Ser360 and Ser364 in the Kinase Domain of Tomato SlMAPKKKα are Critical for Programmed Cell Death Associated with Plant Immunity
Article information
Abstract
SlMAPKKKα, a tomato (Solanum lycopersicum) mitogen-activated protein kinase kinase kinase, is a positive regulator of Pto-mediated effector-triggered immunity, which elicits programmed cell death (PCD) in plants. In this study, we examined whether putative phosphorylation sites in the conserved activation segment of the SlMAPKKKα kinase domain are critical for eliciting PCD. Three amino acids, threonine353, serine360 (Ser360), or serine364 (Ser364), in the conserved activation segment of SlMAPKKKα kinase domain were substituted to alanine (T353A, S360A, or S364A), and these variants were transiently expressed in tomato and Nicotiana benthamiana plants. Two alanine substitutions, S360A and S364A, completely abolished SlMAPKKKα PCD-eliciting activity in both plants, while T353A substitution did not affect its PCD-eliciting activity. SlMAPKKKα wild type and variant proteins accumulated to similar levels in plant leaves. However, SlMAPKKKα protein with the largest size was missed when either S360A or S364A substitutions were expressed, whereas proteins with the smaller masses were more accumulated than those of full-length of SIMAPKKKα and T353A. These results suggest that phosphorylation of SlMAPKKKα at Ser360 and Ser364 is critical for PCD elicitation in plants.
Introduction
Plants are naturally exposed to a variety of pathogens, including bacteria, fungi, viruses, nematodes, insects, and mammals. Plant diseases caused by pathogen attack result in major economic losses in many important crop plants worldwide. To protect themselves against pathogen infection, plants have developed a sophisticated multi-tiered immune system, parts of which are similar to the human immune system and help fight off these pathogens (Eulgem, 2005; Katagiri, 2004). The plant defense responses against pathogen infection are triggered by the ability of plants to detect pathogen- or microbe-associated molecular patterns (PAMPs or MAMPs) or pathogen species/strain-specific factors known as effector proteins, resulting in pattern-triggered immunity and effector-triggered immunity, respectively (Jones and Dangl, 2006; Zipfel, 2008). The latter type of plant immunity is accompanied by a localized programmed cell death (PCD), a form of the hypersensitive response that occurs upon recognition of a pathogen effector protein by a corresponding plant resistance (R) protein (Coll et al., 2011). For example, in tomato (Solanum lycopersicum), PCD is triggered upon recognition by the host Pto protein, a serine/threonine kinase, of either one of two effector proteins, AvrPto or AvrPtoB, from Pseudomonas syringae pv. tomato, the causative agent of bacterial speck disease (Kim et al., 2002; Lin and Martin, 2007). Pto-mediated resistance to P. syringae pv. tomato, including PCD induced by interaction between Pto and AvrPto/AvrPtoB requires Prf, a coiled coil-nucleotide binding site-leucine rich repeat (CC-NBS-LRR) protein (Salmeron et al., 1996). This fast and powerful immune system of plants prevents the growth and spread of pathogens into healthy tissues of plants (Coll et al., 2011).
Mitogen-activated protein kinase (MAPK) cascades are highly conserved and common mechanisms in eukaryotes. The cascades are composed of MAPKK kinases, MAPK kinases and MAPKs (Asai et al., 2002; He et al., 2006). Many environmental changes and conditions MAPK cascades, which are involved in signaling triggered by biotic and abiotic stimuli and in processes such as hormonal and developmental signaling (Group, 2002; Jonak et al., 2002). These protein kinase cascades are linked in various ways to upstream receptors and downstream targets to activate corresponding pathways. MAPKKKs activate MAPKKs through phosphorylation on two serine/threonine residues and subsequently, MAPKKs phosphorylate MAPKs on threonine and tyrosine residues. Activated MAPKs phosphorylate a wide variety of substrates, such as protein kinases, transcription factors, and enzymes, to make cellular physiological changes (Colcombet and Hirt, 2008; Group, 2002). MAPK cascade family genes have been identified in plants, including Arabidopsis, tomato, rice, soybean, maize and Nicotiana benthamiana. For example, 89 MAPKKKs, 5 MAPKKs and 16 MAPKs exist in tomato (Group, 2002; Kong et al., 2012). The presence of so many MAPKKKs, but a few MAPKKs and MAPKs in a single plant suggests that signals might converge on specific MAPKKs after recognition of certain stimuli by MAPKKKs and then signals diverge to certain MAPKs by MAPKKs (Kong et al., 2012; Wu et al., 2014).
