Development of Multiplex PCR for Simultaneous Detection of Citrus Viruses and the Incidence of Citrus Viral Diseases in Late-Maturity Citrus Trees in Jeju Island
Article information
Abstract
Satsuma dwarf virus (SDV) or Citrus mosaic sadwavirus (CiMV) were not consistently detected in RT-PCR assay with the primer sets based on gene of Japan isolates. SDV and CiMV isolates were distinctively divided into two groups based on phylogenetic analysis of PP2 gene cloned from 22 Korean isolates, and the Korean CiMV and SDV isolates shared 95.5–96.2% and 97.1–97.7% sequence identity with Japanese isolate, respectively. We developed PP2-1 primer set based on the PP2 gene sequence of Korean isolates to simultaneously and effectively detect SDV and CiMV. And CTLV-2013 and CTV-po primer sets were newly designed for detection of Citrus tatter leaf virus (CTLV) and Citrus tristeza virus (CTV), respectively. Using these primer sets, a new multiplex PCR assay was developed as a means to simultaneously detect 4 citrus viruses, CTV, CTLV, SDV, and CiMV. The degree of detection by the multiplex PCR were consistent with those of uniplex RT-PCR for detection of each of the viruses. Therefore, the new multiplex PCR provides an efficient method for detecting 4 citrus viruses, which will help diagnose many citrus plants at the same time. We verified that 35.2% and 72.1% of 775 trees in 155 orchards were infected with SDV or CiMV (SDV/CiMV) and CTV by the multiplex-PCR assay, respectively, and CTLV was not detected in any of the trees tested.
Introduction
Citrus is one of the most important fruit crops worldwide. Cultivation of late-maturity citrus cultivars including ‘Setoka,’ ‘Kanpei,’ ‘Ehimekashi No. 28,’ and ‘Shiranuhi’ is increasing, and these cultivars are to becoming economically important in the Korean citrus industry. Most citrus cultivars are grown as grafted plants. Viral pathogens transmitted by grafting as well as insect vectors can cause economic problems. It has been reported that approximately 30 viruses or virus-like agents were found in citrus trees worldwide (Ito et al., 2002; Korkmaz et al., 2000), and 4 viruses, Citrus tristeza virus (CTV), Citrus tatter leaf virus (CTLV), Citrus mosaic sadwavirus (CiMV), and Satsuma dwarf virus (SDV) were found in Korean citrus trees (Hyun and Hwang, 2015).
CTV is a member of the genus Closterovirus, and is a monopartite ssRNA virus of approximately 19.3 kb. Most isolates of CTV cause stem-pitting and reduce tree vigor (Fig. 1A). Various detection methods have been used for CTV including RT-PCR (Kano et al., 1998; Mehta et al., 1997; Sekiya et al., 1991), semi-nested RT-PCR (Yamamoto and Fuji, 2008), and real-time PCR (Bertolini et al., 2008; Saponari et al., 2008). CTLV is the type species of the genus Capillovirus. CTLV is a 600–700 nm long flexuous filamentous particle, with an ssRNA genome consisting of 6,500 nt. It can induce bud union abnormality and stunting (Fig. 1B). Current detection methods include biological indexing on Rusk citrange (Citrus sinensis [L.] Osb. × Poncirus trifoliata), serological diagnosis with enzyme-linked immunosorbent assay (ELISA), and molecular diagnosis with RT-PCR (Miyakawa and Ito, 2000; Shim et al., 2004; Yoshikawa et al., 1996). SDV, the type species of the genus Sadwavirus, has polyhedral particles that are ca. 26 nm in diameter and has two separate positive-strand RNA genome components, RNAs 1 and 2 (Iwanami and Koizumi, 2000). In their 3-terminal regions, RNAs 1 and 2 encode RNA-dependent RNA polymerase (RdRp) and coat protein (CP), respectively (Iwanami, 2010). The affected trees are stunted, develop typically boat-, or spoon-shaped leaves (Fig. 1C). CiMV, Navel orange infectious mottling virus (NIMV), Natsudaidai dwarf virus (NDV), and Hyugagatsu virus (HV) have been described as SDV-related virus (SDV-RV) group (Ito et al., 2004; Iwanami, 2010). Historically, these viruses have been recognized as causal agents of dapples on rinds of satsuma mandarin fruits (CiMV) (Fig. 1D), mottling and curling of new leaves of C. natsudaidai (NDV), chlorotic spots on navel oranges (NIMV), and brown growth rings on Hyuganatsu (HV) (Ito et al., 2004).

Typical symptoms of citrus viral infection. (A) Citrus tristeza virus (CTV) on yuju (Citrus junos). (B) Citrus tatter leaf virus (CTLV) on early satsuma mandarin (Citrus unshiu ‘Miyagawa Wase’). (C) Satsuma dwarf virus (SDV) on ‘Setoka’ ([Citrus unshiu × C. sinensis] × C. reticulate). (D) Citrus mosaic sadwavirus (CiMV) on very early satsuma mandarin (Citrus unshiu ‘Miyamoto Wase’).
