The Plant Cellular Systems for Plant Virus Movement
Article information
Abstract
Plasmodesmata (PDs) are specialized intercellular channels that facilitate the exchange of various molecules, including sugars, ribonucleoprotein complexes, transcription factors, and mRNA. Their diameters, estimated to be 2.5 nm in the neck region, are too small to transfer viruses or viral genomes. Tobacco mosaic virus and Potexviruses are the most extensively studied viruses. In viruses, the movement protein (MP) is responsible for the PD gating that allows the intercellular movement of viral genomes. Various host factors interact with MP to regulate complicated mechanisms related to PD gating. Virus replication and assembly occur in viral replication complex (VRC) with membrane association, especially in the endoplasmic reticulum. VRC have a highly organized structure and are highly regulated by interactions among the various host factors, proteins encoded by the viral genome, and the viral genome. Virus trafficking requires host machineries, such as the cytoskeleton and the secretory systems. MP facilitates the virus replication and movement process. Despite the current level of understanding of virus movement, there are still many unknown and complex interactions between virus replication and virus movement. While numerous studies have been conducted to understand plant viruses with regards to cell-to-cell movement and replication, there are still many knowledge gaps. To study these interactions, adequate research tools must be used such as molecular, and biochemical techniques. Without such tools, virologists will not be able to gain an accurate or detailed understanding of the virus infection process.
Introduction
Viruses cannot be classified as living or nonliving organisms based on the current classification system. Outside the host cell, viruses do not exhibit basic biological characteristics like homeostasis, organization, metabolic function, response to stimuli, replication and growth, and adaptation. Therefore, outside host cells they are considered as nonliving organisms. However, although viruses lack their own reproduction system, they can replicate within their hosts’ cells and move to neighboring cells. Under these conditions, they can be considered living organisms (Hull, 2009; Walsh and Mohr, 2011).
Like all viruses, plant viruses are too small to be observed with a light microscope. Most plant viruses consist of nucleic acids as the genetic information and a coat protein (CP) that protects the nucleic acids. Since plant viruses do not have systems to produce their own components, they hijack the host’s cellular machinery for replication and intracellular/intercellular movement to achieve systemic infection (Gergerich and Dolja, 2006; Hull, 2009).
Plasmodesmata (PDs) are unique structures that occur in plant but not animal cells. PDs are specialized intercellular channels that are gateways between adjacent plant cells. These channels pass through plant cell walls and allow adjacent cells to communicate through chemical signal exchange. They also allow cells to exchange nutrients and other compounds that are critical for normal cell fate (Ding, 1998; Lucas, 1995; Maule, 2008; Roberts and Oparka, 2003).
The endomembrane system and the cytoskeleton are two major cellular systems in eukaryotic cells. In the plant cell cytoplasm the endomembrane system consists of many inter-related membranes and organelles such as chloroplasts, endoplasmic reticulum (ER), endosomes, the Golgi apparati, vacuoles, mitochondria, nuclear envelopes, peroxisomes, plasma membranes (PM), and vesicles (Chen et al., 2012; Morita and Shimada, 2014). The actin filament and the microtubule are two cytoskeleton components that are critical for maintaining the proper function of the cell. They play a role in positioning of the endomembrane system and the movement of constituents among them (Toivola et al., 2010; Wang and Hussey, 2015).
Systemic infection of plant cells by viruses requires several critical steps. After virus replication at initial infection foci, plant virus particles spread to non-infected adjacent cells as virions or viral ribonucleoprotein (vRNP) complexes by cell-to-cell movement (local movement). Then, viruses are transported to other organs by long distant movement (systemic movement) within plant host (Dolja et al., 1992; Zwart et al., 2012). These processes are closely coupled with cellular processes in association with the PD, endomembrane, and cytoskeleton (Harries et al., 2010; Heinlein, 2015; Ju et al., 2005; Novoa et al., 2005). While all steps are required for the development of the viral disease, local or systemic movement of the virus can be a critical event. If the virus loses the ability to move, infection will be restricted to an initial infection site or infected organ. Therefore, this review describes intra/intercellular virus trafficking via PD, and the interactions of cellular systems with viral factors to facilitate virus movement and replication within plants.
Structure of PDs and Selective Transport of Proteins through PDs
Plant cells communicate each other via PD which are specialized intercellular channels that allow the exchange of various molecules including sugars, vRNP complexes, transcription factors, and mRNA (Crawford and Zambryski, 2000; Heinlein and Epel, 2004; Zambryski and Crawford, 2000). There are two types of PDs; simple and complex. Simple or primary PD is a single pored channel, whereas complex PD or secondary PD consists of more than two pored channels with branches (Oparka et al., 1999). Simple PD is formed during cytokinesis and is found in immature tissue, whereas complex PD is developed after cell division. Regardless of structural difference, all PDs start as simple PD and may develop into complex PD by fusion of adjacent simple PD during tissue development (Lucas et al., 1993; Oparka et al., 1999; Roberts et al., 2001).
Based on green fluorescent protein (GFP) trafficking studies, non-specific movement occurs in immature tissue, where simple PDs are more common than in mature tissue with high size exclusion limits (SEL). However, GFP movement through PD is restricted in mature tissue, where complex PDs are common with low SEL (Imlau et al., 1999; Itaya et al., 1998, 2000; Oparka et al., 1999; Roberts et al., 2001). These observations indicate that the developmental stage of plant tissues affects trafficking specificity of PD.
Electron microscopic studies showed that the PM, ER, and cytoplasm are continuous between cells through the PD (Ding et al., 1992; Tilney et al., 1991). In addition, immune-electron micrographs showed that actin and myosin localize to PDs (Blackman and Overall, 1998; White et al., 1994) and these two structures play a role in intercellular trafficking via PDs (Aaziz et al., 2001). The central structure of the PD is the desmotubule (DT) which consists of the ER surrounded by globular proteins (Ding et al., 1992; Tilney et al., 1991). Globular proteins also line the PM. Spoke-like extensions extend from the DT to either the PM or to globular proteins lining the PM (Ding et al., 1992; Tilney et al., 1991). The cytoplasmic sleeve is the space between the PM and the DT and its diameter is estimated to be approximately 2.5 nm in the neck region of the PD. Exchange of molecules between cells occurs through this very narrow cytoplasmic sleeve (Ding et al., 1992).