Downstream signaling pathways after Pto recognition have been intensively studied and many components have been identified, including MAPK cascades (Oh and Martin, 2011). Tomato SlMAPKKKα and SlMAPKKKɛ function as positive regulators of Pto-mediated PCD (del Pozo et al., 2004; Melech-Bonfil and Sessa, 2010). A homolog of SlMAPKKKα, NbMAPKKKα, also functions as a positive regulator of PCD during pathogen infection (Jin et al., 2002). Similarly, overexpression in leaves of the full length or kinase domain only of NbMAPKKKβ or NbMAPKKKγ causes cell death (Hashimoto et al., 2012). In particular, overexpression of the full length or kinase domain only of SlMAPKKKα in the N. benthamiana plant triggers autonomous cell death, which requires the function of downstream SlMKK2 (Oh et al., 2013). In the pathway, involved in plant defense signaling, tomato SlMKK2 could interact with upstream SlMAPKKK and downstream MAPK, via typical 14-3-3 binding sites and/or two conserved leucines in the N-terminal MAPK-docking site of tomato SlMKK2. These two leucines of SlMKK2 are critical for the interaction with SlMPK3 to induce PCD (Li et al., 2014). Recently, it was found that SlMKK2 contributes to disease resistance against P. syringae pv. tomato, Xanthomonas campestris pv. vesicatoria and Botrytis cinerea and functions as a positive regulator of defense response (Li et al., 2014; Melech-Bonfil and Sessa, 2011). Our previous studies also have shown that, as a modular protein for efficient signal transfer or as a scaffolding protein, tomato 14-3-3 protein (TFT7) interacts with a C-terminal serine residue (Ser535) of SlMAPKKKα, leading to stabilization of SlMAPKKKα abundance and activity by its phosphorylation and also it interacts with a N-terminal threonine residue (Thr33) of downstream SlMKK2, which phosphorylates and activates a SlMAPK1/3 (Oh et al., 2010). Among 16 SlMPKs in tomato, SlMPK1, SlMPK2, and SlMPK3 play a role in regulating defense response against pathogen infection (Pedley and Martin, 2004). The cascade in tomato involving SlMAPKKKα-SlMKK2-SlMPK1/SlMPK3 was shown to participate in Pto-mediated PCD in defense response (Oh and Martin, 2011; Oh et al., 2010).
Although C-terminal phosphorylation of SlMAPKKKα has been shown to be critical for interaction with TFT7 to stabilize itself, the phosphorylation status of the SlMAPKKKα kinase domain and its function has not been studied well. Because ectopic overexpression of the full length or kinase domain only of SlMAPKKKα triggers strong PCD, its phosphorylation status in the kinase domain might be important. Therefore, here, we examined if putative phosphorylation sites in the activation segment of SlMAPKKKα kinase domain are required for PCD induction in plants and found that two serine residues in the activation segment are critical for triggering PCD in plants.
Materials and Methods
Plant materials and growth conditions
Tomato (S. lycopersicum) cv. Rio Grande and N. benthamiana plants were grown in a greenhouse under the following conditions: 16/8 h light/dark cycle, 26/22°C day/night, 70% to 80% humidity. Eight-week-old tomato and 6-week-old N. benthamiana plants were used for Agrobacterium-mediated transient assay and for immunoblotting experiments.
Amino acid substitution
For substitution of threonine-353 (Thr353), serine-360 (Ser360), or serine-364 (Ser364) to alanine in SlMAPKKKα, PCR-based site-directed mutagenesis was performed as described previously (Ho et al., 1989).
Agrobacterium-mediated transient assay
The SlMAPKKKα gene and its derivatives were cloned into pER8, which is the Agrobacterium tumefaciens binary vector (Zuo et al., 2000), to provide expression in plants with an N-terminal double HA tag, and its expression was controlled by an estradiol-inducible system. Plasmid constructs including empty vector were transformed into A. tumefaciens strain GV2260. For induction of Agrobacterium harboring pER8-SlMAPKKKα, -SlMAPKKKα (K231M), -SlMAPKKKα (T353A), -SlMAPKKKα (S360A), and -SlMAPKKKα (S364A), cells were resuspended in induction media (10 mM MES, pH 5.6, 10 mM MgCl2, and 150 μM acetosyringone) and incubated overnight at 28°C before infiltration. Then, Agrobacterium strain carrying each construct was infiltrated at a final OD600 of 0.4 for analysis. To induce expression of SlMAPKKKα gene and its derivatives, 2 μM of β-estradiol with 0.005% silwet was applied on top of the agroinfiltrated area of leaves 2 days after agroinfiltration.