ELISA has been the most commonly employed procedure for the detection of citrus viruses because of its sensitivity, efficacy, and relatively low cost (Garnsey and Cambra, 1991). However, many citrus viruses may be difficult to detect using ELISA because of their low titer or uneven distribution. PCR is a powerful technique that is sensitive and highly specific and has been used to detect citrus viruses. Simple PCR detects one target per reaction and requires different PCR reactions to detect multiple targets. Multiplex PCR is a useful technique since different RNA and DNA viruses may infect a single host, such as a citrus tree. This sensitive technique would be very useful in citrus budwood certification programs (Roy et al., 2005). Several multiplex PCR systems have already been developed for the detection of six viroids and one citrus virus (Ito et al., 2002), and seven citrus viruses (Roy et al., 2005).
In this study, we designed a set of three specific primer pairs for the detection of four citrus viruses and developed a multiplex PCR method that enables the simultaneous detection of four citrus viruses. Viral infection was surveyed on late-maturity citrus cultivars using this multiplex PCR technique.
Materials and Methods
Viral isolates
The citrus viruses, SDV, CiMV, CTV, and CTLV were used in this study. Scions were collected from 22 satsuma mandarin and ‘Setoka’ trees, which showed positive reactions by ELISA for SDV in a preliminary assay. The scions were grafted on trifoliate and maintained in a greenhouse. Most of samples were co-infected with 3 or 4 viruses (Table 1).
Nucleic acid extraction and uniplex PCR
Total RNA was extracted from approximately 20 mg of young citrus leaves using TRIzol Reagent (catalog no. 15596-026; Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. The RNA was dissolved in 100 μl FORMAzol solution (Molecular Research Center, Cincinnati, OH, USA) for stabilization of RNA solubilization and stored in −20°C or −70°C. RT-PCR was performed as a two-step procedure, which included RT, using a TOPscript cDNA synthesis kit (catalog no. EZ105S; Enzynomics, Daejeon, Korea) for synthesis of cDNA according to the manufacturer’s protocol. PCR was carried out with primer sets (Table 2) using Accupower Hotstart PCR pemimix (catalog no. K-5051; Bioneer, Daejeon, Korea). Amplification was performed in a programmable thermocycler (model T-Gradient Thermoblock; Biometra, Göttigen, Germany), using an initial denaturation step of 95°C for 2 min, followed by 35 amplification cycles of denaturation at 94°C for 30 s, annealing at 50–55°C for 1 min, and extension at 72°C for 2 min, and a final extension at 72°C for 10 min. After PCR, 12 μl of the product was electrophoresed in a 1.0% agarose gel in 1× TAE buffer for 30 min at 3 V/cm and visualized by ethidium bromide staining. A 100-bp DNA Ladder (catalog no. 3407A; TaKaRa, Tokyo, Japan) was used for molecular weight markers.
Sequence comparison and phylogenetic analyses
To sequence the PP2 gene for the polyprotein of the SDV or CiMV, general procedures for plasmid DNA purification, cloning, and transformation were used, according to each of manufacturer’s recommendations. The amplified DNA fragments were recovered from agarose gels and purified using the AccuPrep Gel Purification Kit (catalog no. K-3035; Bioneer) and then cloned and transformed using the Topo TA Cloning Kit (Invitrogen). Sequencing grade plasmid DNA was obtained using the AccuPrep Nano-Plus Plasmid Mini Extraction Kit (catalog no. K-3111; Bioneer). For each isolate, DNA was sequenced in both directions using M13 forward and SP6 reverse primers for three subclones. Sequencing was done automatically using dyelabeled dideoxy nucleotides and DNA polymerase in an Applied Biosystems 3730XL DNA sequencer (Applied Biosystems, Foster City, CA, USA). The consensus sequence for the three subclones was used for phylogenetic analysis. Sequence alignments were done with the Clustal X 2.0 program (Larkin et al., 2007). The region corresponding to base pairs 2,287 to 5,230 of CiMV genomic RNA, segment RNA 2, complete sequence (Gen-Bank AB465581.1), and that corresponding to base pairs 2,235 to 5,311 of SDV genomic RNA (RNA2) for polyprotein, complete cds (GenBank AB009959.2) were used as reference for sequence analysis.
The nucleotide sequences were aligned using Clustal W and pairwise sequence comparisons, and phylogenetic analysis was performed with MEGA 6 (Tamura et al., 2013). Neighbor-joining trees were constructed according to the maximum-likelihood method in MEGA 6. A heuristic search employing 1,000 random stepwise addition replicates and all gaps and missing data were completely deleted. Bootstrap values were based on 1,000 repetitions.
