.
DNA products from RT-PCR were purified using Axy-Prep DNA Gel Extraction Kit (Axygen Biosciences, New York, NY, USA). Thirty microliters of elution buffer were used to elute the DNA which was later stored at −80°C. PCR was performed using the product of purified RT-PCR as template for confirmation of the specific bands and amplification of DNA. KAPA2G Fast Ready Mix kit (KAPA Biosystems, Wilmington, MA, USA) was used for PCR amplification. Reaction setup for PCR contained 1 μl of purified DNA, 1.25 μl of each primer, 1.5 μl of DMSO, 7.5 μl of RNase-free water, and 12.5 μl of 2X KAPA 2G Fast Ready Mix in 25 μl of final volume. Advanced-Primus 96 machine (PEQLAB Ltd.) was used for PCR and the amplification program started with an initial denaturation step at 95°C for 2 min, followed by 35 cycles of 95°C for 30 s, 50°C for 15 s, 72°C for 15 s, and a final extension at 72°C for 1 min. PCR products were analyzed on ethidium bromide stained agarose gel and stored at −20°C.
Sequence analysis
Purified DNA samples were sent to the “Advanced Genetic Technologies Center (AGTC)”, College of Agricultural Sciences, University of Kentucky, USA for sequencing. Bidirectional nucleotide sequencing was performed using forward and reverse primers. Sequences from the bidirectional sequencing were blasted on the NCBI BLAST program for nucleic acids and the sequences were identified and compared with other sequences available on NCBI website. Sequences were aligned, cleaned and then imported to DNASTAR to check identity percentage between isolates, and construct a homology tree. Sequences for all six Saudi isolates of LTSV were submitted to GenBank with the following accession numbers: KT715530 (R1), KT735162 (R2), KT735163 (Q1), KT735164 (Q2), KT735165 (H1), and KT735166 (H2).
All Saudi isolates of LTSV were also compared with ten other sobemoviruses reported worldwide based on their coat protein gene sequences that were obtained from NCBI. Viruses included in comparison were: Southern bean mosaic virus (SBMV), Turnip rosette virus (TRoV), Subterranean clover mottle virus (SCMoV), Cocksfoot mottle virus (CfMV), Rice yellow mottle virus (RYMV), Southern cowpea mosaic virus (SCPMV), Sesbania mosaic virus (SsbMV), Imperata yellow mottle virus (IYMV), Ryegrass mottle virus (RGMoV), and Sowbane mosaic virus (SoMV) (Callaway et al., 2013; Dwyer et al., 2003; Eastwell et al., 2010; Lee and Anderson, 1998; Ling et al., 2013; Yassi et al., 1994). DNASTAR was used to get the identity percentages and phylogenetic relationships between these isolates of sobemoviruses.
Host range
A pure LTSV isolate detected in a recent study (Al-Shahwan et al., 2014) was provided by Prof. Ibrahim M. Al-Shahwan to carry out the host range study. Chenopodium quinoa was used for virus propagation (Forster, 1982; Forster and Jones, 1980; Green, 1971). A group of plant species representing several families were used for this experiment. These plant species included: Pisum sativum (Pea)-Arveja, Trifolium alexandrinum (Egyptian clover), Vicia sativa (Common Vetch), Vicia faba (Broad Bean)-Lara, Vigna unguiculata (four cultivars; Cowpea, California Black Eyed Pea, Black Eyed Pea, and Strike), Lathyrus sativus (Grass Pea), Chenopodium glaucum, C. amaranticolor, C. album, five lots of C. quinoa, Datura stramonium, Nicotiana occidentalis, Solanum nigrum (Black nightshade), and Gomphrena globosa (Globe amaranth) (Forster, 1982; Forster and Jones, 1980).
Mechanical inoculation was performed according to a procedure adopted by Hill (1984). Seeds of each plant species were germinated in pots filled with soil mix of sand and peat moss in 1:1 ratio. Seedlings of each plant species were later on transplanted to new pots. Leaves of alfalfa plants infected with LTSV from the above mentioned isolate were ground using mortar and pestle using 3 ml of 10 mM potassium phosphate buffer, pH 7 for each gram of leaf tissues. Eight layers of sterile cheesecloth were used to filter the homogenate. Four seedlings of each species were inoculated with LTSV sap after dusting with carborundum (400 mesh) while four other seedlings were mechanically inoculated with buffer only to serve as control. The inoculated and control plants were kept in the greenhouse at temperature between 25–27°C and were examined for symptoms expression. Symptoms were recorded two weeks post inoculation. Infection of the inoculated plants was confirmed by testing samples from these plants with DAS-ELISA. Dot blot hybridization was also performed to verify the results. This experiment was repeated twice.