Protein extraction and immunoblotting
For protein extraction, leaf samples from N. benthamiana were collected 42 h after induction of protein expression by β-estradiol. Total proteins were extracted using 500 μl of extraction buffer (GTEN buffer [10% glycerol, 25 mM Tris pH 7.5, 1 mM EDTA, 150 mM NaCl], 2% w/v polyvinylpolypyrrolidone [PVPP], 10 mM DTT, 1× plant protein protease inhibitor and 0.1% Triton X-100). Total 15 μl of extracts were loaded on 10% SDS-PAGE gel and transferred onto a polyvinylidene fluoride membrane. Transferred membranes were hybridized with anti-HA (1:4,000) (Sigma, St. Louis, MO, USA) and anti-SlMAPKKKα (1:2,500) (Oh et al., 2010), respectively. Signals were detected by Amersham ECL-PLUS Western blotting detection system (GE Healthcare, Milwaukee, WI, USA). The putative sizes of SIMAPKKKα and SIMAPKKKα-KD are approximately 67 kDa and 29 kDa, respectively. Rubisco proteins were stained with Coomassie Brilliant Blue (CBB) to test for equal loading.
Results
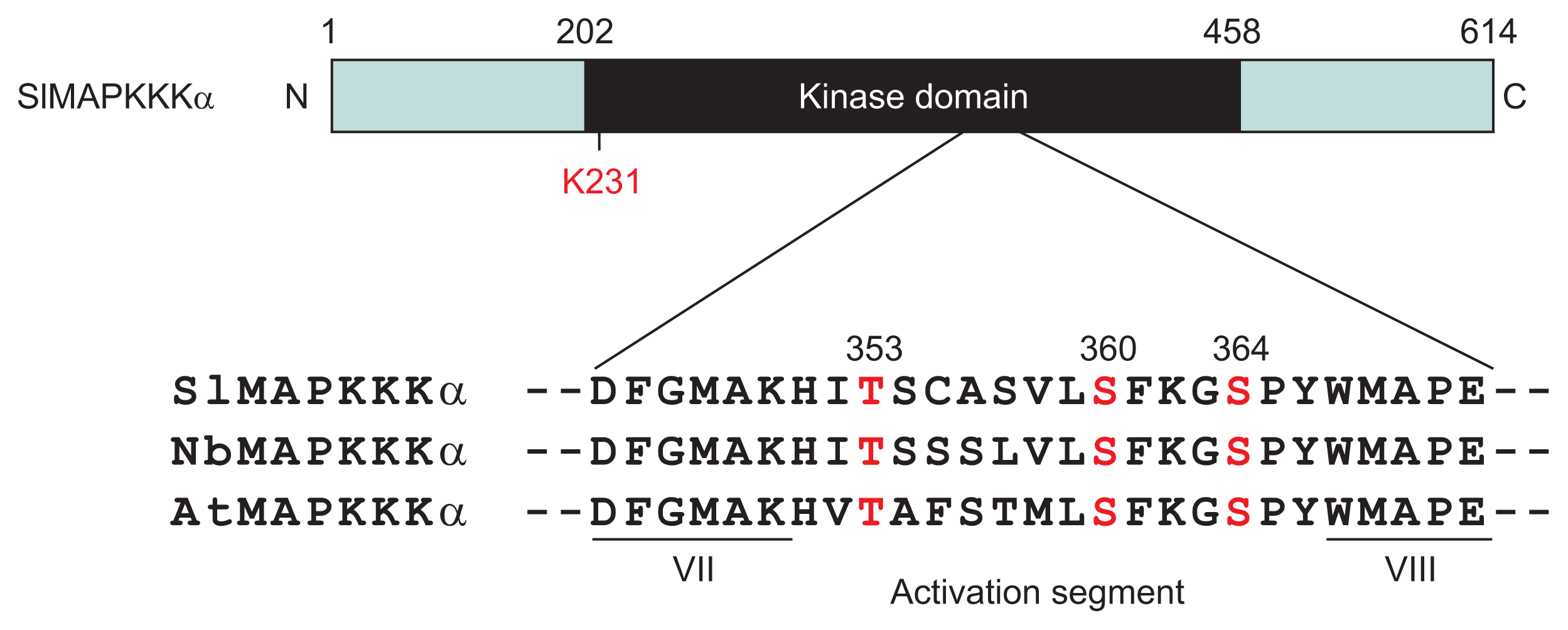
Three putative phosphorylation sites in the activation segment in the kinase domain of SIMAPKKKα are conserved
Previous studies have shown that PCD, which is triggered by the interactions of tomato Pto with either AvrPto or AvrPtoB from P. syringae pv. tomato required SlMAPKKKα as a positive regulator in tomato and in N. benthamiana plants (del Pozo et al., 2004). In the MAPK signaling cascade, the MAPKKs are activated by MAPKKKs through phosphorylation on serine and serine/threonine residues in their activation motif (S/TXXXXXS/T), located between kinase subdomains VII and VIII (Group, 2002). Multiple sequence alignment of the predicted amino acid residues of MAPKKKα genes revealed that SIMAPKKKα contains this typical conserved motif (Fig. 1) and that it is conserved in tomato, tobacco and Arabidopsis. The motif contains three putative phosphorylation sites, which are Thr353, Ser360, and Ser364. Based on this sequence analysis, we substituted these three residues to alanine (Ala) in both full-length SlMAPKKKα and kinase domain only (SlMAPKKKα-KD) proteins for transient ectopic expression in plant leaves.