Multiplex PCR
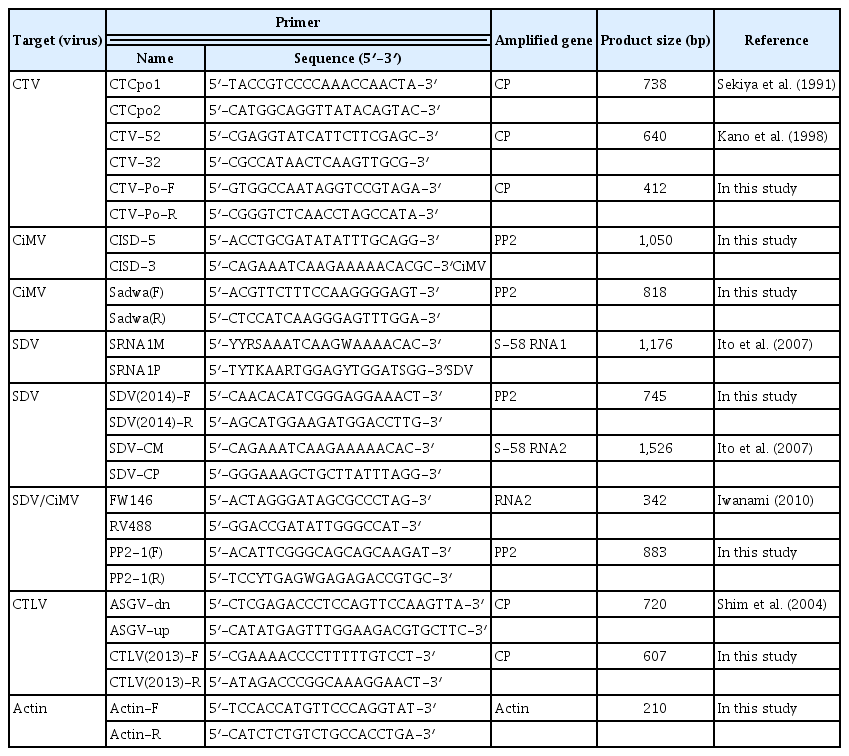
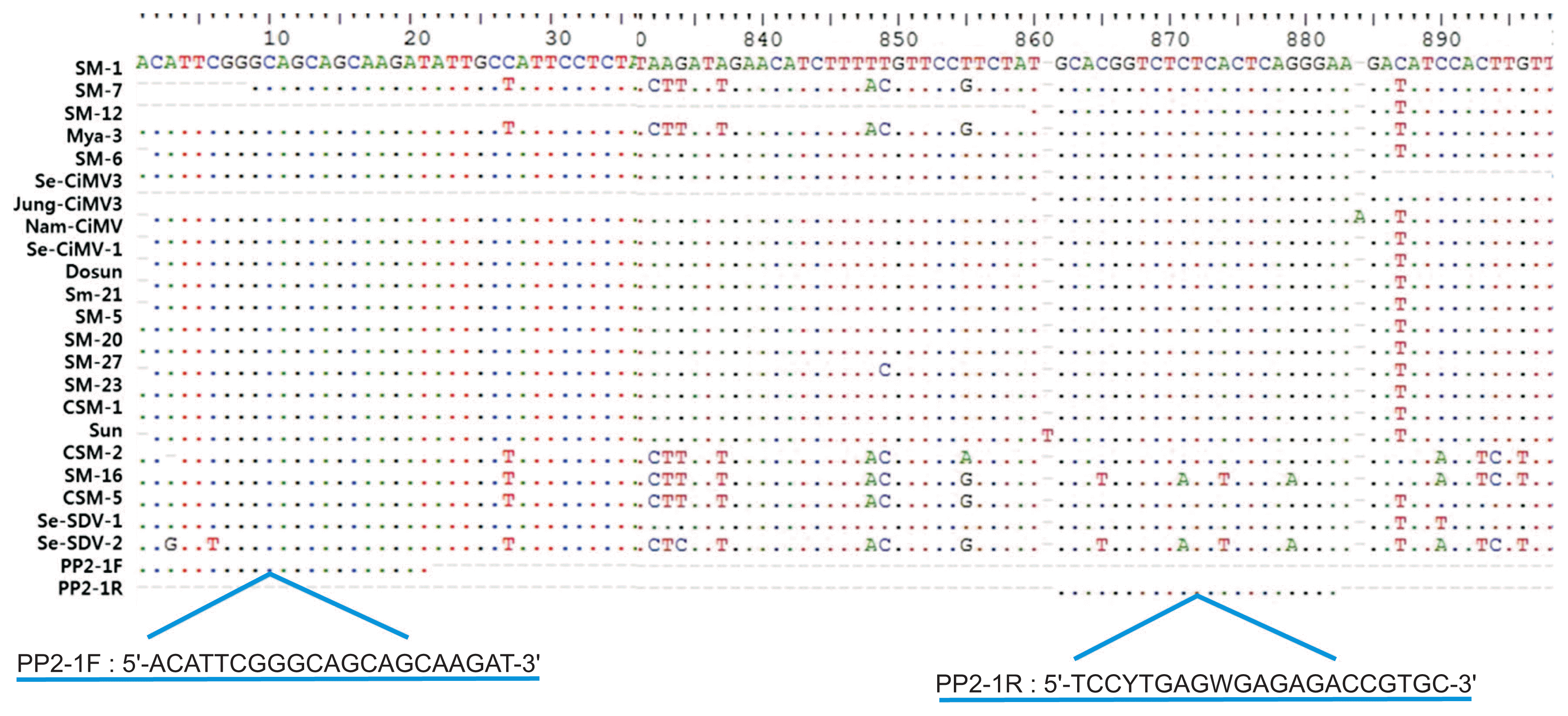
Four primer sets were designed for multiplex PCR using Primer 3 version 0. 4.0 (http://bioinfo.ut.ee/primer3-0.4.0/primer3/); (i) conserved areas within the CP gene for both CTV and CTLV, (ii) the PP2 gene for polyprotein for the SDV and CiMV, and (iii) the actin gene as a positive control. Specific nucleotide regions were selected in the PP2 gene to design the multiplex PCR primers for simultaneous detection of SDV and CiMV (Fig. 2). Multiplex PCR was performed using Accupower Multiplex PCR Premix (catalog no. K-2111; Bioneer). The amplifications were performed in a programmable thermocycler (model T-Gradient Thermoblock), using an initial denaturation step of 95°C for 2 min followed by 35 amplification cycles of denaturation at 94°C for 30 s, annealing at 60°C for 1 min, and extension at 72°C for 2 min, and a final extension at 72°C for 10 min. After PCR, 12 μl of the product was electrophoresed in a 1.0% agarose gel in 1× TAE buffer for 30 min at 3 V/cm and visualized by ethidium bromide staining. A 100-bp DNA Ladder (catalog no. 3407A; TaKaRa) was used as a molecular weight markers.
