Occurrence and Epidemics of Bacterial Canker of Kiwifruit in Korea
Article information
Abstract
Bacterial canker is the largest limiting factor in the cultivation and production of kiwifruit worldwide. Typical symptoms comprise necrotic spots on leaves, canker and dieback on canes and trunks, twig wilting, and blossom necrosis. Pseudomonas syringae pv. actinidiae (Psa), which is the causal agent of kiwifruit bacterial canker, is divided into four biovars based on multilocus sequence analysis of different genes, additional PCR testing of pathogenic genes (argKtox cluster, cfl, and various effector genes), and biochemical and physiological characterization. Bacterial canker caused by Psa biovar 2 designated Psa2 was detected for the first time on the green-fleshed kiwifruit cultivar Hayward in 1988 and the yellow-fleshed kiwifruit cultivar Hort16A in 2006 in Korea. Psa biovar 3 designated Psa3, responsible for the current global pandemics of kiwifruit bacterial canker, began to appear in Korea in 2011 and caused tremendous economic losses by destroying many vines or orchards of yellow-fleshed kiwifruit cultivars in one or several growing seasons. Bacterial canker epidemics caused by both Psa2 and Psa3 are prevalent in Korea in recent years. In this review, we summarize the symptomatology, etiology, disease cycle, diagnosis, and epidemiology of kiwifruit bacterial canker in Korea.
Introduction