Serines at position 360 and 364 of SlMAPKKKα are required for eliciting cell death in plants
To test whether overexpression of the full length or kinase domain only versions of SlMAPKKKα and their alanine substitution variants cause cell death, we used Agrobacterium to transiently express them under an estradiol-inducible system in tomato and N. benthamiana leaves. As positive controls, SlMAPKKKα and SlMAPKKKα-KD were used for the expression of the full-length and KD alone, respectively. As negative controls, we expressed an empty vector and the inactive form of the SlMAPKKKα-KD (K231M) that has a substitution at the essential lysine in the ATP-binding site (Oh et al., 2010). PCD was strongly triggered by overexpression of SlMAPKKKα or SlMAPKKKα-KD at 36 to 48 h after estradiol application in tomato plants (Fig. 2A). In contrast, no cell death was observed when an empty vector- or the SlMAPKKKα-KD (K231M) variant was infiltrated. These results with positive and negative controls were consistent with previous reports (Oh et al., 2010). As shown in Fig. 2A, the substitution of Ala at Thr-353 (T353A) did not affect the PCD-eliciting ability of SlMAPKKKα or SlMAPKKKα-KD. Strong cell death was observed in leaf areas expressing SlMAPKKKα (T353A) and SlMAPKKKα-KD (T353A) as well as in leaf areas expressing positive controls. In contrast, leaf areas expressing SlMAPKKKα (S360A) or SlMAPKKKα (S364A), which are variant forms of SlMAPKKKα with Ala substitution at two putative serine residues (Ser360 and Ser364), did not cause PCD. These results suggest that these Ser360 and Ser364 positions of SlMAPKKKα are important for PCD-eliciting ability of SlMAPKKKα. The pattern of PCD induction by SlMAPKKKα and its variants in N. benthamiana plants was identical to that in tomato leaves (Fig. 2B) with Agrobacterium strains expressing SlMAPKKKα (S360A) and SlMAPKKKα (S364A) not causing PCD, and SlMAPKKKα (T353A) inducing cell death as well as SlMAPKKKα in N. benthamiana leaves. Under the identical conditions using kinase domain only proteins, results of analysis of the cell death-inducing ability by expressing SlMAPKKKα-KD and its derivatives in N. benthamiana plants were qualitatively similar to those of full length of SlMAPKKKα. Based on these results and previous reports (del Pozo et al., 2004), Ser360 and Ser364 positions, but not Thr353, of SlMAPKKKα appear to play a role in PCD responses in plants.
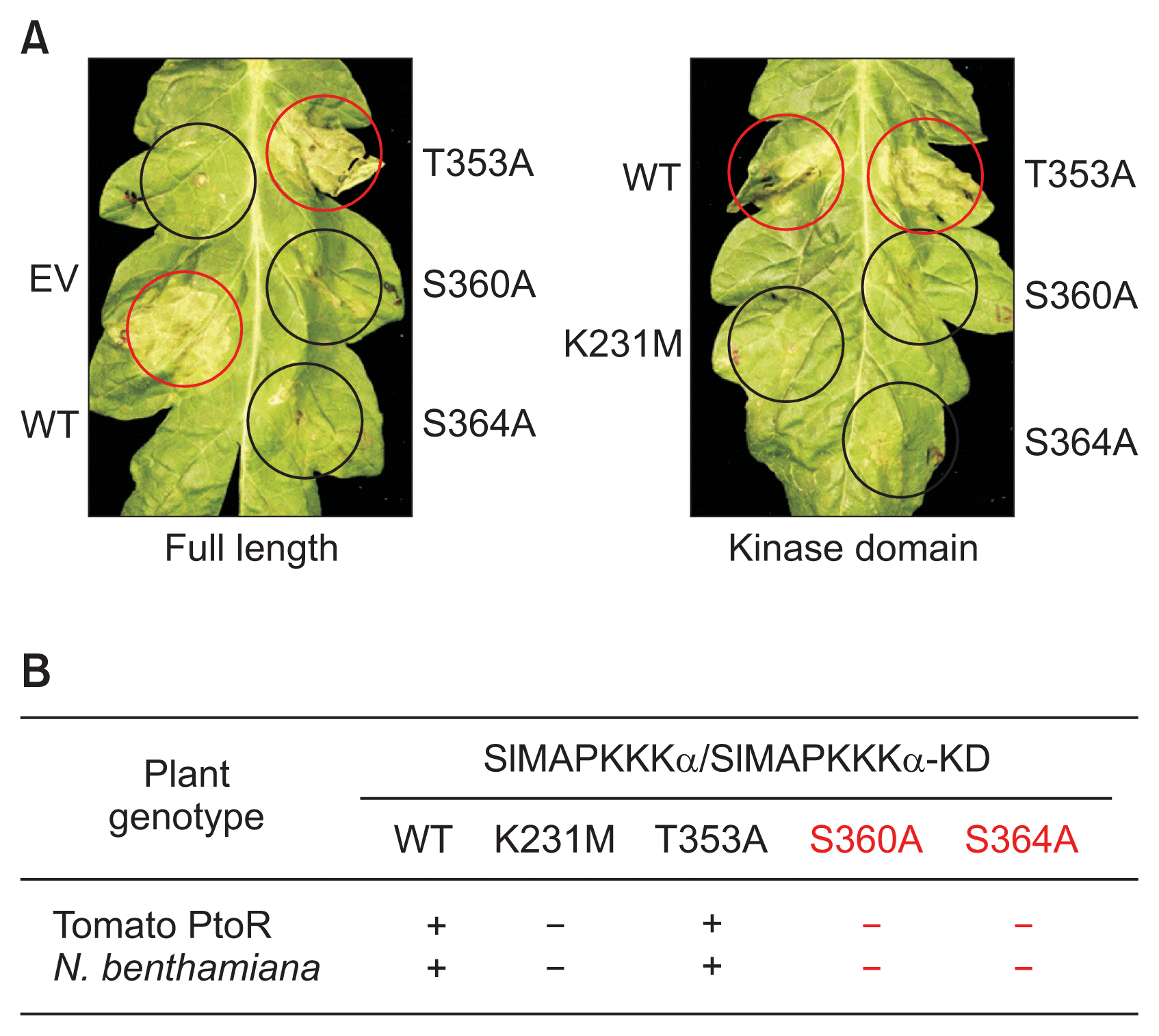