Sensitivity of the multiplex PCR
We used CSM-1 isolate co-infected with CiMV, CTV and CTLV to assay the detection sensitivity. The cDNA from CSM-1 isolate was diluted 10, 50, 100, 500 and 1,000 times with DEPC-DW, and then multiplex PCR and uniplex PCR were performed by procedure described above to simultaneously detect 3 viruses and each of CiMV, CTV and CTLV, respectively.
Survey on orchards
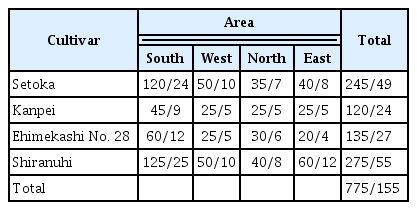
Leaf samples were collected from total of 775 trees in 155 orchards in Jeju Island. Of them, 245 trees in 49 orchards were the ‘Setoka’ cultivar, 120 trees in 24 orchards were the ‘Kanpei’ cultivar, 135 trees in 27 orchards were the ‘Ehimekashi No. 28’ cultivar, and 275 trees in 55 orchards were the ‘Shiranuhi’ cultivar (Table 3). The collected samples were assayed using multiplex PCR for detection of SDV/CiMV, CTLV, and CTV. The RNA extraction and multiplex PCR were performed by procedure described above.
Results
Uniplex PCR
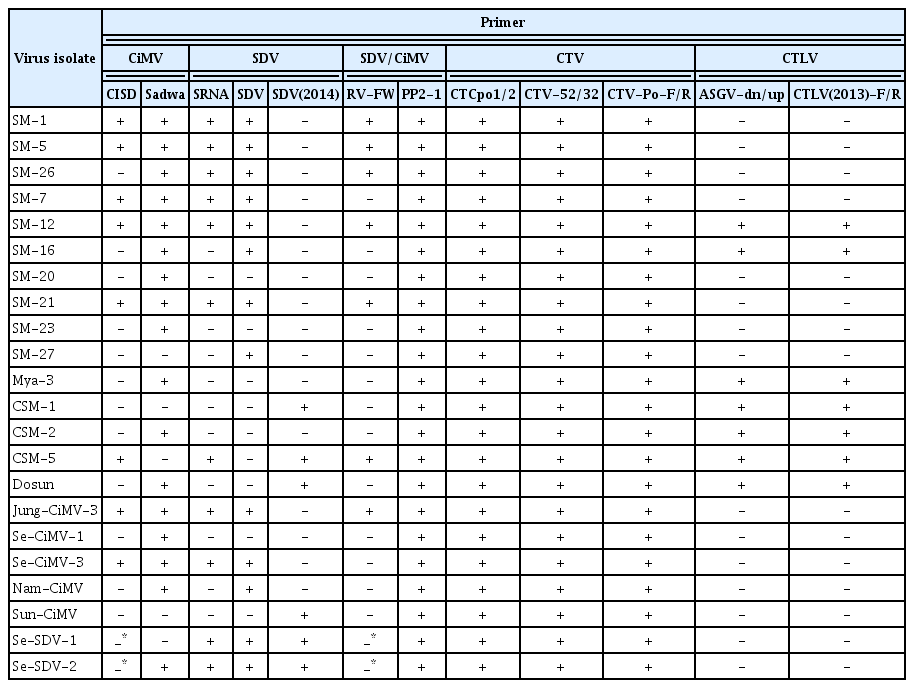
Twenty-two isolates were assayed for CTV, CTLV, SDV, and CiMV using uniplex PCR with the primer sets shown in Table 2. CISD and Sadwa primer sets were used to detect CiMV. SRNA, SDV, and SDV(2014) primer sets were for SDV, and RV-FW primer set was for simultaneous detection of SDV and CiMV. Nine of the 22 isolates were detected with CISD, and 19, 12, 4, 9, and 8 isolates were detected with the Sadwa, SDV, Iwanami, SRNA, and RV-FW primer sets, respectively. SDV/CiMV were detected with PP2-1 primer sets in all isolates tested. CTV was detected in all isolates with the CTCpo1/2 and CTV52/32 primer sets. CTLV was detected in 7 isolates with the ASGV-dn/up primer set (Table 1).