Dot blot hybridization
PCR DIG Probe synthesis kit from Roche Diagnostics (Mannheim, Germany) was used for this purpose. Hybridization machine, Amersham Biosciences (Pittsburgh, PA, USA), was used and the procedure from Roche Diagnostics GmbH (Mannheim, Germany) was followed. Labelling of nucleic acid with digoxigenin was performed with the abovementioned reaction setup. Each reaction contained 2 μl of template DNA, 2.5 μl of each primer, 0.35 μl of enzyme mix, 12.65 μl of PCR Grade water, 2.5 μl of PCR Buffer with MgCl2 and 2.5 μl of PCR DIG Labeling Mix in 25 μl of final volume.
Fresh samples from the host range experiment were collected and three disks were cut by 1.5 ml Eppendorf tube (about 200 mg). Two hundred microliters of AMES buffer was added into the Eppendorf for grinding (El-Ela et al., 2006). Dry nitrocellulose membrane was cut according to the desired size. Layout was planned and each sample with positive controls were assigned to a specific place on this membrane. Five microliters of supernatant were added on specific allocated space from each sample and control ones. One positive control from PCR product was used for confirmation. PCR grade water was added as negative control.
Results
Field survey and analysis
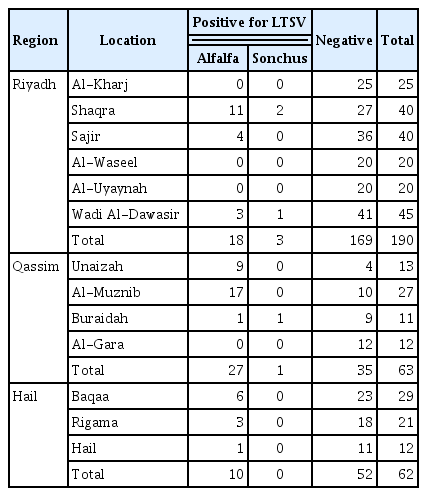
Mosaic, mottling, stunting, interveinal chlorosis or streaks, crinkling, yellowing, and small leaves were the symptoms mostly observed on the collected plant samples during the survey (Fig. 1). Fiftyfive samples out of the 308 alfalfa samples collected from different locations in all regions (Table 1), were found positive against LTSV with a percentage of 17.9%, whereas four out of seven S. oleraceus samples were found positive to this virus with a percentage of 57% by DAS-ELISA. These results revealed that 18.7% of the total collected samples from all regions were found infected with LTSV.
Symptoms observed on infected alfalfa and Sonchus oleraceus plants during the survey. (A) Mosaic, mottle, streaks and crinkling on alfalfa. (B) Yellow mosaic on S. oleraceus. (C) Crinkling, small leaves and mottling on alfalfa. (D) Stunting on alfalfa.
RT-PCR products analysis
Agarose gel electrophoresis of RT-PCR products of samples: 56-R, 83-R (from Riyadh region), 22-Q, 25-Q (from Qassim region), 4-H, and 23-H (from Hail region) revealed formation of specific bands on ethidium bromide stained agarose gel (Fig. 2). The size of these bands were approximately 1,033 bp as indicated from the Gene Direx 1 kb DNA marker which confirmed the presence of LTSV in the tested samples (Fig. 2). Results of RT-PCR were re-confirmed by PCR after purification of the RT-PCR products.
Sequence analysis
Percentage identity data between isolates revealed that all the LTSV isolates detected in Saudi Arabia were closely related (98.6–100%); however, the isolates from Qassim samples (22-Q and 25-Q) were 100% identical compared to those from Riyadh region (56-R and 83-R) which were 98.9% identical and those from Hail (4-H and 23-H) which were 98.6% identical (Fig. 3). Saudi isolates of LTSV were also found to be closely related to isolates reported in the two countries in which it was so far characterized (95.8–97.6%) (Ling et al., 2013); however, they seem to be more closely related to the Canadian isolate-JQ782213 (97.1–97.6%) than the New Zealand isolate-U31286 (95.8–97.1%) (Fig. 3).