Several researchers proposed that constriction or expansion of the PD neck region could regulate transport of molecules between cells (De Storme and Geelen, 2014; Demchenko et al., 2014; Ding, 1998; Lucas, 1995; Maule, 2008; Robards and Lucas, 1990; Thomas et al., 2008; Zavaliev et al., 2011). One of these proposals explained that the mechanism that modulates molecular movement across the cell wall through the PD is related to callose (β-1,3-glucan [GLU]) located in the near-cell wall (De Storme and Geelen, 2014; Zavaliev et al., 2011). The accumulation and degradation of callose between the cell wall and the PM around the neck region of PDs constrict and dilate, respectively (Zavaliev et al., 2011). Although the mass of callose increased at the PD neck region when plants encounter abiotic/biotic stresses (Bolwell et al., 2002; Stass and Horst, 2009; Ueki and Citovsky, 2002; Voigt and Somerville, 2009; Zavaliev et al., 2010), there were many reports that virus infection induces callose deposition to slow/inhibit virus movement as a defense reaction. Beffa et al. (1996) showed that GLU suppression by antisense transformation reduced disease severity, which might be due to callose accumulation. Callose induction by tissue treatment with salicylic acid reduced Tobacco mosaic virus (TMV) spread to neighboring cells (Krasavina et al., 2002). When other viruses, including Maize dwarf mosaic virus, Potato virus X (PVX), Potato virus Y, Soybean mosaic virus infected experimental hosts resistant to viral infection, callose deposition was stimulated and accumulated in cell walls around local lesions. This callose deposition was regardless of PD association but due to the plant defense reaction (Li et al., 2012; Zavaliev et al., 2010). When resistant hosts were inoculated with TMV, high levels of callose deposition at PDs and increased pathogenesis-related (PR) protein 1 expression were observed, suggesting that callose deposition at PDs may be coupled with PR expression to turn on the resistant mechanism (Kathiria et al., 2010; Zavaliev et al., 2010). In addition, cadmium treatment of plants also triggered callose deposition at PDs of the vasculature, which hindered systemic infection by plant viruses (Ueki and Citovsky, 2002).
In addition to PD regulation by callose deposition/degradation, other host factors are involved in PD gating. One example is centrin, a calcium-binding protein and responsible for forming calcium-sensitive contractile nanofilaments localized in the PD neck region. Centrin facilitates PD gating by calcium-induced regulation (Blackman et al., 1999). Centrin is contracted by dephosphorylation, which occurs when cytoplasmic calcium increases or heat shock is applied (Martindale and Salisbury, 1990). This dephosphorylation may involve closing the PD (Blackman et al., 1999; Martindale and Salisbury, 1990; Roberts and Oparka, 2003). Although a centrin is one of host factors that regulate cell-to-cell communication, there is little evidence for its role in virus trafficking through PDs.
A second host factor that regulates callose deposition is calreticulin (CRT) which is a calcium-sequestering protein that is localized to PDs (Baluška et al., 1999, 2001; Bayer et al., 2004; Chen et al., 2005; Laporte et al., 2003; Michalak et al., 1999; Ostwald and MacLennan, 1974; Roberts and Oparka, 2003; Tomenius et al., 1987). CRT is also commonly found in the ER (Michalak et al., 1999; Roberts and Oparka, 2003). Several studies reported that CRT is involved in the cell-to-cell movement of plant viruses (Chen et al., 2005; Laporte et al., 2003; Roberts and Oparka, 2003; Shen et al., 2010; Ye et al., 2013). CRT is induced by abiotic/biotic stress and increases calcium-sequestering activity in the cell. Once calcium levels decrease by CRT expression, PD permeability increases by increasing SEL of PDs. However, CRT expression does not always promote virus trafficking via PDs (Chen et al., 2005; Laporte et al., 2003; Roberts and Oparka, 2003; Shen et al., 2010; Verchot, 2014; Ye et al., 2013). CRT expression may facilitate or inhibit virus movement (Chen et al., 2005; Laporte et al., 2003). Chen et al. (2005) showed that overexpression of TMV movement protein (MP) hindered TMV movement, whereas Laporte et al. (2003) reported that overexpression of CRT promoted cell-to-cell movement of Grapevine fanleaf virus (GFLV). Interestingly, MPs of both viruses interact with CRT. This difference may due to different modes of CRT action. With TMV, TMV MP may not be able to reach PD due to the accumulation of CRT in the PD region targeted by the MP (Chen et al., 2005). However, CRT interacts with GFLV MP and redirects MP to the tubular network instead of occupying the PD (Laporte et al., 2003). The tubular network across PD serves as a trafficking routefor virion between cells (Laporte et al., 2003).
Another host factor is the non-cell-autonomous protein (NCAP) pathway. There are two types of NCAP pathways; the gate-open (GO)-NCAP model and PDs-selective NCAP model. PDs-selective NCAP is similar to the GO-NCAP pathway in that the PD dilates to allow movement of macromolecules between cells (Lucas and Lee, 2004). The PDs-selective NCAP pathway differs from the GO-NCAP pathway in that it requires specific proteins to interact with the PD to trigger dilation of the pore. In other words, the PDs-selective NCAP is more selective than the GO-NCAP. A cellular factor (GO protein) binds to a gating receptor at the mouth of the PD to form a gating complex. This complex triggers dilation of the PD and allows marcromolecules to move between cells. Some macromolecules may be anchored to chaperones and/or carriers, which transfer them to the docking area around PDs. The induction of PD dilation in this manner leads selective intercellular transport. There is evidence that movement of the vRNP complex into neighboring cells is selectively regulated by the interactions of virus protein with endogenous proteins. For example, movement of the TMV MP-RNA complex into adjacent cells was regulated by the interaction of TMV MP with the Non-Cell-Autonomous Pathway Protein 1 (NCAPP1) (Lee et al., 2003).