Sequence comparison of SDV and CiMV isolates
All tested isolates were positive for SDV by ELISA in a preliminary assay (data not shown). However, the detection level was different in PCR assay with different primer sets, and SM-20, SM-23, Mya-3, CSM-1, CSM-2, Dosun, Se-CiMV-1, and Sun-CiMV isolate were not detected in uniplex PCR with CISD, SRNA, SDV, and RV-FW primer sets based on gene of Japan isolates (Table 1). Therefore, we partially cloned PP2 gene for polyprotein and used it for sequence comparison of the SDV and CiMV isolates to design more specific and common primer for SDV and CiMV. The cloned genomes consisted of from 2,931 to 3,063 nt. CiMV and SDV isolates were distinctively divided into two groups based on phylogenetic analysis and pairwise sequence comparisons using the sequence of the PP2 gene (Fig. 3). The Korean CiMV isolates except for the CSM-5 isolate had 99.1–100.0% nucleotide sequence identity with one another and shared 95.5–96.2% sequence identity with Japanese isolate. Two Korean isolates of SDV showed 99.3% identity with one another, and 97.1–97.7% sequence identity as compared with Japanese isolate (Table 4).
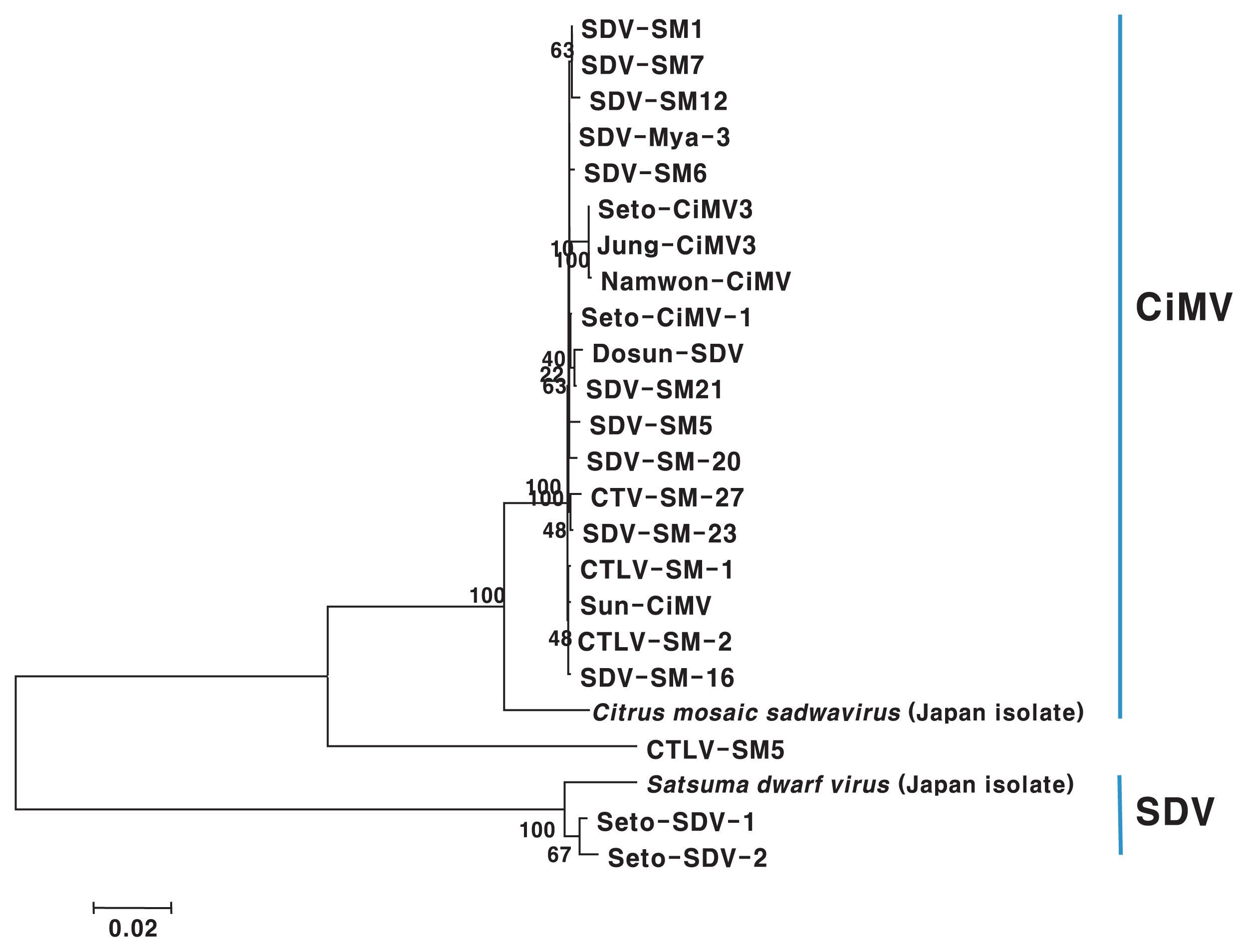
Phylogenetic tree resulting from maximum parsimony analysis of sequences of the PP2 gene. Bootstrap values are based on 1,000 replicates.
Specific primer design and multiplex PCR
We designed specific primers for multiplex PCR to simultaneously detect 4 citrus viruses. The CTV-Po primer set was designed from a gene amplified with CTCpo1/2 primers for CTV, and the CTLV(2013) primer set was used for CTLV (Table 2). PP2-1 primer set was designed from specific nucleotide regions in the PP2 gene for simultaneous detection of SDV and CiMV (Fig. 2). RT-PCR assay using the PP2-1 primer set detected all isolates tested in this study including 19 CiMV isolates and 2 SDV isolates identified by PP2 gene analysis (Table 2). The actin gene was amplified with an actin (F/R) primer set as a positive control. Four amplified products of the expected sizes (412 bp for CTV, 607 bp for CTLV, 835 bp for SDV and CiMV, and 210 bp for actin) were observed in infected samples (Fig. 4). We verified that each of the PCR products were specifically amplified corresponding the gene of the expected virus by sequence analysis (data not shown).