The cell death eliciting activity of SlMAPKKKα in tomato and Nicotiana benthamiana. (A) Programmed cell death (PCD) assay with the wild type SlMAPKKKα and alanine substituents at the designated amino acids in tomato cv. Rio Grande-PtoR leaves. The indicated forms of SlMAPKKKα were expressed transiently. Photos were taken 48 h after induction. EV, empty vector; WT, wild type; red circles, PCD; black circles, no PCD. (B) Summary of PCD assay in tomato and N. benthamiana plants. +, PCD; −, no PCD. These assays were repeated three times.
Phosphorylation at Ser360 and Ser364 might be required for cell death induction in plants
Multiple residues within SlMAPKKKα are important for the PCD-inducing function of SlMAPKKKα. In particular, Ser535 is known as a putative phosphorylation site of SlMAPKKKα that elicits cell death by its interaction with tomato 14-3-3 protein 7 (TFT7) (Oh et al., 2010). Besides Ser535, we showed here that Ser360 and Ser364 may be the primary phosphorylation targets of SlMAPKKKα and/or SlMAPKKKα-KD. To determine if any of the Ala substitutions affected the mobility or the accumulation of SlMAPKKKα proteins in our experiments, the full length and kinase domain only proteins were examined using immunoblot analysis. HA-tagged SlMAPKKKα and other variant forms of SlMAPKKKα were expressed in N. benthamiana plants and total proteins were extracted from leaf samples at 42 h after induction of protein expression. To detect the full length proteins and HA-tagged SlMAPKKKα-KD, anti-SlMAPKKKα and anti-HA were used, respectively. Immunoblot analysis presented in Fig. 3A shows that at least three different protein bands were observed in the case of full length SlMAPKKKα. The kinase-inactive variant of SlMAPKKKα, SlMAPKKKα (K231M), migrated as a single low-abundance protein probably due to the instability of this protein in plants. The multiple species of SlMAPKKKα may be due to phosphorylation of the protein in the plants. Consistent with this possibility, immunoblotting revealed that SlMAPKKKα protein with the largest mass was missed when either the S360A or S364A substitutions are present in the protein. The three protein bands are observed with SlMAPKKKα (T353A), which induces cell death as well as the SlMAPKKKα wild type protein. Next, we examined SlMAPKKKα-KD using HA antibody by immunoblot analysis (Fig. 3B). All of the HA tagged SlMAPKKKα-KD proteins were detectable in plants except for SlMAPKKKα-KD (K231M) probably due to its instability. Together, these experiment suggest that phosphorylated on Ser360 and Ser364 of SlMAPKKKα is likely required for cell death induction in plants.