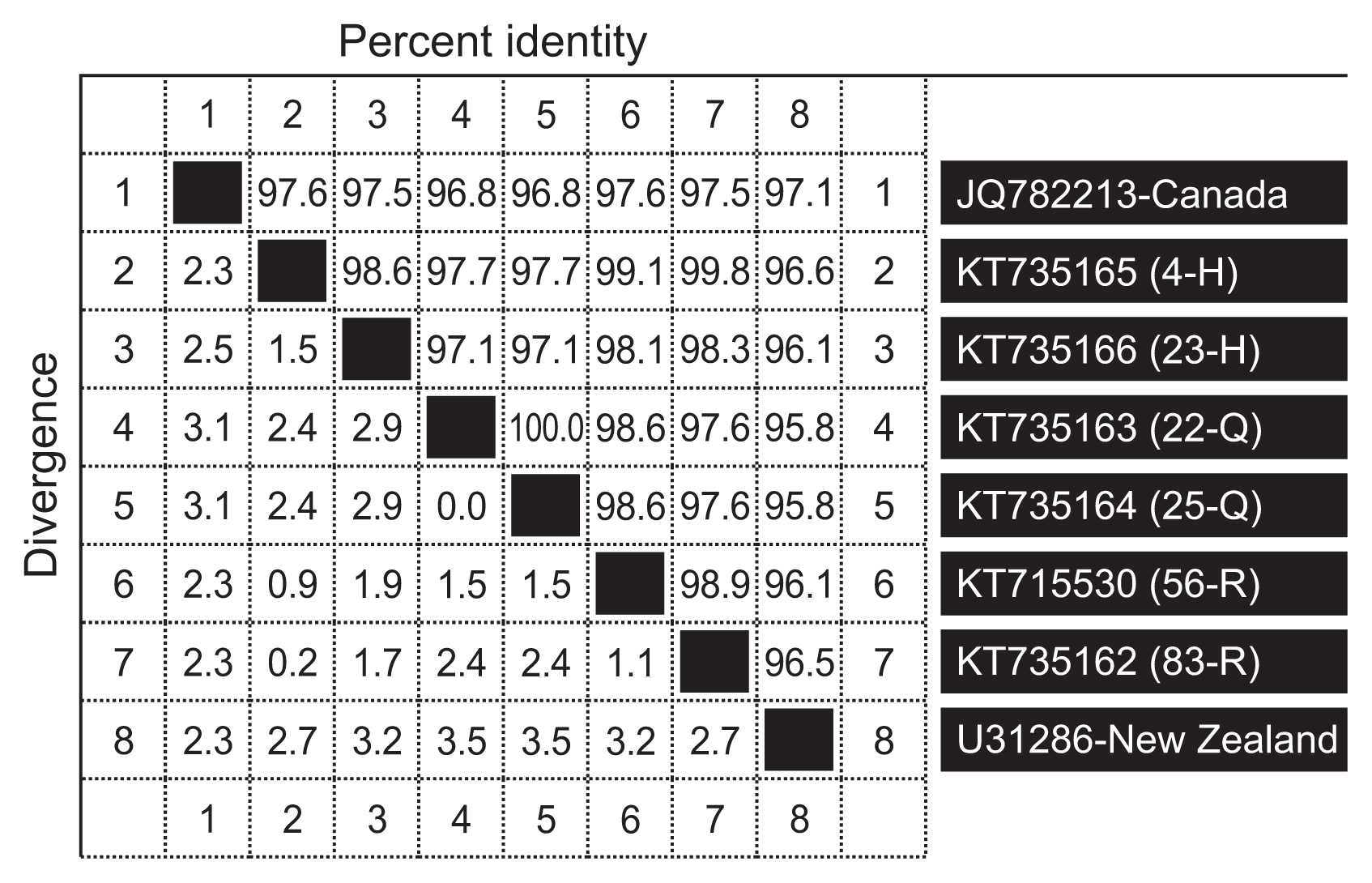
Percentage identity between six Saudi isolates of Lucerne transient streak virus, one isolate from Canada and one isolate from New Zealand. H, Hail; Q, Qassim; R, Riyadh.
Analysis of the homology tree constructed from coat protein gene sequences of Saudi and GenBank isolates of LTSV confirmed the percentage similarity between them (Fig. 4). In fact, Fig. 4 shows 100% identity between Qassim isolates and their close relationship to one of the Riyadh isolates (R1). It also shows the close relationship between Hail isolates (H1 and H2) and their close relationship to the other Riyadh isolate (R2). It furthermore, shows the close relationship between Saudi, New Zealand, and Canadian isolates of this virus, although the Saudi isolates were more closely related to Canadian than New Zealand isolates.