Sensitivity of the multiplex PCR
Detection sensitivity of multiplex PCR was compared with uniplex PCR for each of CiMV, CTV, and CTLV. CiMV and CTLV were enoughly detected until 1:500 dilution of cDNA and CTV was detected 1:50 dilution in multiplex PCR. CiMV and CTLV were also detected until 1:500 dilution and CTV was detected in 1:10 and 1:50 dilution (Fig. 5).
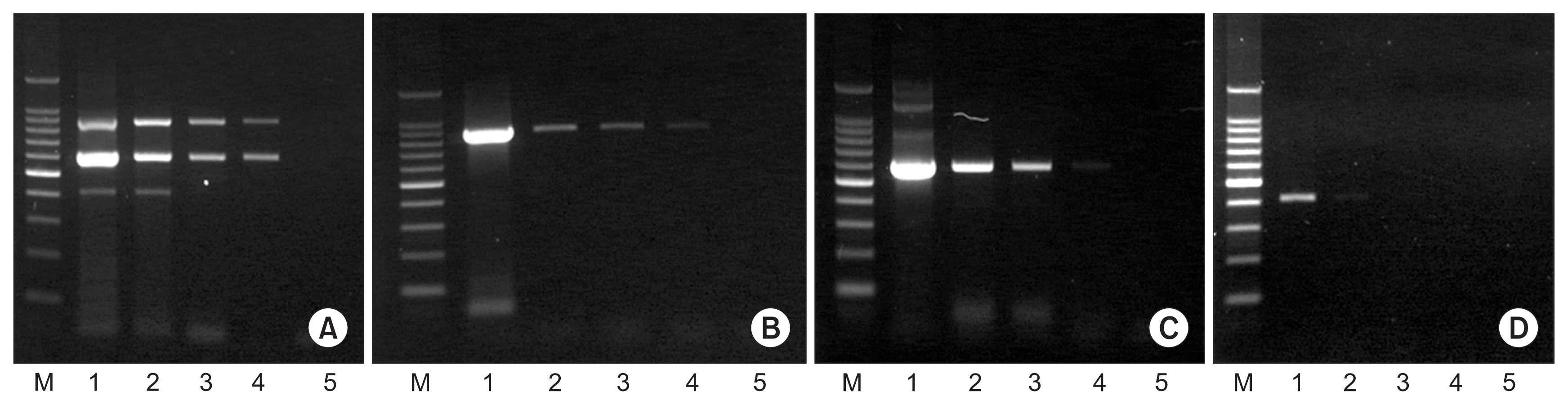
Comparison of detection sensitivity between multiplex PCR and uniplex PCR for each of Citrus mosaic sadwavirus (CiMV), Citrus tristeza virus (CTV), and Citrus tatter leaf virus (CTLV). (A) Multiplex PCR. (B–D) Uniplex PCR for each of CiMV, CTLV, and CTV, respectively. Lane M: 100-bp DNA ladder; lanes 1–5: 10, 50, 100, 500, and 1,000 times dilution of cDNA from CSM-1 isolate.
Survey on orchards
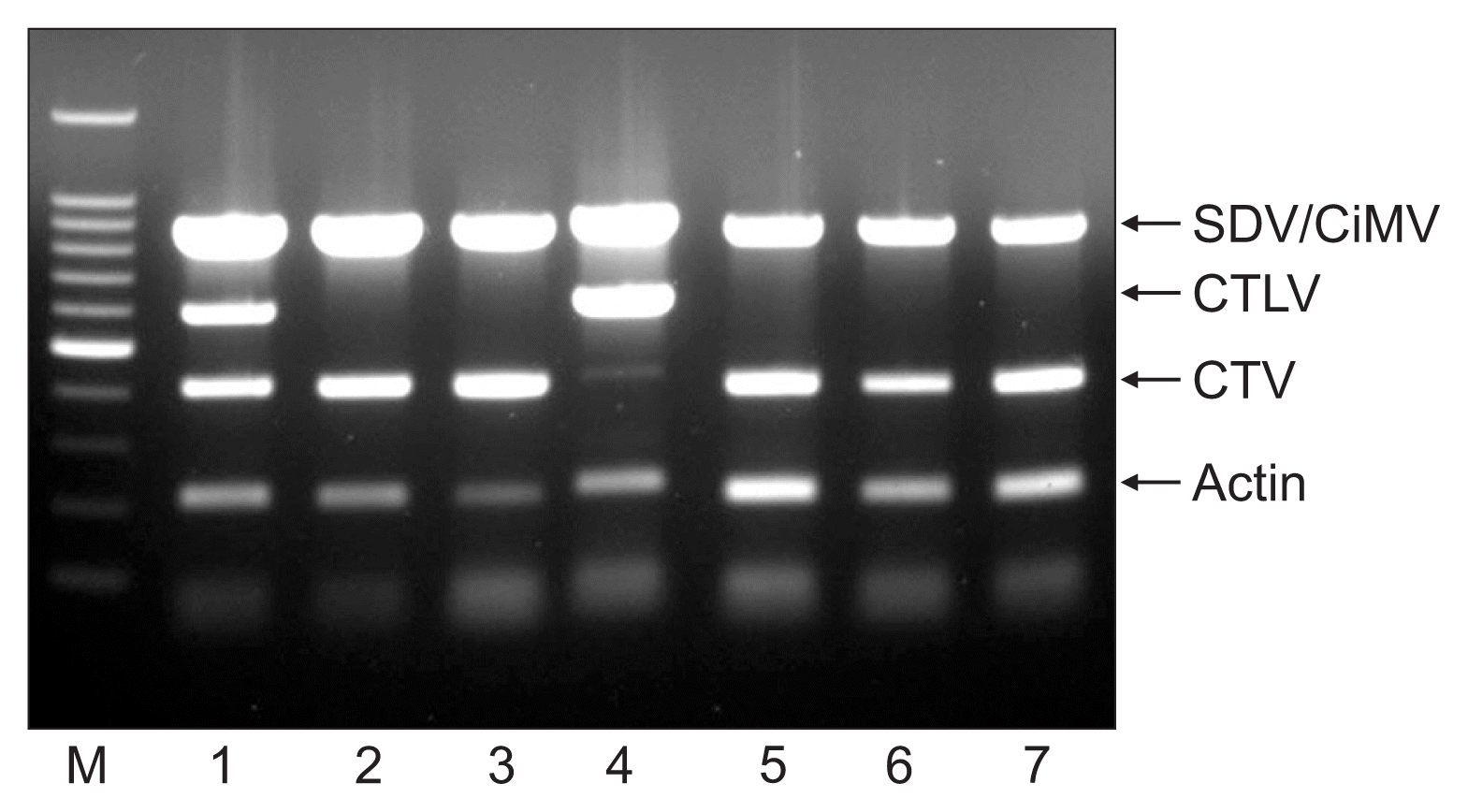
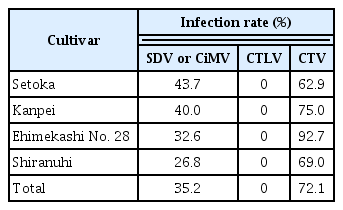