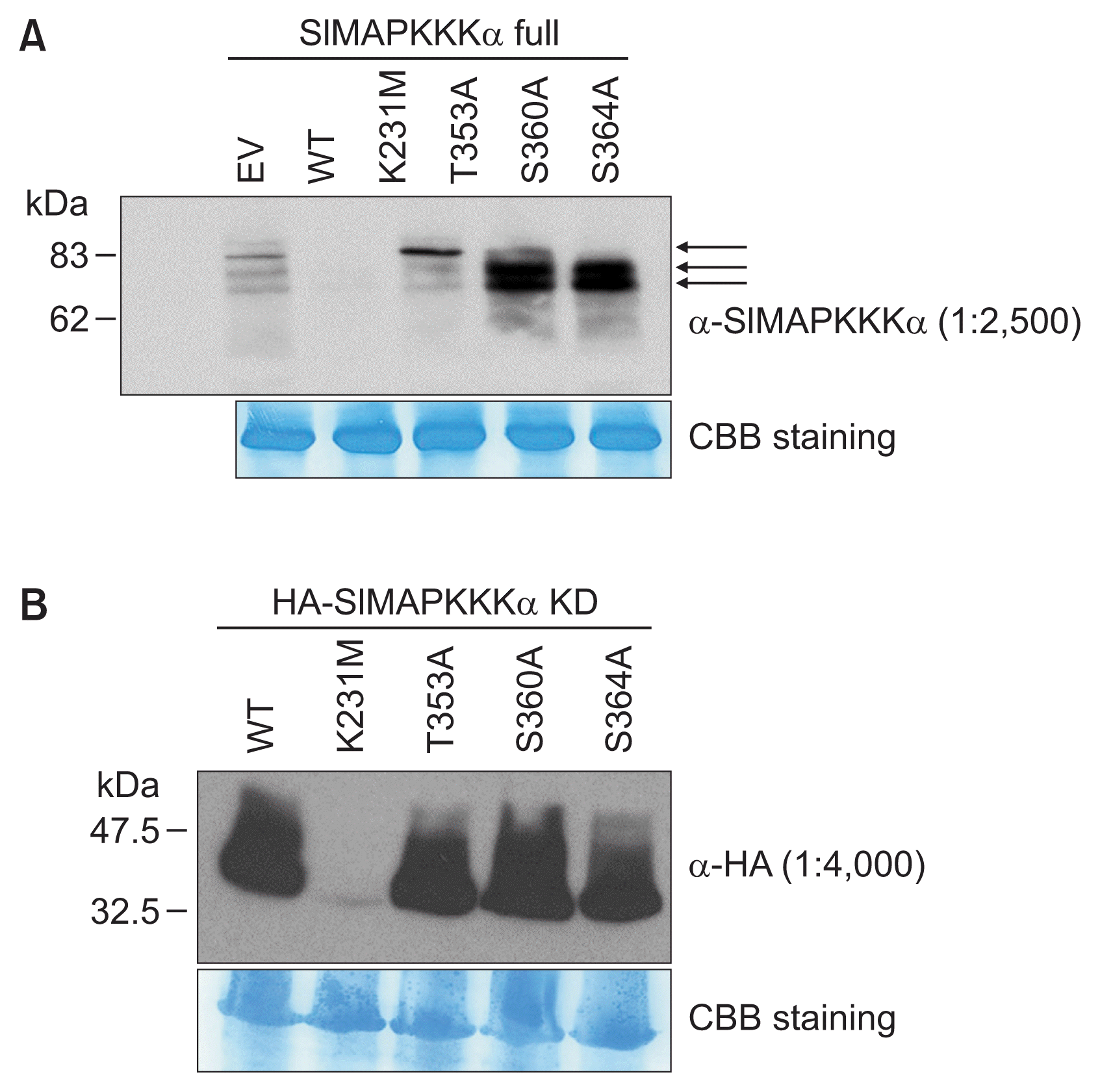
Expression of the full length (A) and kinase domain only (KD) (B) of HA-tagged SlMAPKKKα in Nicotiana benthamiana. The indicated forms of SlMAPKKKα protein were transiently expressed. An antibody of SlMAPKKKα raised in rabbits (α-SlMAPKKKα) was used to detect the full length proteins, and an HA antibody (α-HA) was used to detect HASlMAPKKKα KD. Rubisco proteins were stained with Coomassie Brilliant Blue (CBB) as loading controls. EV, empty vector; WT, wild type. Arrows in Fig. 3A indicate different forms of full length SlMAPKKKα proteins.