The Replication of Plant Viruses and Viral Replication Complex
It is generally accepted that TMV can only enter plant cells by mechanical manner (Shaw, 1999). Like other plant viruses, the systemic movement of TMV has two requirements; the replication of the viral RNA within a cell and trafficking of vRNA into adjacent cells through PD (Kawakami et al., 2004; Liu et al., 2005; Maule, 2008; Peña and Heinlein, 2012) (Fig. 1). Wu et al. (1994) observed that TMV started uncoating for translation, and formed small granules containing CP and vRNA within 5 min after inoculation into protoplasts. Christensen et al. (2009) showed that TMV vRNA without the 5′ cap lost the ability to form granules, indicating that the formation of granules with vRNA is actin-dependent.
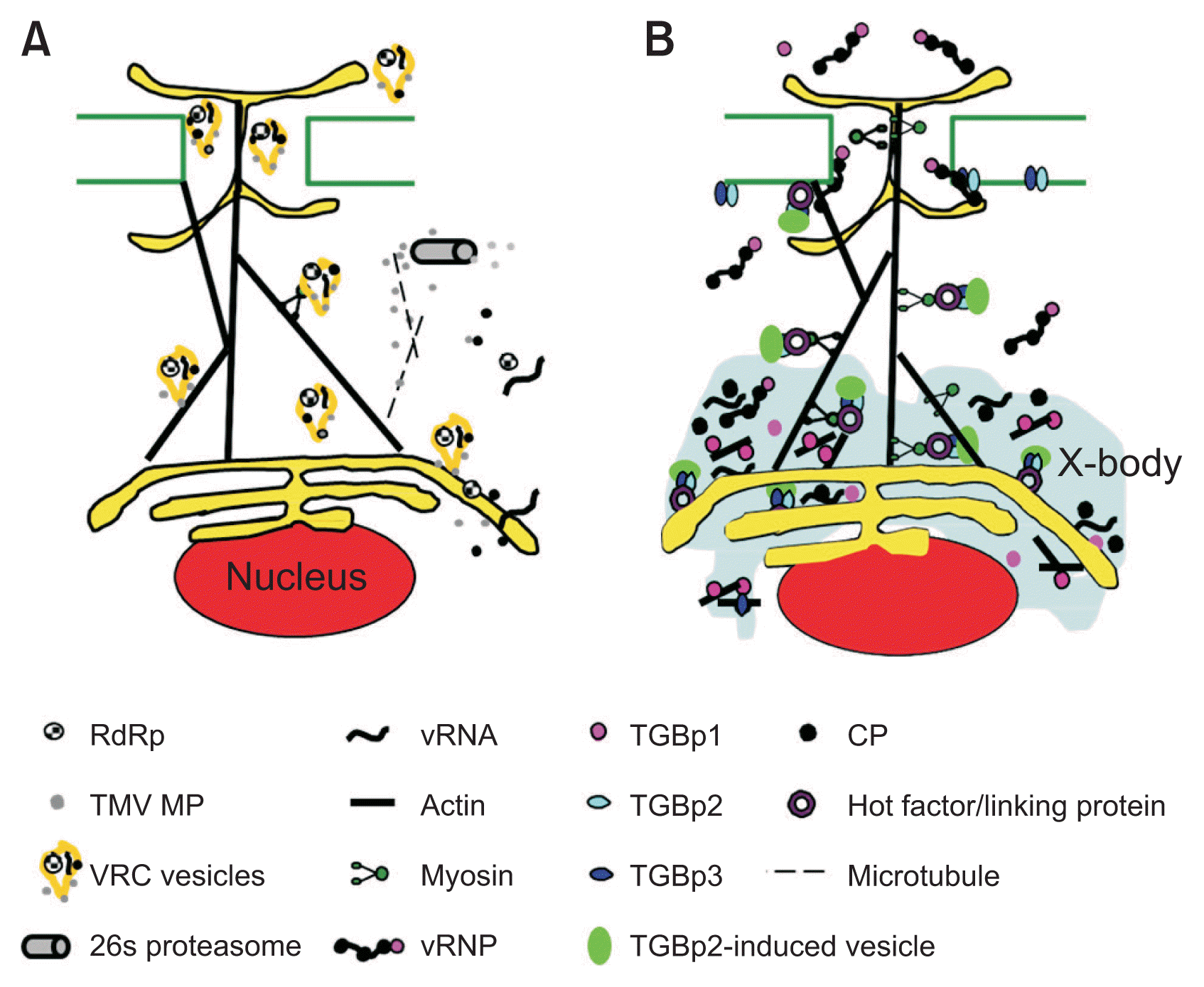
Models for cell-to-cell movement of Tobacco mosaic virus (TMV) and Potexvirus. (A) Model describing the TMV cell-to-cell movement. Viral ribonucleoprotein (vRNP) complexes contain RNA dependent RNA polymerase (RdRp), movement protein (MP) viral RNA (vRNA), and coat protein (CP) in the early TMV infection. Phosphorylated MP is localized on perinuclear and involves in conversion vRNP complexes into virus replication complexes (VRCs) vesicles in association with endoplasmic reticulum (ER) membrane. These vesicles move along actin filaments in association with myosin to the plasmodesmata (PD) and across the PD at the leading infection front. Microtubules (dotted line) transport TMV MP to a 26s proteasome for the degradation at the infection center. (B) Model describing movement of Potexvirus. Triple gene block protein 1 (TGBp1)-derived X-body (induced inclusion structure due to virus infection) contributes to the compartmentalization of viral gene products during the viral replication to protect them from cytosolic nuclease. The CP and TGBp1 bind to the vRNA, forming the vRNP complex in X-body and move to the PD. Triple gene block protein 2 (TGBp2) induces the ER-derived small granular vesicles and colocalizes with triple gene block protein 3 (TGBp3). Vesicles containing both TGBp2 and TGBp3 are associated with ER and move along actin filament to the PD. The RNP complex may not bind to TGBp2 and TGBp3 complex. The RNP complex and the vesicle containing TGBp2 and TGBp3 move independently to the PD. The RNP complex moves across the PD but the TGBp2 and TGBp3 complexes may not move to the adjacent cells. TGBp1-derived X-body (induced inclusion structure due to virus infection) contributes to the compartmentalization of viral gene products during the viral replication to protect them from cytosolic nuclease.