Kiwifruit is native to China and surrounding Asian countries. There are more than 70 individual species within the Actinidia genus, and most of them grow as rampant vines in forests (Ferguson, 2010). Among commercially cultivated Actinidia species, A. deliciosa and A. chinensis are major species. The most popular cultivars Hayward and Hort16A, which were bred in New Zealand, belong to A. deliciosa and A. chinensis, respectively. The green-fleshed kiwifruit cultivar Hayward was introduced into Korea in 1970s while the yellow-fleshed kiwifruit cultivar Hort16A has been cultivated on Jeju Island since 2004. Several yellow-fleshed kiwifruit cultivars such as Haegeum, Jecygold, and Halla-gold were bred in Korea, and other cultivars were imported from China and New Zealand.
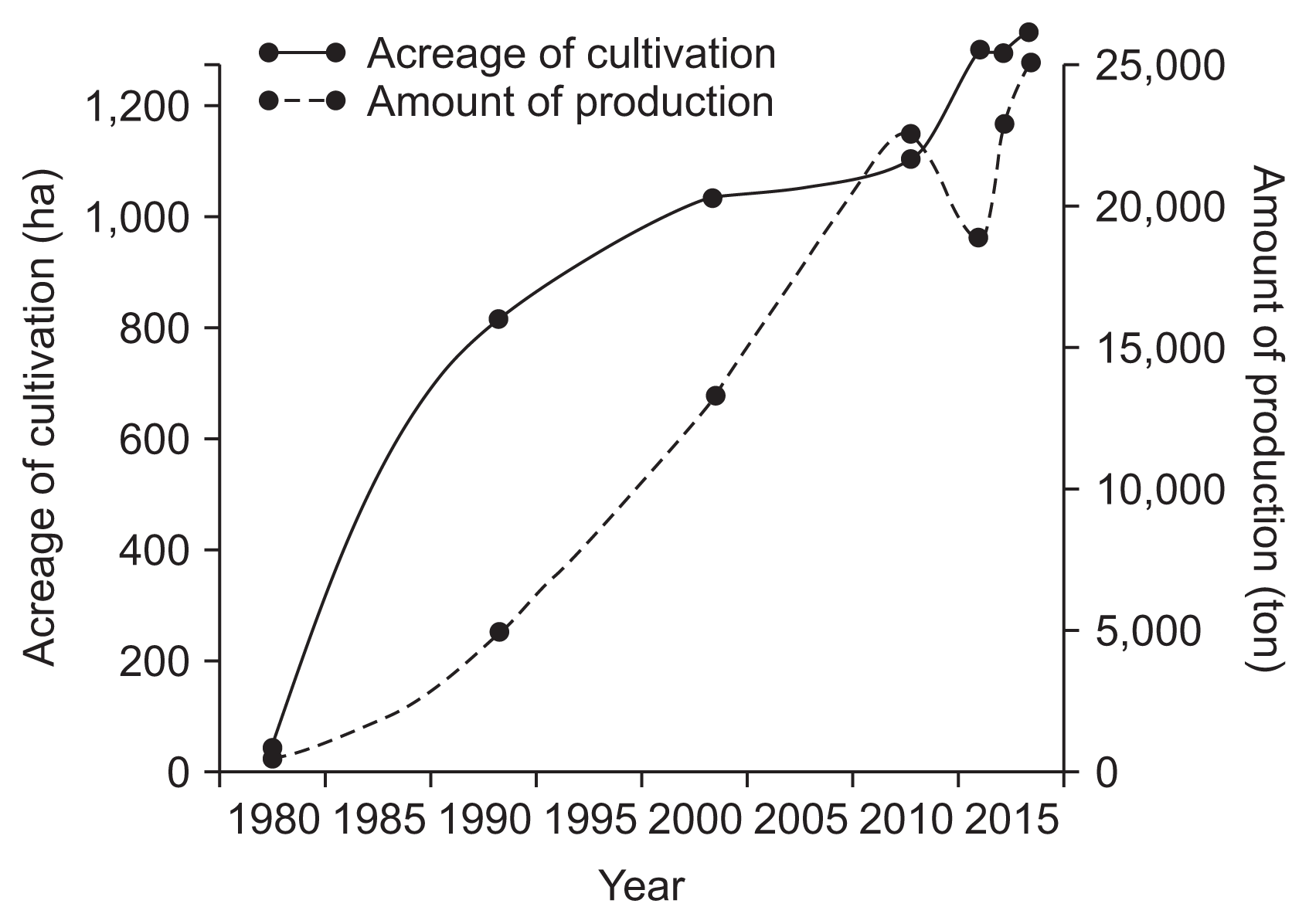
The cultivation and production of kiwifruit are rapidly skyrocketing due to the strong market demand of consumers in recent years (Fig. 1). The cultivation acreage for kiwifruit in 2015 reached up to approximately 1,300 ha, comprising 45%, 31%, and 24% located in Jeonnam, Gyeongnam, and Jeju provinces, respectively. Major cultivation regions with their acreages are shown in Fig. 2. Green-fleshed kiwifruit cultivars occupy 65% of the acreage, followed by 33% for yellow-fleshed kiwifruit cultivars and 2% for red-fleshed kiwifruit cultivars.
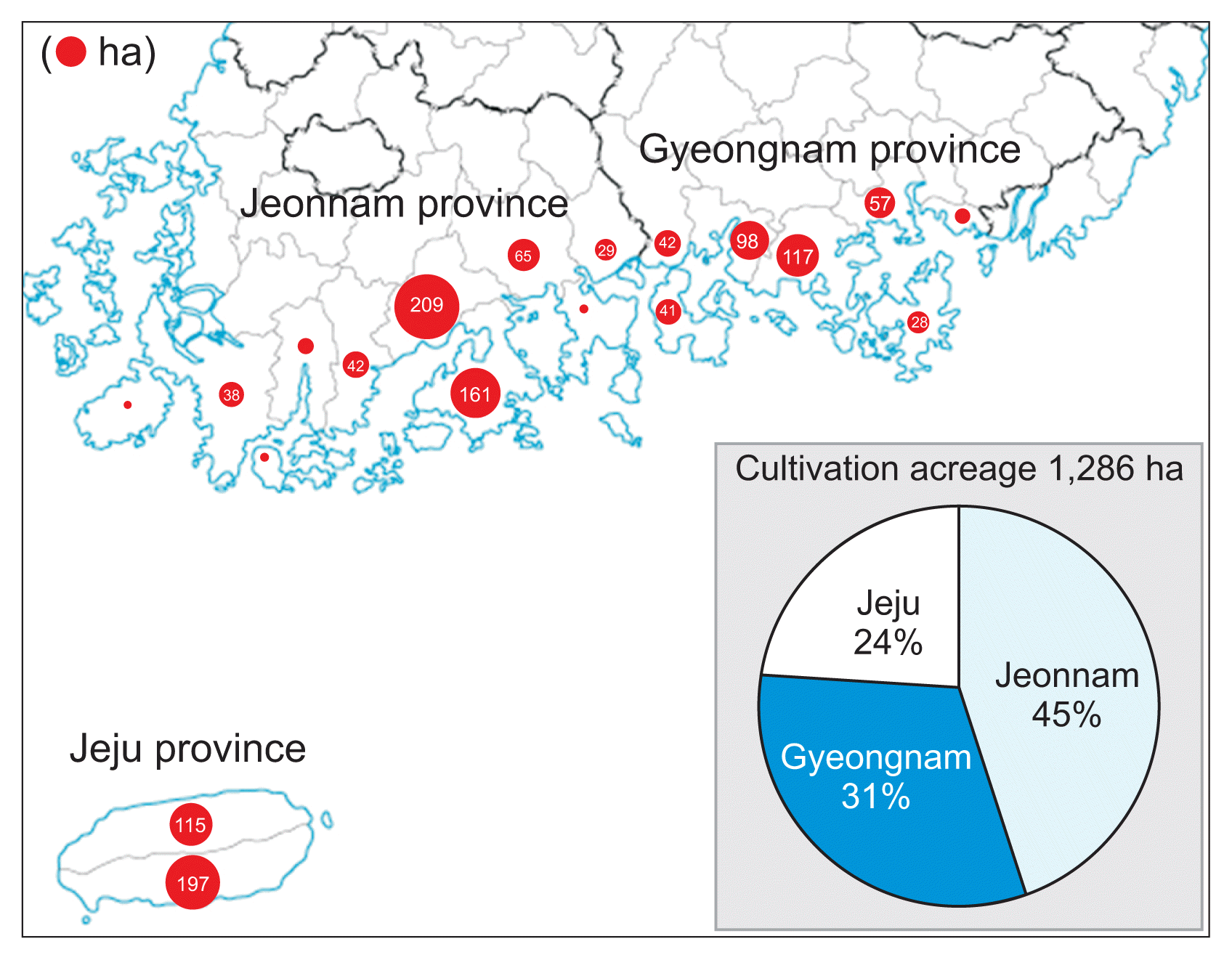
Cultivation acreage of kiwifruit in major kiwifruit-growing regions in Korea. Area of circle represents cultivation acreage in each region in Korea.