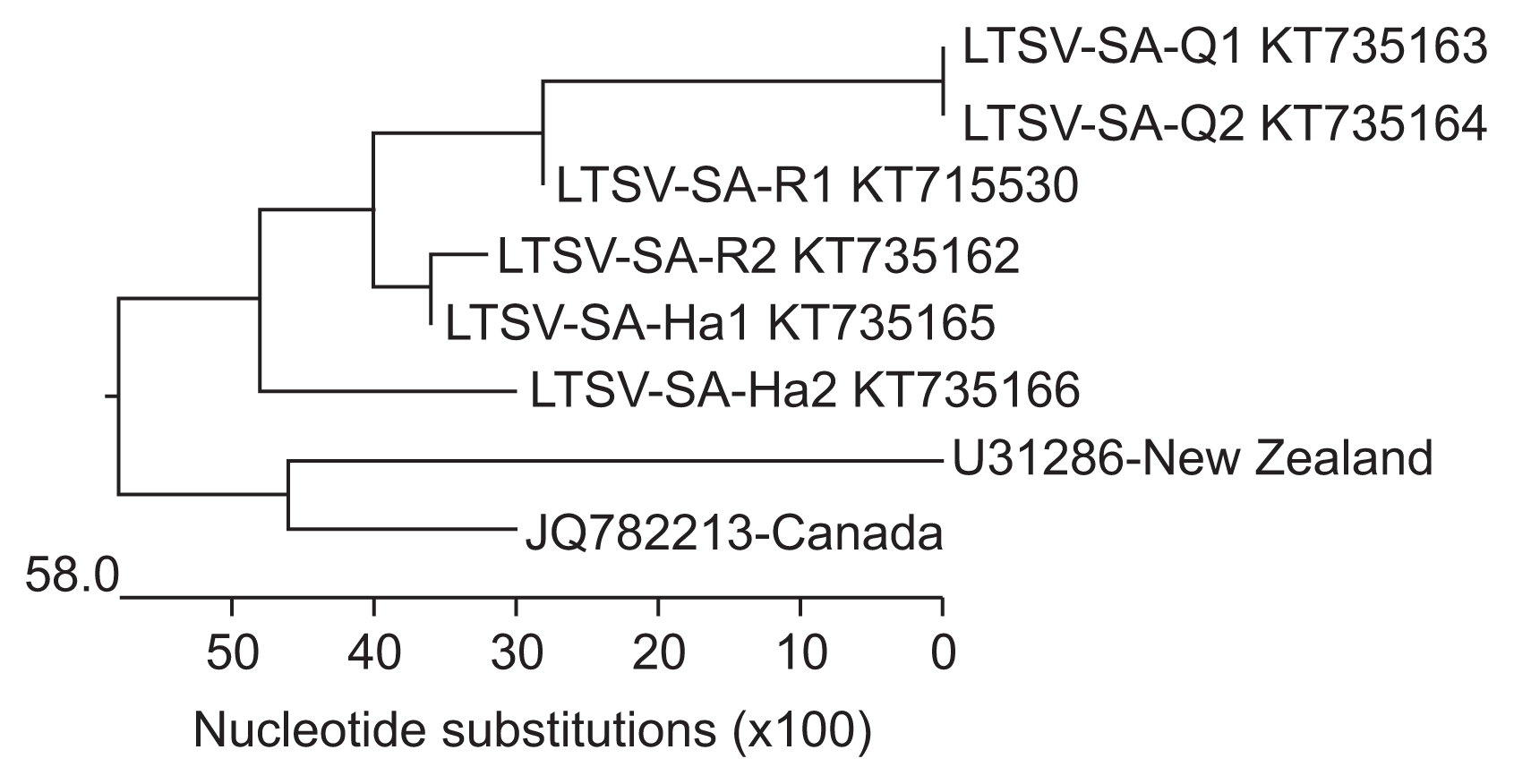
Homology tree based on multiple coat protein gene sequence alignment between six isolates of Lucerne transient streak virus (LTSV) from Saudi Arabia and one each from Canada and New Zealand obtained from the National Center for Biotechnology Information.
All the LTSV isolates reported worldwide fell in one cluster indicating their close relationships (Fig. 5). Comparing the Saudi isolates of LTSV with 10 other sobemoviruses indicated distant relationship between them which is in agreement with what was reported earlier (Opalka et al., 2000). The highest identity percentage between coat protein gene sequence of Saudi isolates of LTSV and SCMoV was 30%. The lowest identity percentage was for CfMV which shared only 23.1% identity with Saudi isolates of LTSV.