Triple gene block protein 1 (TGBp1) destabilizes PVX virions and promotes translation of virion-derived RNA. While PVX virions are not translatable in vitro, in the presence of wheat germ extracts, TGBp1 stimulates translation of virion-derived RNA (Atabekov et al., 2000; Karpova et al., 2006; Kiselyova et al., 2003; Lukashina et al., 2009; Rodionova et al., 2003). TGBp1 ATPase activity and helicase activity may contribute to the unwinding of virions for translation. However, there is no data to indicate this is an in vivo mechanism for translation of virions. Karpova et al. (2006) and Rodionova et al. (2003) presented atomic force microscopy and electron microscopy that visualized the localization of the TGBp1 at one end of the virion. Based on these studies, the PVX virion can be remodeled by binding of TGBp1 to 5′ end of the virus particle, leading to particle destabilization and RNA exposure for the translation. Since TGBp1 is not present at the beginning of infection, it was suggested that TGBp1 might act after the virus has moved into neighboring cells as a factor that promotes translation of virion-derived RNAs (Atabekov et al., 2000; Karpova et al., 2006; Kiselyova et al., 2003; Rodionova et al., 2003). It is worthy to note that the level of CP phosphorylation is involved in transition of PVX RNA between translatable or untranslatable forms (Karpova et al., 2006). However, it is not known how CP phosphorylation modulates binding of the CP to PVX RNA in the virion (Atabekov et al., 2001). There is evidence that phosphorylation of viral protein affects translatability of virus RNA. Phosphorylation of TMV MP occurred during movement via PD plays a role in the translation of TMV vRNA resulting in conversion nontranslatable form into translatable form (Karpova et al., 1999). In addition, this phosphorylation activity seems to regulate the intracellular trafficking of TMV by regulating SEL of PD in a host-dependent manner (Waigmann et al., 2000).
During virus replication, different plant viruses induce membrane-bound vesicles from various organelles, including ER, mitochondria, the nucleus, and chloroplasts (Kopek et al., 2007). Modifications of the membrane or organelle structures during the replication of plant viruses may occur by increasing the proteins that originate from viral genomes to promote further replication and movement (Grangeon et al., 2012a). It is believed that these unique vesicles may prevent the degradation of the viral genome by intracellular enzymes, or may allow the plant virus to avoid host defense mechanisms by confining virus replication intermediates to vesicles (Bamunusinghe et al., 2009, 2011; Diaz and Ahlquist, 2012; Diaz and Wang, 2014). The viral RNA genomes for Carnation Italian ringspot virus, PVX, Tomato bushy stunt virus, and Turnip yellow mosaic virus associate with chloroplasts, ER, peroxisomes, and mitochondria, respectively (Hwang et al., 2008; McCartney et al., 2005; Prod’homme et al., 2003; Verchot, 2011, 2014). TMV also replicates in a membrane-associated complex that contains replicase, MP, vRNA, and host factors (de Castro et al., 2013; Grangeon et al., 2012a; Hagiwara et al., 2003; Más and Beachy, 1999; Peña and Heinlein, 2012). The ER was considered the site of TMV replication and viral replication complex (VRC) formation (de Castro et al., 2013; Heinlein et al., 1998; Más and Beachy, 1999; Reichel and Beachy, 1998). During TMV replication, ER aggregation was observed in the cortical region with less membrane tubule structure. However, ER aggregates were turned over, becoming normal ER at the end of replication, indicating an association of TMV replication with ER (Reichel and Beachy, 1998). In addition to ER association, TMV replicase associates with two host proteins, TOM1 and TOM2, that are related to tonoplast, even though there is no evidence that TMV replication occurs at the tonoplast (Ishikawa et al., 1993; Tsujimoto at al., 2003). These studies suggest that host cell membrane modification by virus replication and the role of replicase are more complicated than previously thought.
The Movements of Plant Viruses across PDs
Because virus movement complexes are larger than PD opening, TMV cannot move to neighboring cells without a special mechanism to overcome this physical barrier (Citovsky et al., 1992; Deom et al., 1987). TMV encodes a single MP that is 30 kDa in size. After the effect of TMV MP on PD dilation was discovered (Deom et al., 1987), MPs of many other viruses were described and have been employed to describe cell-to-cell movement of a wide range of plant viruses. The TMV MP cooperatively binds single-strand RNA (Citovsky et al., 1992). It also dilates PD and traffics the viral RNA into neighboring cells (Deom et al., 1987; Wolf et al., 1989, 1991). The TMV MP interacts with the ER and colocalizes with the viral replicase and vRNA early in virus infection (Heinlein et al., 1998; Hirashima and Watanabe, 2003; Más and Beachy, 1999). Several studies showed the association of replicase with TMV movement. The helicase domain of the TMV replicase is required for TMV cell-to-cell movement (Hirashima and Watanabe, 2003), suggesting that a replicase may also be an important component of the complex that is transported between cells. Immunofluorescence labeling by Liu et al. (2005) showed that the TMV replicase colocalized with the MP in granular bodies in tobacco protoplasts and leaves. Liu et al. (2005) also reported that mutations in the replicase domain altered the subcellular accumulation pattern of the TMV MP. Netherton et al. (2007) reported that TMV uses the membrane structure for the viral RNA replication and intracellular trafficking. These studies suggest that the subcellular targeting of TMV MP is related to the TMV replicase. Kawakami et al. (2004) reported an interesting phenomenon related to the movement of the inclusion body induced by TMV infection. Their time-lapse study showed that the first virus movement to non-infected adjacent cells is closely related to replication proteins. Based on their study, MPs were localized to small inclusion bodies at 12 h after inoculation (hai). At this time small inclusion bodies were not associated with specific structures and these small bodies showed no movement. After that, they observed MP accumulation and association with these bodies and VRC between 12 and 16 hai. Small inclusion bodies were enlarged for the formation of VRC and started to move around 14 hai when MP was colocalize with replication proteins (Kawakami et al., 2004). Both MP and VRC interact with actin filaments and this association was essential for their intracellular movement (Kawakami et al., 2004; Liu et al., 2005). In addition to actin filament involvement in virus movement, MP interacts with the microtubules for intercellular movement of TMV via PD (Brandner et al., 2008).