Discussion
We found previously that SlMAPKKKα is required for cell death associated with plant immunity and functions as a positive regulator of Pto-mediated PCD induced by interaction with AvrPto or AvrPtoB (del Pozo et al., 2004). Moreover, expression of not only the full-length SlMAPKKKα, but its kinase domain only also caused strong cell death in leaves of tomato and N. benthamiana plants (Fig. 2). Numerous studies have shown that MAPKKKs phosphorylate MAPKKs through phosphorylation of conserved serine and/or threonine residues in the T-loop of their kinase domains (Asai et al., 2002; Group, 2002; Mishra et al., 2006). Based on these reports, three putative phosphorylation residues (Thr353, Ser360, and Ser364) in the conserved activation residues between kinase subdomains VII and VIII of SlMAPKKKα were identified and characterized (Fig. 1). Substitutions of two serine residues to alanine in both full length and kinase domain only forms of SlMAPKKKα abolished its ability to trigger PCD in both tomato and N. benthamiana plants. The SlMAPKKKα protein species with the largest was missing in these variants. Similar to SlMAPKKKα, overexpression of SlMAPKKKα (T353A), which caused cell death, also appeared as three protein species in our immunoblotting assay. These multiple bands of SlMAPKKKα and SlMAPKKKα (T353A) are possibly due to posttranslational modification, particularly phosphorylation, occurring on the proteins in plants. Previously, it was also shown that Ser535 residue in C-terminal region of SlMAPKKKα, which is a putative phosphorylation site, is required for its interaction with TFT7 (14-3-3 protein) and affects PCD-eliciting activity via interaction with TFT7 in plants (Oh and Martin, 2011; Oh et al., 2010).
Although SlMAPKKKα is a positive cell death regulator activated by AvrPto/Pto, its mode of action for defense response is still unknown. However, the fact that altered protein expression pattern of variant forms (S360A and S364A) were detected in leaves overexpressing SlMAPKKKα (S360A) and SlMAPKKKα (S364A) allows us to propose the following explanation. A fully phosphorylated form of SlMAPKKKα might be required for cell death induction triggered by AvrPto/Pto or by other stimuli. In particular, Ser360 and Ser364, but not Thr353, are essential for the full phosphorylation of SlMAPKKKα.
The present and previous studies suggest that at least three serine residues at the 360, 364, and 535 positions of SlMAPKKKα are phosphorylated by elicitation of Pto-mediated cell death signaling in tomato and N. benthamiana plants. After phosphorylation, a TFT7 dimer recruits two kinases by physical interaction and then SlMKK2 is phosphorylated by SlMAPKKKα, of which ATP is bound at the Lys231 site (Oh and Martin, 2011) (Fig. 4). Our simplified model also suggests that these two serine (Ser360 and Ser364) residues, which are located in the kinase domain, might participate in a cell death signaling pathway mediated by AvrPto/Pto interaction through other receptor classes due to the fact that the kinase domain only could not interact with TFT7, whereas Ser535, which is present in the C-terminal region of SlMAPKKKα is involved in both its interaction with TFT7 in yeast and plant cells and has the ability to elicit cell death (Oh et al., 2010).