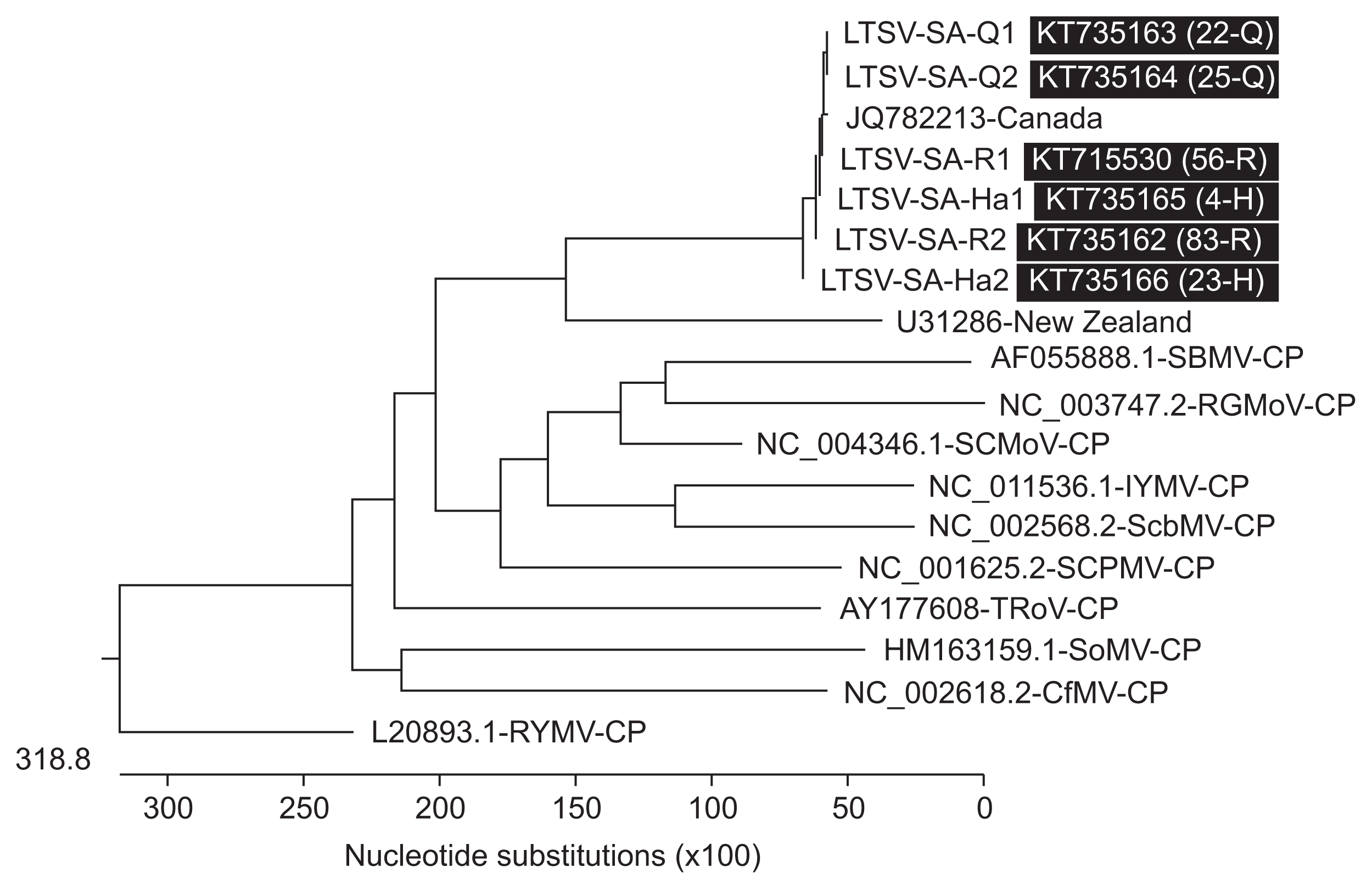
Homology tree based on multiple coat protein gene sequence alignment between six isolates of Lucerne transient streak virus (LTSV) from Saudi Arabia, one LTSV isolate from each of Canada and New Zealand and ten other sobemoviruses obtained from National Center for Biotechnology Information. Q, Qassim; R, Riyadh; H, Hail.
Host range
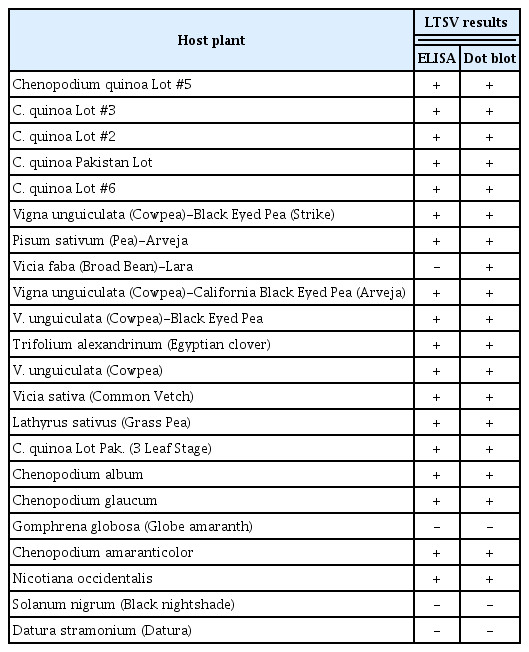
Symptoms were observed on 10 out of 14 mechanically inoculated plant species. These were chlorotic local lesions on C. quinoa Lot #2, C. quinoa Lot #6, C. album, and C. amaranticolor, necrotic local lesions on P. sativa, L. sativus, C. quinoa Pakistan Lot, C. amaranticolor, mottle on C. quinoa Pakistan Lot, C. quinoa Lot #6, V. unguiculata-California Black Eyed Pea, and C. album, mosaic on C. quinoa Pakistan Lot, V. unguiculata-Strike, and N. occidentalis, marginal chlorosis on T. alexandrinum, stunting on C. glaucum, and chlorotic streaks on V. sativa as shown in Fig. 6 and Table 2. DAS-ELISA confirmed the infection of the 10 plant species that showed symptoms in the host range experiment with LTSV. DAS-ELISA results for the host range experiments is given in Table 3.