Using electron microscopy, the PVX CP was shown to reside inside PDs, but it does not trigger PD dilation (Oparka et al., 1996). Fibrillar material containing CP was seen near the mouth of the PDs, leading some researchers to suggest that either the PVX virion or an intermediate form contributes to virus cell-to-cell movement (Oparka et al., 1996). The 5′ non-translational region (NTR) of the PVX genome contains a sequence motif recognized by the CP and is required for transport of viral RNA across PD. Lough et al. (2006) proposed that the CP first binds to this sequence motif and recruits PVX TGBp1 to the viral RNA, forming the TGBp1/CP/vRNA complex. This complex was confirmed by atomic force microscopy and electron microscopy (Karpova et al., 2006). In vitro RNA binding assays also indicated that TGBp1, CP, and vRNA can form a complex (Atabekov et al., 2000; Lough et al., 1998). Mutations in the Potexvirus CP that disrupt virion formation have no impact on the ability of the virus to move from cell-to-cell (Lough et al., 2000), supporting the hypothesis that TGBp1/CP/vRNA is a vRNP complex that traffics through PD.
The PVX TGBp1 is a multifunctional protein. TGBp1 has an RNA helicase and two canonical ATPase motifs that are G-K-S and D-E-Y tripeptides. Both motifs are necessary to provide ATP hydrolysis essential to unwind double-stranded RNA (Kalinina et al., 1996, 2002; Leshchiner et al., 2006). In addition to RNA helicase activity, the TGBp1 proteins are involved in gating PD, although the role of TGB1p RNA helicase activity for PD transport has not been determined. It is certain that the TGBp1 protein provides PD permeability of 10-kDa molecules, resulting in trafficking between cells (Angell et al., 1996; Howard et al., 2004; Lough et al., 1998). Microinjection and bombardment experiments showed that TGBp1 can increase PD permeability for virus transport (Angell et al., 1996; Howard et al., 2004; Yang et al., 2000). Using GFP fusion with TGBp1 of PVX, researchers have shown that in addition to having RNA-binding and helicase activities, TGBp1 protein increases the PD SELs for viral cell-to-cell movement (Angell et al., 1996; Tamai and Meshi, 2001). TGBp1-mediated X-body (induced inclusion structure due to virus infection) reorganization contributes to the compartmentalization of viral gene products during infection (Tilsner et al., 2012; Yan et al., 2012). A TGBp1 mutant that lost movement function could not form rodlike structures, whereas mutants that still supported cell-to-cell movement formed rod-like structures. This indicated that TGBp1’s movement function is closely associated with the formation of rod-like structures (Yan et al., 2012). In addition, Tilsner et al. (2012) showed that PVX TGBp1 reorganizes actin and endomembranes into the X-body as a VRC.
Deleting PVX triple gene block protein 2 (TGBp2) or triple gene block protein 3 (TGBp3) from the potexvirus PVX genome restricted virus movement, indicating that these factors contribute to virus export from the cell (Lough et al., 2000). Microinjection experiments delivering large fluorescent dextrans or fluorescein isothiocyanate-labeled TGBp1 proteins to transgenic plants expressing TGBp2 and TGBp3 showed that TGBp2 and TGBp3 promote cell-to-cell movement of TGBp1, but do not directly increase PD SEL (Lough et al., 1998). These data suggested that TGBp2 and TGBp3 play a role in promoting virus transport to the PD rather than across the PD (Lough et al., 1998). Further experiments showed that the TGBp2 and TGBp3 proteins associate with ER (Krishnamurthy et al., 2003; Mitra et al., 2003). TGBp2 has two transmembrane domains, while the PVX TGBp3 has one (Krishnamurthy et al., 2003; Mitra et al., 2003). Mutations that disrupted ER-association of TGBp2 or TGBp3 also inhibited virus movement, suggesting that ER association is essential for PVX cell-to-cell transport (Krishnamurthy et al., 2003; Mitra et al., 2003). TGBp2 interacts with a host factor named TGBp2 interacting protein 1, which regulates the activity of GLU, a key enzyme involved in callose degradation (Fridborg et al., 2003). GLU expression also facilitates TMV systemic movement (Bucher et al., 2001). A GLU-deficient mutant showed more callose deposition, inhibiting movement of TMV, PVX and Cucumber mosaic virus 3a protein (Iglesias and Meins, 2000).
Voinnet et al. (2000) showed that PVX TGBp1 acts a suppressor of RNA silencing. Mutations that inhibitted silencing suppressor activity also blocked virus cell-to-cell movement, suggesting that silencing suppression is linked to virus transport (Bayne et al., 2005; Voinnet et al., 2000). Mutations impeding TGBp1 suppressor activity also inhibited virus movement, indicating that these processes are coordinated (Bayne et al., 2005). Whether the TGBp1 blocks amplification of the 21 nt small interfering RNA or acts downstream of RNA-dependent RNA polymerase 6 is still uncertain. However, it is likely that its role as a silencing suppressor protects the replicating virus from RNA silencing. This may be sufficient to promote cell-to-cell spread of viruses, or there may be additional machinery that has yet to be discovered that further links the viral PD transport machinery with RNA silencing.