SDV or CiMV, and CTV were detected in 35.2% and 72.1% of 775 trees in 155 orchards using multiplex PCR, respectively. Forty-three point seven percent of 245 ‘Setoka’ cultivar trees in 49 orchards were infected with SDV/CiMV, and 62.9% were infected with CTV. Forty percent and 75.0% of ‘Kanpei’ cultivar trees were infected with SDV/CiMV and CTV, respectively. SDV/CiMV and CTV were found in 32.6% and 92.7% of trees of the ‘Ehimekashi No. 28’ cultivar, respectively. In the ‘Shiranuhi’ cultivar, 26.8% and 69.0% of 275 trees in 55 orchards were infected with SDV/CiMV and CTV (Fig. 6, Table 5). CTLV was not detected in any of the trees tested.
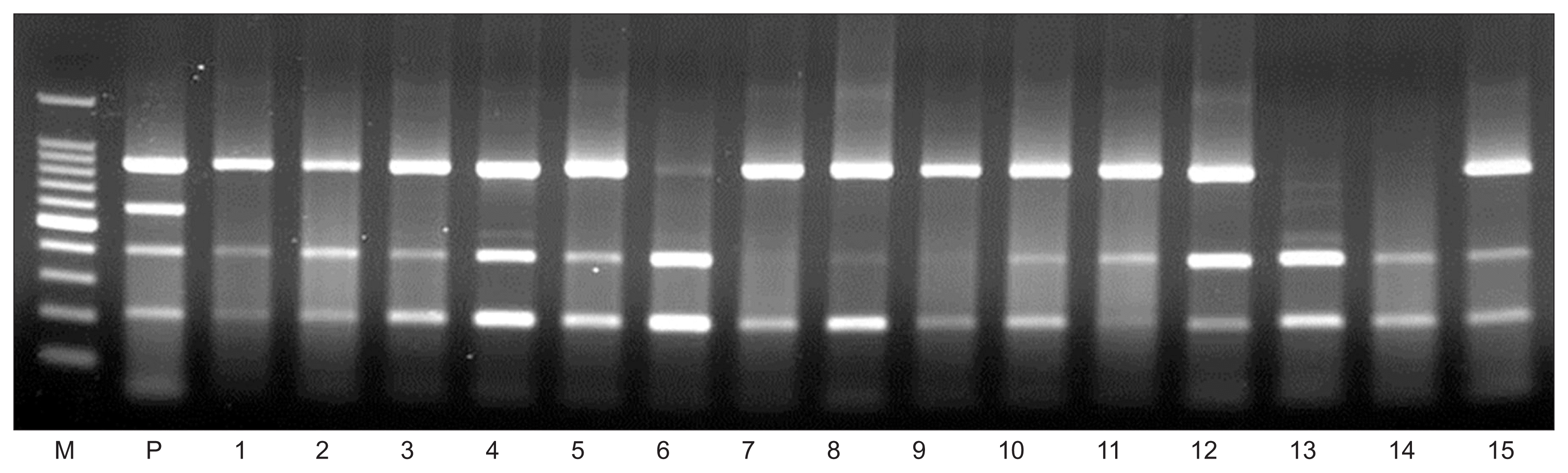
Detection of 4 citrus viruses from ‘Kanpei’ citrus trees in orchards by multiplex PCR assay. Lane M: 100-bp DNA ladder; lane P: positive control; lanes 1–15: isolates from ‘Kanpai’ citrus trees.