Symptoms observed on some of the host range plants inoculated with the Saudi isolate of Lucerne transient streak virus. (A) Chlorotic local lesions and mottling on Chenopodium quinoa Pakistan Lot. (B) Chlorotic local lesions on Chenopodium amaranticolor. (C) Mosaic on Nicotiana occidentalis. (D) Mosaic on Chenopodium album.

Symptoms expression of host range plants three weeks post inoculation with Lucerne transient streak virus
Dot blot hybridization
Dot blot hybridization indicated that 11 species were positive to LTSV (Fig. 7). V. faba which was negative to all tests turned out to be positive with dot blot hybridization. All the five lots of C. quinoa were positive to LTSV infection by both DAS-ELISA and dot blot hybridization. Similarly, all four cultivars of V. unguiculata namely California Black Eyed Pea, Black Eyed Pea, Black Eyed Pea-Strike, and Cowpea were also positive against LTSV infection by both DAS-ELISA and dot blot hybridization (Fig. 7).
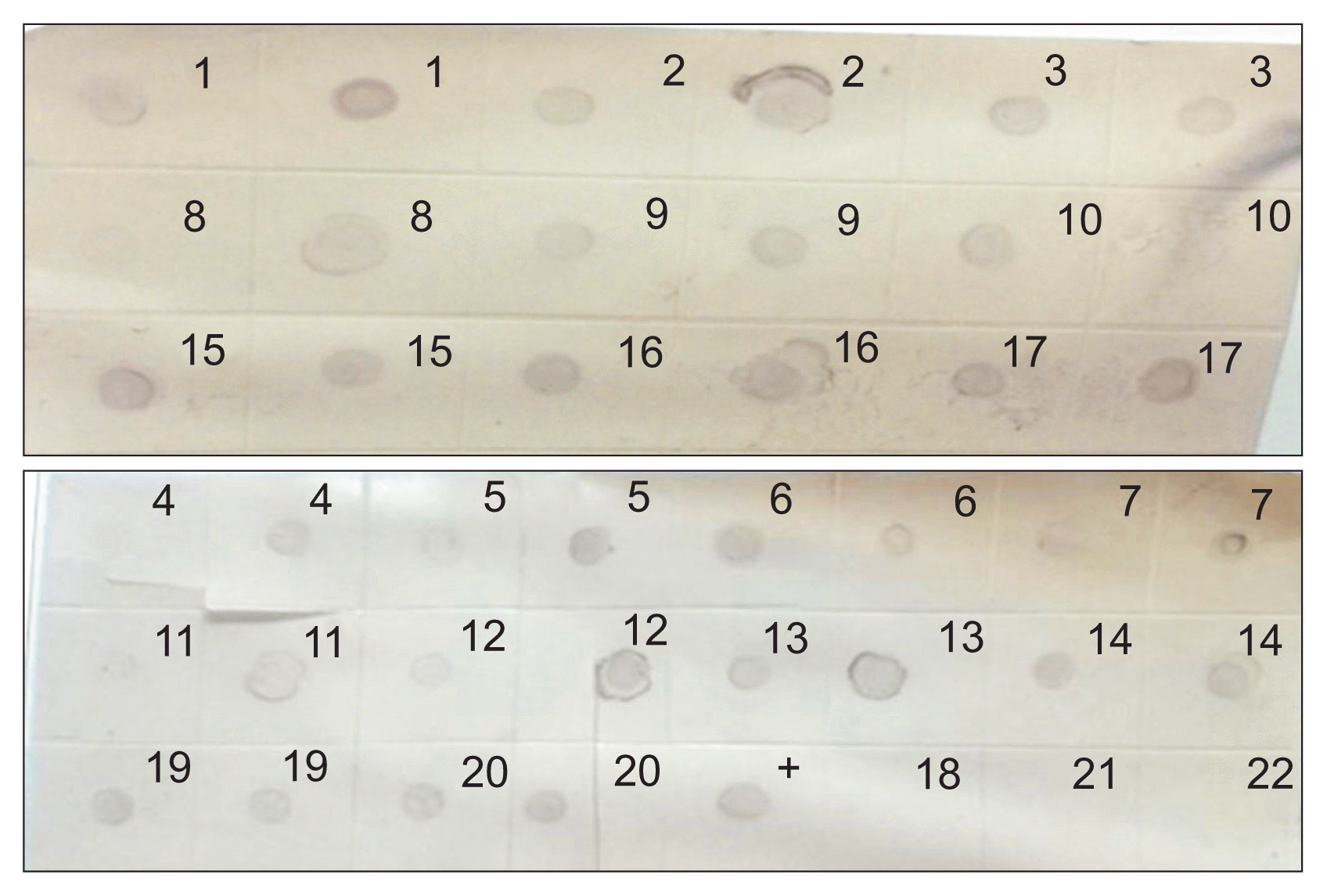
Dot blot hybridization with DIG labelled cDNA probe for 22 samples collected from the host range experiments. Note: 1, Chenopodium quinoa Lot #5; 2, C. quinoa Lot #3; 3, C. quinoa Lot #2; 4, C. quinoa Pakistan Lot; 5, C. quinoa Lot #6; 6, Vigna unguiculata (Cowpea)-Black Eyed Pea (Strike); 7, Pisum sativum (Pea)-Arveja; 8, Vicia faba (Broad Bean)-Lara; 9, Vigna unguiculata (Cowpea)-California Black Eyed Pea; 10, V. unguiculata (Cowpea)-Black Eyed Pea; 11, Trifolium alexandrinum (Egyptian clover); 12, V. unguiculata (Cowpea); 13, Vicia sativa (Common Vetch); 14, Lathyrus sativus (Grass Pea); 15, C. quinoa Lot Pak; 16, Chenopodium album; 17, Chenopodium glaucum; 18, Gomphrena globosa (Globe amaranth); 19, Chenopodium amaranticolor; 20, Nicotiana occidentalis; 21, Solanum nigrum (Black nightshade); 22, Datura stramonium (Datura); +, positive control.
In this investigation, it was found that four plant species were reported for the first time, to our knowledge, as hosts for LTSV namely; Egyptian clover (T. alexandrinum), N. occidentalis, C. glaucum, and Grass Pea (L. sativus). N. occidentalis showed mosaic symptoms against the Saudi isolate of LTSV and found to be positive for the virus by both DAS-ELISA and dot blot hybridization; hence, it can serve as a new potential propagative host for LTSV.
Discussion
LTSV has not been studied at all in Saudi Arabia prior to this study except for the work performed by Al-Shahwan and co-workers (Al-Shahwan et al., 2013, 2014). So far, this virus has only been reported from three countries worldwide, namely; Australia (Blackstock, 1978), New Zealand (Forster and Jones, 1979), and Canada (Paliwal, 1983). The survey carried out in this study was planned to determine the status of this virus in three of the most important alfalfa producing regions in Saudi Arabia. Symptoms on alfalfa and S. oleraceus were similar in Riyadh, Qassim, and