Lucas (2006) described TMV MP/NCAP model to explain TMV movement across PD by showing that the MP/vRNA-vRNP complex interacts with a cellular carrier, which then interacts with the docking complex at the PD. Binding of the MP/vRNA/carrier to the docking complex triggers PD dilation. Then the MP/vRNA/carrier complex moves across the PD to adjacent cells. In the receiving cell, the carrier protein dissociates from the MP/vRNA/carrier complex. The viral RNA is then translated in the receiving cell, initiating virus replication in that cell (Lucas, 2006). The NCAP model indicates that the MP/vRNA complex moves through the cytoplasm and across the PD.
The cytoskeleton system is a highly dynamic network involving in intra-/intercellular trafficking (Wang and Hussey, 2015). The plant cytoskeleton (actin and tubulin) is involved with virus replication and with virus movement (Christensen et al., 2009; Ju et al., 2005; Liu et al., 2005). However, the role of actin filament in virus movement cannot be explained by the NCAP model.
Actin Filament and Membrane Association
As mentioned above, cellular membranous systems and cytoskeletal elements are involved in developing VRC, which are highly organized structures. Studies with many plant viruses revealed that VRC is generated from various regions in cells and their movement is highly associated with PDs. Therefore, it is certain that cytoskeleton-associated processes involved in remodeling of the subcellular membranous structures regulate virus replication and movement.
Intercellular movement of TMV via PDs did not occur when microfilaments were disrupted by pharmacological means or by gene silencing (Kawakami et al., 2004; Liu et al., 2005). Kawakami et al. (2004) mentioned that granules or membrane-bound VRC containing the TMV MP, move along the actin network toward and through the PD. Liu et al. (2005) showed that the VRC move along the actin network and then dock at the PD. The VRC in this model is the vRNP complex, not membrane-bound granules, which remain associated with the ER network as they move through the PD (Liu et al., 2005). Both studies reported that TMV movement is associated with actin filament, but despite Turnip vein clearing virus (TVCV) belonging to the same genus as TMV, the intercellular movement of TVCV was unaffected by the disruption of microfilament association (Harries et al., 2009b). It is worthy to note that TVCV and TMV infect different groups of the hosts. While TVCV infects Cruciferae, TMV infects Solanaceae. This may suggest that TVCV utilizes a different infection strategy than TMV to adapt to different host species. It may utilize different host components in intracellular movement, and possibly intercellular movement.
It was suggested that granular bodies containing the TMV replicase, MP and viral RNA to move from the perinuclear region to the PD along actin filaments (Christensen et al., 2009; Kawakami et al., 2004; Liu et al., 2005). Heinlein et al. (1995) and McLean et al. (1995) showed that TMV MP interacted with the cytoskeleton network for trafficking along actin filaments to reach the PDs. The chemical inhibitors, cytochalasin D or latrunculin B, which disrupted the actin networks, did not affect the formation of initial granules, but inhibitted intracellular movement of these granular bodies and TMV cell-to-cell movement. This indicated that vRNA movement is actin-dependent (Christensen et al., 2009).
As mentioned above, the successful transport of viruses across PDs is essential for the systemic infection (Kotlizky et al., 2001). The vRNP complex locates or recruits specific sites within a cell to form membranous VRC, where new vRNAs are synthesized. Then, the new vRNA are transported to the PD with/without subcellular organelle association in the form of vRNPs or in the vesicle form of VRC. Cytoplasmic streaming may be involved in the intracellular movement of plant virus (Verchot-Lubicz and Goldstein, 2010). Myosin motors move along with actin filaments to provide the driving force for cytoplasmic streaming. Myosin motors transport vesicles, which are significantly bigger than them. Myosin motors also are involved in moving and orienting subcellular organelles (Kachar, 1985). Although cytoplasmic streaming appears chaotic, cytoplasmic streaming is highly regulated by the interaction of myosin motors with “cargo”. Initially formed vRNP may use cytoplasmic streaming to recruit binding site for the virus replication. Numerous studies revealed that plant virus proteins are associated with various organelles (Bamunusinghe et al., 2009, 2011; Hwang et al., 2008; Kawakami et al., 2004; Liu et al., 2005; Netherton et al., 2007; Sanfaçon, 2005). It is noteworthy that the ER network passes through PDs in a continuous manner and actin and myosin localize to PDs (Blackman and Overall, 1998; Ding et al., 1992; Maule, 2008; Tilney et al., 1991; White et al., 1994).
The dynamic membrane network and the cytoskeleton are associated with virus replication and movement (Harries et al., 2009a; Heinlein et al., 1995; Hofmann et al., 2009; Kawakami et al., 2004; Liu et al., 2005; Wright et al., 2007). Based on numerous studies, it is reasonable that actin filament is connected with the ER network intracellularly/intercellularly to form an ER-associated actin system. Since the neighboring cells connect to each other by the ER network through PD (Ding et al., 1992; Maule, 2008) and vRNA replicates on remodeled ER membrane (Más and Beachy, 1999; Netherton et al., 2007). Like other membrane systems, the ER membrane is comprised of fluid lipid molecules and embedded proteins. In other words, the ER membrane system is flexible and diffusible. In fact, photoactivation studies conducted with photoactivatable GFP anchored to the ER membrane showed the movement of this special protein by ER diffusion (Runions et al., 2006). The authors observed that ER remodeling was stopped by cytoskeleton depolymerization due to latrunculin B treatment and recognized that the movement speed of photoactivated GFP was decreased compared to the non-treated sample. This indicated that the actin filament actively modulates the ER remodeling. Considering that the actin filament is closely associated with the ER network, and the many viral proteins move with membrane-association, it is reasonable to propose that endomembrane system plays a role in plant virus movement. Several studies reported a similar phenomenon describing the intracellular trafficking of vRNA of the Turnip mosaic virus (TuMV) along the ER/microfilaments (Grangeon et al., 2012b; Wan et al., 2015; Wei et al., 2010). It is certain that VRC should move intercellularly to non-infected adjacent cells for successful infection. However, it was not clear how VRC crossed between cells before the study conducted by Grangeon et al. (2013). This is because there was only one study describing the intercellular movement of vesicle-associated TMV not the general phenomenon, but a specific event for TMV under an unusual condition. However, Grangeon et al. (2013) clearly showed that the membrane-bound 6K2 of TuMV remodeled organelles to form vesicles associated with the VRC and this VRC-vesicle moved to non-infected neighboring cells. This indicated that vesicle movement across PD is not specific for TMV.