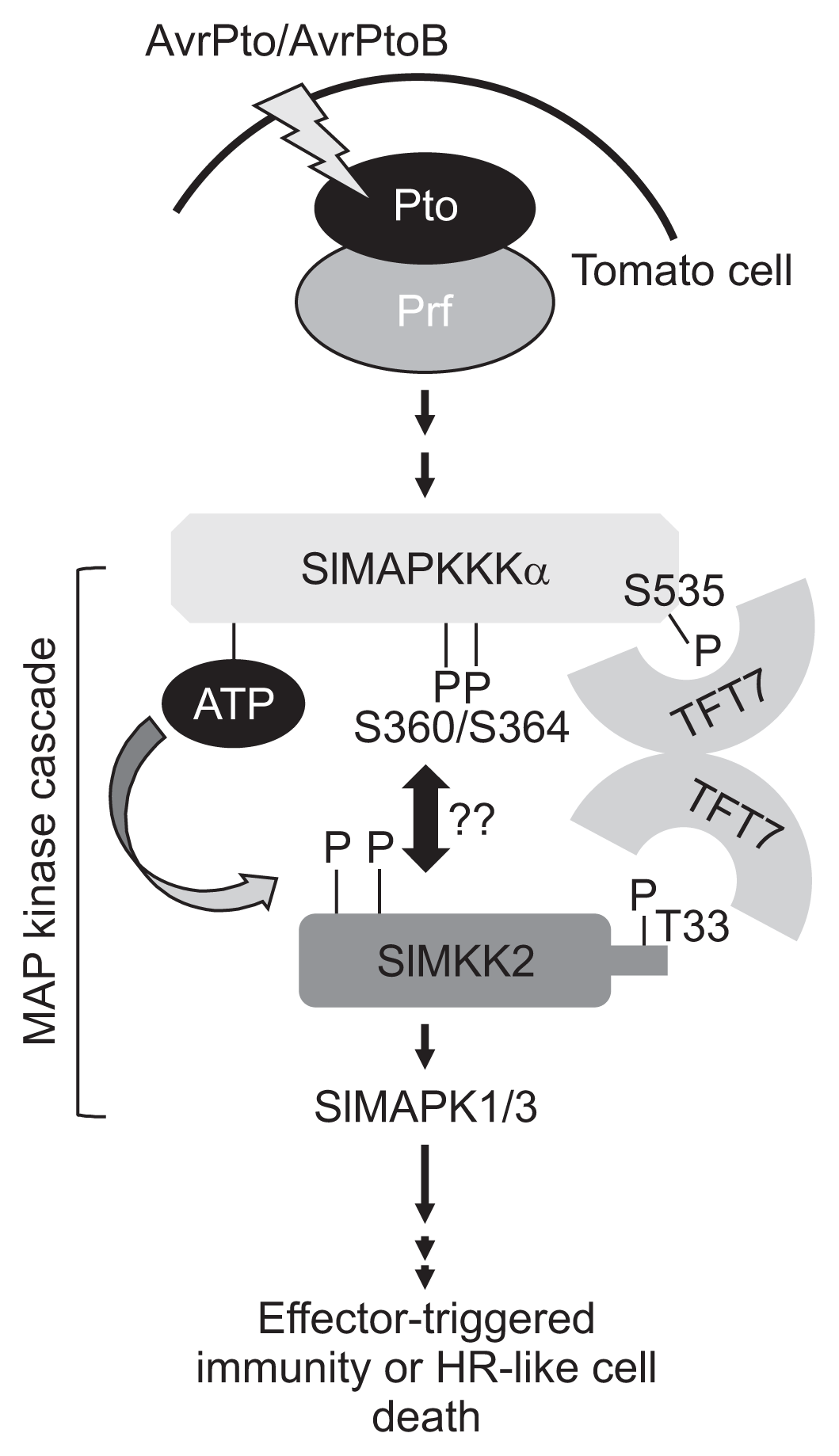
A model for MAPK signaling pathway regulating Pto-mediated effector-triggered immunity in plants. The bacterial effector/plant resistance (R) protein combinations trigger the MAPK cascade, which can lead to induction of cell death in plants. SlMAPKKKα phosphorylates and activates a SlMKK2, and SlMKK2 then phosphorylates and activates a SlMAPK1/3, resulting in cell death in plants. In this signaling pathway, TFT7 play an important role in signal transduction by formation of a stable homodimer that regulates efficient signal transfer from SlMAPKKKα to SlMKK2 in a Ser535 phosphorylation-dependent manner. Two other residues (Ser360 and Ser364) are also required for induction of cell death in plants, suggesting that at least these three residues (Ser360, Ser364, and Ser535) of SlMAPKKKα are phosphorylated and needed for kinase activation to trigger cell death via binding with TFT7 and/or other proteins.
In this study, we showed that alanine substitutions at the Ser360 and Ser364 positions of SlMAPKKKα significantly interfered with PCD induction and potentially affected formation of a fully phosphorylated form of the protein. Therefore, it is likely that, besides TFT7 protein, SlMAPKKKα also recruits a target protein to promote phosphorylation of Ser360 and Ser364 positions during activation of MAPK cascades regulating defense responses such as PCD. Identification of the target proteins remains to be determined. These findings provide a range of new possibilities for understanding cell death signaling regulated by phosphorylation event of MAPKKKs.
Acknowledgments
This work was supported by the Golden Seed Project (Center for Horticultural Seed Development No. 213003-04-4-SBG20), Ministry of Agriculture, Food and Rural Affairs (MAFRA), Ministry of Oceans and Fisheries (MOF), Rural Development Administration (RDA), and Korea Forest Service (KFS) to CSO and the National Science Foundation (IOS-1451754) to GBM.