Hail regions, but few differences were observed. Since only 55 out of 308 alfalfa samples and four out of seven S. oleraceus samples were positive to LTSV from the symptomatic plant samples collected in this study, this suggests that other viruses or pathogens or environmental conditions might be responsible for the symptoms that were observed on the negative samples. In fact, other viruses have been lately reported from alfalfa in Saudi Arabia (Al-Shahwan et al., 2014) but probably more work should be done to identify causal agents that might have emerged more recently. The reason that different number of samples were collected from different regions was because these regions differ in their area and importance with regards to alfalfa production. Although LTSV was detected in all three surveyed regions, its percentage of detection varied from region to region. From Table 1, 18 alfalfa samples were only found positive against LTSV by DAS-ELISA among 185 samples from Riyadh region (9.7%) with the highest percentage of detection (48%) observed in Shaqra samples. The percentage of detection of LTSV in alfalfa in Qassim region was very high (44% of the 61 alfalfa collected samples) with Governorate of Unaizah had the highest percentage of detection (69%). The percentage of detection of LTSV in alfalfa in Hail was lower (16%) with the highest virus detection in Baqaa samples (21%). These differences in percentage of LTSV detection are probably due to well establishment of the virus in some areas (i.e., Unaizah) while absent so far in other areas (i.e., Al-Kharj). Detection of LTSV in weed (S. oleraceus) that was encountered during this study may indicate the importance of these weeds in harboring and playing an important epidemiological role of the disease caused by this virus. Overall, the survey indicated that LTSV had a percentage of detection of 17.9 in alfalfa, and 57% in S. oleraceus, which is probably alarming for the alfalfa production in some of the visited locations in Saudi Arabia. LTSV infection seems to be increasing in the Kingdom of Saudi Arabia as a recent study reported a percentage of infection of only 2.9% in alfalfa, and 2.3% in S. oleraceus and other weeds in 2012–2013 surveys (Al-Shahwan et al., 2014). In this study, it was clear that the percentage of infection with LTSV was higher in both alfalfa and S. oleraceus compared to the previous studies (Al-Shahwan et al., 2014). Qassim region had even more than 50% of infected samples in some locations, which suggests its well establishment in these locations (i.e., Unaizah). To conclude this part, it can be said that LTSV was detected in alfalfa in all surveyed regions in central Saudi Arabia and the percentage of detection varied from region to region and from location to location in the same region. S. oleraceus was found to be infected with LTSV for the first time in Qassim region.