Microtubule Association
There are numerous studies regarding microtubule involvement in virus movement and replication. Upon animal virus infection, virions and VRC move from the infection site to the replication site and then complexes containing viral proteins and viral genomes are transported to the virus assembly site and the PM for further egress.
The utilization of the cytoskeleton network by plant viruses is intricate and remains to be elucidated (Boyko et al., 2000b, 2007; Harries et al., 2009b; Wright et al., 2007). However, like animal viruses, plant viruses actively employ the microtubule cytoskeleton systems for virion and VRC movement from the replication site to PDs (Boyko et al., 2000a, 2007; Haikonen et al., 2013; Harries et al., 2009b; Heinlein et al., 1995, 1998; Liu and Nelson, 2013; Martinière et al., 2009; McLean et al., 1995; Niehl et al., 2013; Padgett et al., 1996).
Boyko et al. (2000a, 2000b, 2007) reported the importance of TMV MP interaction with microtubules for the intracellular movement of vRNA to PDs. They showed that the microtubule-MP association disappeared with inactivation of MP by high temperature. They also reported that microtubule association with MP is temperature-dependent. The mutants of MPs unable to associate with microtubules showed dramatically decreased movement of MP (Boyko et al., 2007). Boyko et al. (2000a) also showed that TMV MP has a conserved sequence domain that is similar to a sequence motif of the microtubule. When point mutations were introduced into this motif, TMV showed decreased cell-to-cell movement and there was no interaction between MP and microtubules, suggesting that the MP-microtubule association is essential for the movement of TMV (Boyko et al., 2000a). MPs bind to microtubules to stabilize them by forming the microtubule-associated protein complex (Ashby et al., 2006; Boyko et al., 2000a; Ferralli et al., 2006).
Several studies showed that MPB2C (MP-binding protein 2C), known as a tobacco microtubule-associated protein, interacted with MP resulting in a change in the spread MP to adjacent cells (Curin et al., 2007; Gillespie et al., 2002; Kragler et al., 2003). This reduced movement may be due to no MPs’ associated with microtubules because MPB2C preoccupy the site on MP where microtubules bind. When NtMPB2C was coexpressed, the cell-to-cell movement of MPR3, a point mutant of MP, increased but there was no association with microtubules (Gillespie et al., 2002). This suggests that NtMPB2C acts as an adverse regulator for the cell-to-cell movement of MP that operates at the level of microtubules. These two studies showed opposite results regarding cell-to-cell movement and the relationship between MPB2C and microtubules. Considering this complicated relationship between MPB2C, microtubules and movement of MP, the function of MPB2C for the intercellular movement of MP is ambiguous. Curin et al. (2007) investigated the role of MPB2C in movement of MP and its interaction with microtubules. They observed that MPB2C is not essential for the movement of MP, but subcellular localization of MP clearly changed toward PDs. MPB2C also acts as a modulator for intercellular trafficking of PVX (Cho et al., 2012).
The MP interacts with other intracellular targets with microtubule association (Ferralli et al., 2006; Heinlein et al., 1998). MP-microtubule complexes accumulate in ER-derived membrane structure (Ferralli et al., 2006). They suggested that this association may play an important role for the MP to recruit ER membranes and for remodeling of ER during virus infection.
The MP contains hydrophobic domains that are more likely transmembrane domains that may be involved in ER association with MP (Brill et al., 2000, 2004; Fujiki et al., 2006; Reichel and Beachy, 1998). This ER-MP association occurs at an early MP expression stage to form ER-VRC, in which the viral RNAs (Christensen et al., 2009). Analysis of intracellular distribution showed that microtubule-MPs were distributed to the cortical cytoplasm, whereas nonfunctional mutant MPs remained with perinuclear replication bodies (Más and Beachy, 2000). Regardless of mutation, the wild type MP and the mutant MP interacted with the ER and the viral RNA (Más and Beachy, 2000). Upon TMV infection with MP:GFP, microtubule-associated small particles form in cells at the infection front. The study indicates that the small VRC is anchored to microtubule early and then mature into large VCRs that act as viral factories (Más and Beachy, 2000). The microtubule-associated MPs are variable. This phenomenon relies on time after infection, host properties, types of virus, and environmental factors (Boyko et al., 2000b; Gillespie et al., 2002; Heinlein et al., 1998; Padgett et al., 1996). Regardless of the situation, MPs accumulate intracellularly along with microtubules for microtubule stabilization, intracellular movement of VRC, and virus spread via PDs (Ashby et al., 2006; Boyko et al., 2007; Curin et al., 2007; Niehl et al., 2013). Also, this microtubule complex association may inhibit cell-to-cell movement of viruses at the infection site. When there is sufficient accumulation of microtubule-MP complex, MPs evanesce during late infection. Thus, in the central region of the infection sites, MP is degraded by the 26S proteasome induced by this association.