Discussion
It was suggested that SDV was distinct from the Comovirus, Fabavirus, and Nepovirus genera, and needed to be separated into a new genus, probably within the family Comoviridae (Iwanami et al., 1999). SDV is currently only one species in the genus Sadwavirus in the family Secoviridae, in the order Picornavirales (International Committee on Taxonomy of Viruses; http://ictvonline.org/virusTaxonomy.asp).
The all virus isolates used in this study showed positive reaction in ELISA using monoclonal antibody of SDV in preliminary assay. Nineteen isolates were identified CiMV and two isolates were SDV by sequence analysis of PP2 gene (Fig. 3). This might be caused by that CiMV was serologically related to SDV. This result showed that most isolates in Jeju Island were CiMV rather than SDV or SDV-related other virus species though co-infections with SDV and CiMV might be there. The sequence analysis of PP2 gene were not consistent to RT-PCR analysis using Sadwa and SDV(2014) primer sets based on gene of Korean as well as Japan isolates (Table 1, Fig. 2). This might be mostly caused by variation in isolates or coinfection. Any primer sets used in this study did not detect all isolates tested. However, the PP2-1 primer set designed from PP2 gene of Korean isolates successfully detected all isolates tested in this study including 19 CiMV and 2 SDV isolates. Therefore, we think the PP2-1 primer set was more effective than already designed primer sets to detect SDV or CiMV isolates. SDV or CiMV were differently detected in RT-PCR assay according to each of all primer sets tested for SDV or CiMV (Table 1). Some isolates didn’t even positively react in any primer sets CISD, SDV, Iwanami, SRNA and RV-FW designed based on gene of Japan isolates (Table 1). This might be caused by variation among isolates, and relatively low similarity between Korean and Japan isolates because the primer sets, CISD, SRNA, SDV and RV-FW were designed based on gene of Japan isolates. The CP of CiMV, NIMV, and HV shared 77–82% amino acid sequence identity with that of SDV (Ito et al., 2004). Nucleotide sequence identity of PP2 gene was 70.4–71.1% between SDV and CiMV isolates in Korea (Table 4). The Korean isolates of CiMV except CSM-5 isolate had 99.1–100.0% nucleotide sequence identity and 95.5–96.2% between Korean isolates and Japan isolate. Two Korean isolates of SDV showed 99.3% identity, and 97.1–97.7% among Korean and Japan isolates. Analysis of 1,139 nucleotide sequences of 3′-terminal RNA 1 gene revealed three CiMV variants that differed by up to 5.4% (Ito et al., 2007).
SDV or CiMV were infected in ‘Setoka’, ‘Kanpei’, ‘Ehimekashi No. 28’ and ‘Shiranuhi’ cultivar of late-maturity citrus by 43.7%, 40.0%, 32.6%, and 26.8%, respectively (Table 5). It have been reported that total fresh weight of the whole plants and fruit yield in citrus tree infected with SDV were decreased by approximately 60% and 25–45%, respectively, as compared with healthy plants (Imada et al., 1980). Therefore, late-maturity citrus cultivars in Jeju might be affected by SDV or CiMV. CTV was highly infected, the infection rates were 62.9–92.7%. However, we did not observe severe symptoms by CTV, and most of CTV isolates were mild strain in Jeju Island in our preliminary ELISA test with antiserum for CTV mild strain (data not shown). By these results, we think CTV is not damage severely to late-maturity citrus cultivars tested in this study though infection rate was very high. Kim et al. (1999) reported CTLV was detected in 9.3% of trees tested and the infection rate was relatively high in very early satsuma mandarin (Citrus unshiu ‘Iwasaki Wase’ etc.) by ELISA. However, CTLV was not detected at all in this survey. These results might be caused by that which the trees showing CTLV symptom were constantly removed and late-maturity citrus cultivars were grafted on early satsuma mandarin rather than very early satsuma mandarin mainly infected with CTLV.
CiMV, NIMV, NDV, and HV have been described as SDV-RV (Ito et al., 2004; Iwanami, 2010). CSM-5 isolate had approximately 85.5% and 69.0% of nucleotide sequence identity of PP2 gene by isolates of CiMV and SDV in Korea, respectively. And there was 79.2% of nucleotide sequence identity between CSM-5 and NDV isolate in Japan. These results suggest that CSM-5 isolate might be other species of SDV-RV.
The all isolates of SDV/CiMV, CTLV and CTV detected by uniplex PCR in tested isolates were equally verified in multiplex PCR assay developed in this study though some isolates were more strongly detected in uniplex PCR than multiplex PCR. Detection limit was not different in multiplex PCR and uniplex PCR assay for each of SDV, CiMV, CTV, and CTLV. Therefore, this multiplex PCR method may be successfully used for simultaneously detecting 4 citrus viruses as well as multiple virus infections in the same plant because it was a simple, reliable, rapid and effective diagnostic method.
Acknowledgments
We are thankful to Golden Seed Project (Center for Horticultural Seed Development, no. 213003-04-4-SBU10), Ministry of Agriculture, Food and Rural Affairs (MAFRA), Ministry of Oceans and Fisheries (MOF), Rural Development Administration (RDA) and Korea Forest Service (KFS).