Coat protein of LTSV falls in ORF4 of genomic RNA and it is located by the 3′ proximal end (Tamm and Truve, 2000). The reason for using coat protein gene in this study was because it was proved to be a handy tool for plant virus characterization including sobemoviruses (Tamm and Truve, 2000). Results of BLAST obtained here, indicated that Saudi isolates of LTSV also paired with coat protein of available sequences from NCBI in ORF4. Alignment of all Saudi isolates with other isolate of LTSV from Canada (Ling et al., 2013) and New Zealand (NCBI) showed that all isolates of LTSV characterized anywhere in the world are closely related. This can be noticed from the small differences among Saudi isolates of LTSV that range between 0–2.9% and a little higher between Canadian and New Zealand isolates (Fig. 3) which might suggest low genetic variability among LTSV isolates identified so far based on coat protein gene sequencing. Only the two Qassim isolates of this virus were 100% identical with each other. The small differences found between other LTSV isolates elsewhere were probably giving each isolate its own identity.
The distant relation found between LTSV and other sobemoviruses was in agreement with what was reported earlier (Lapierre and Signoret, 2004; Opalka et al., 2000). Opalka et al. (2000) reported that LTSV coat protein had percentage identity of 22.2%, 23.8%, 23%, and 26.4% with RYMV, SBMV, SCPMV, and SsbMV, respectively and is almost similar to what was observed in this study. This is also true for the amino acid similarity of LTSV with RGMoV, which was found to be 25.3% in the coat protein region of both viruses (Lapierre and Signoret, 2004). Falling of all LTSV isolates in one cluster indicates their similarity on the basis of coat protein gene sequences (Fig. 5). Likewise, the occurrence of sobemoviruses in different clusters suggest their differences from each other and from LTSV on the same basis as well. Overall, it can be said that all of the Saudi isolates of LTSV were found to be more closely related to the Canadian isolate-JQ782213 (97.1–97.6%), as compared to New Zealand isolate-U31286 (95.8–97.1%). The Saudi isolates of this virus had the highest similarity (30%) with SCMoV and lowest similarity (23.1%) with CfMV confirming that the Saudi isolate of LTSV is also distantly related to other sobemoviruses based on coat protein gene sequence which is in agreement with what was described in literature.
The use of a pure LTSV isolate in the inoculation of the host range experiments and confirmation of infection with both DAS-ELISA and dot blot hybridization allowed observations of the symptoms of this virus on the ten different infected plants. It is interesting that dot blot hybridization result showed that the symptomless V. faba was positive to LTSV which is in agreement with the work done with LTSV-Au (Australian) isolate where it was reported to infect V. faba without inducing symptoms after inoculation, but LTSV-NZ (New Zealand) doesn’t infect this plant species (Dall et al., 1990). Also, it can be mentioned that the obtained host range results were in agreement with the previous results for this virus (Forster, 1982; Forster and Jones, 1980); however, four plant species used in this investigation that turned out to be new hosts for this virus. These were Egyptian clover (T. alexandrinum), N. occidentalis, C. glaucum, and Grass Pea (L. sativus). Since N. occidentalis showed clear mosaic symptoms against this isolate of LTSV which were not reported before, it might be a potential new propagative host for this virus. Production of chlorotic local lesions on C. amaranticolor and Pisum sativum by the Saudi isolate of LTSV which later developed into necrotic local lesions were in agreement with LTSV-NZ isolate results but it differed from the Australian isolate (LTSV-Au), which didn’t infect these two plant species (Dall et al., 1990). To summarize it can be said that four plant species namely, C. glaucum, L. sativus, N. occidentalis, and T. alexandrinum, were reported for the first time as hosts for LTSV, whereas N. occidentalis was found to be a good systemic and propagative host for this virus.
The use of nucleic acid hybridization assay confirmed the positive DAS-ELISA results for all the symptomatic plants in the host range experiments and furthermore indicated infection of V. faba with LTSV though it was symptomless and negative to DAS-ELISA. This result also indicated that the sensitivity of dot blot hybridization over DAS-ELISA. Although the host range experiments carried out in this study were limited to dicots only, the natural host range for sobemoviruses was reported to be relatively narrow but not less than 15 different families of both monocot and dicot plant species in general (Tamm and Truve, 2000).
Finally, it is recommended to conduct a national scale survey to determine the exact status of LTSV in the Kingdom of Saudi Arabia, especially in other regions and locations which were not covered in this study. Furthermore, collaborative efforts, between scientists, should be initiated to search for resistance sources against this virus.
Acknowledgments
This project was supported by Agriculture Research Center, at College of Food and Agriculture Sciences, and Deanship of Scientific Research, King Saud University.