Although numerous studies have been conducted, the role of association of TMV MP with microtubules in TMV movement is not yet clarified. Más and Beachy (1999, 2000) observed that vRNAs were colocalized with microtubules and they suggested that TMV movement rely on microtubule in association with MP. However, the role of the microtubule cytoskeleton during TMV movement has been challenged from the study conducted by Gillespie et al. (2002). TMV still moved although microtubule inhibitors were treated in leaves to disrupt microtubule (Gillespie et al., 2002; Kawakami et al., 2004), suggesting that there may be a microtubule-independent pathway for TMV movement to adjacent cells or that not all microtubule species is disrupted by treatment of microtubule inhibitors. Indeed, Seemanpillai et al. (2006) verified that pharmacological agents used in previous studies did not disrupt all microtubule system. α-tubulin was disrupted upon treatment of the microtubule-disrupting agent, colchicine, but β-tubulin was not disrupted by the pharmacological agent (Seemanpillai et al., 2006), indicating that microtubule association is not essential for TMV movement.
Unlike TMV, there is little literature regarding to the microtubule association with MP of other plant viruses. Therefore, further research will be required to fully understand the role of microtubule during the movement of plant viruses.
The Utilization of Secretory Pathway by Plant Viruses
Like other eukaryotic cells, plant cells have a sophisticated secretory system made up of the ER, the Golgi, trans-Golgi network, many different types of endosomes, and vacuoles (Chrispeels, 1991; Hanton et al., 2005). Proteins synthesized in the ER are transported to target areas via the Golgi and proteins outside of the PM can be brought into the cell through endocytosis. It is essential to have this dynamic and well organized secretory system to maintain normal cell life cycle (Hanton et al., 2005). As most of the animal viruses are enveloped, they generally utilize the modified secretory system for virus assembly and virus replication and spread to adjacent cells (Netherton et al., 2007).
Since most of plant viruses are non-enveloped, they may not utilize the secretory pathway during virus infection. However, several studies reported that plant viruses also modify the secretory system for infection, vRNA replication and intracellular/intercellular trafficking of viral factors (Carette et al., 2000, 2002; dos Reis Figueira et al., 2002; Grangeon et al., 2012a; Heinlein et al., 1998; Laliberté and Sanfaçon, 2010; Laliberté and Zheng, 2014; Más and Beachy, 1999; Reichel and Beachy, 1998; Verchot, 2011; Wei and Wang, 2008).
The ER membrane within a cell infected with TMV is modified into irregular aggreosomes early in infection (Reichel and Beachy, 1998). This structure induced by TMV infection, contains various virus and host factors, such as the virus replicases, MP, vRNA, ribosomes, and EF1-α (host translation elongation factor) (dos Reis Figueira et al., 2002; Heinlein et al., 1998; Más and Beachy, 1999; Reichel and Beachy, 1998). When the plant is infected by TuMV, ER-derived small vesicles within cells are induced and rapid movement of these vesicles is associated with the actin filament and ER (Beauchemin et al., 2007; Cotton et al., 2009; Grangeon et al., 2012a). TuMV 6K2 contains the domain involved in formation of the ER-derived motile structures (Beauchemin et al., 2007). These 6K2-induced vesicles contain many viral and host factors, including viral double-stranded RNA and poly-A binding protein, indicating that the virus replicates in these vesicles (Beauchemin and Laliberté, 2007; Beauchemin et al., 2007; Cotton et al., 2009). Unlike TuMV, Tobacco etch virus (TEV) that belongs to the same genus as TuMV, also induces the motile ER-derived 6K2-induced vesicles. However; TEV associates with ER exit sites in a CP complex II- and CP complex I-dependent manner (Wei and Wang, 2008). The other difference compared to TEV is that the TuMV-induced perinuclear globular structure that includes viral 6K2, CP complex II-related structure, ER, Golgi, and chloroplasts are present (Grangeon et al., 2012b). This suggests that 6K2 vesicles may be derived from the globular complex, but not from the ER. Furthermore, many viruses, such as Cowpea mosaic virus, GFLV and PVX induce perinuclear ER-derived structures (Carette et al., 2000, 2002; Ju et al., 2005; Ritzenthaler et al., 2002; Tilsner et al., 2012). Brefeldin A (BFA) treatment resulted in normal replication for Melon necrotic spot virus, but inhibitted intercellular movement of the virus (Genovés et al., 2010). However, when BFA was applied to GFLV-infected cells, the GFLV replication was reduced (Ritzenthaler et al., 2002). These two BFA studies suggest that different viruses employ different strategies for forming membranous structures for efficient virus replication. These studies indicate that utilization of the secretory pathway is not unique to animal viruses.
Conclusions and Perspectives
Numerous studies have been conducted to understand the behavior of plant viruses regarding movement and replication by using biochemical, cell biological, genetical, and molecular methods. Particularly, studies based on cell biology contributed greatly to the elucidation of the PD structure, the intra/intercellular trafficking of viral proteins, and interaction between cellular and viral factors. It is highly likely that there are as yet undetermined, complex linkages between virus replication and virus movement. For example, although MPs are dispensable factors for virus replication, they are involved in the production of VRC (Guenoune-Gelbart et al., 2008; Tilsner and Oparka, 2012). Since plant cellular systems are complex, it is worthy to note that different plant viruses react differently to different hosts or host factors. For example, actin filament influences the movement of TMV, but the movement of TVCV is independent of actin filaments (Harries et al., 2009b). Despite tremendous work to understand virus infection, more research is needed to get answers for many unresolved questions. There are many host factors that must be identified to understand the exact mechanisms used by viruses for replication intracellular/intercellular movement. Although Nicotiana benthamiana is the most used plant model to study plant viruses, it has only been recently used because of the lack of genomic resources (Bombarely et al., 2012; Goodin et al., 2008). Since the N. benthamiana genome became available, more precise genetic studies can now be carried out to identify genes that influence virus transport and the replication.
Acknowledgments
This research was supported by Rural Development Administration fund PJ01004202, Republic Korea. We appreciate Ron Walcott (University of Geogia in Athens, GA, USA) and Seoyeon Ju (Seoul Global High School in Seoul, South Korea).