Bacterial canker is the largest limiting factor in the cultivation and production of kiwifruit. Kiwifruit bacterial canker was detected for the first time in 1984 on a green-fleshed kiwifruit cultivar Hayward in Japan (Serizawa et al., 1989; Takikawa et al., 1989), and the disease also occurred in Korea (Koh and Lee, 1992; Koh et al., 1994) and China (Wang et al., 1992). Many orchards in Japan and Korea were completely destroyed by bacterial canker epidemics during that time, resulting in huge economic losses to Hayward growers. Around the same time, the disease also occurred in Italy (Scortichini, 1994) but did not have as severe an impact as in Japan and Korea. For this reason, bacterial canker had not received worldwide attention until massive outbreaks on the yellow-fleshed kiwifruit cultivar Hort16A in major kiwifruit-growing countries.
Bacterial canker on the yellow-fleshed kiwifruit cultivar Hort16A occurred for the first time in Korea (Koh et al., 2010) in 2006, followed by Italy (Balestra et al., 2009) in 2008. The disease has been reported in Portugal (Balestra et al., 2010), Spain (Abelleira et al., 2011), France (Vanneste et al., 2011b), and Turkey (Bastas and Karakaya, 2012) in Europe as well as New Zealand (Everett et al., 2011) and Chile (EPPO, 2011) on other continents. Currently, bacterial canker has spread to a pandemic scale worldwide and causes tremendous economic losses by destroying kiwifruit vines or orchards in one or several growing seasons (Scortichini et al., 2012).
Etiology
Pseudomonas syringae pv. actinidiae (Psa) is the causal agent of bacterial canker of kiwifruit. This pathogen, belonging to the Family Pseudomonadaceae and Order Pseudomonadales, was first described in Japan in 1989 by Takikawa et al. (1989). It is a Gram-negative, aerobic, motile, and rod-shaped bacterium with polar flagella, oxidase-negative, arginine dihydrolase-negative, opaque, pale-yellowish, and formed circular colonies on peptone sucrose agar media (Fig. 3).

Morphology of Pseudomonas syringae pv. actinidiae causing bacterial canker. (A) Colony of pathogen on peptone sucrose agar medium incubated at 25°C for 3 days. (B) Electron micrograph of cell showing rod-shaped cell and two polar flagella.
High-throughput sequences have led to deep genome analysis based on molecular markers such as single nucleotide polymorphisms (SNPs) or multilocus sequence analysis (MLSA) of different genes such as housekeeping, type III effectors, and phytotoxins. This has allowed a phylogenetic tree to be drawn based on the alignment on many core genome protein sequences as well as an understanding of the phylogenetic connections between different Psa isolates worldwide (Donati et al., 2014). Based on these analyses, global Psa populations were mainly divided into three lineages (biovars 1, 2, and 3) designated Psa1, Psa2, and Psa3, respectively (Chapman et al., 2012).
The first lineage Psa1, isolated in Japan in 1984 (Takikawa et al., 1989) and Italy in 1992 (Scortichini, 1994), possesses the chromosomal phaseolotoxin gene cluster acquired by horizontal gene transfer (Sawada et al., 1997, 1999, 2002). The second lineage Psa2, distributed only in Korea, possesses the coronatine gene cluster instead of the phaseolotoxin gene cluster as phytotoxins (Han et al., 2003). The third lineage Psa3, responsible for the current bacterial canker pandemics worldwide, is characterized by the lack of genes expressing either phaseolotoxin or coronatine and possesses four putative hop genes encoding effector proteins (Scortichini et al., 2012).
Psa3, responsible for the current bacterial canker pandemics worldwide, began to appear from central Italy since 2008 (Ferrante and Scortichini, 2009). This pathogen has rapidly spread to other kiwifruit-producing regions in Italy and has been reported in other European countries (Scortichini et al., 2012). On other continents, it has also been detected in New Zealand (Everett et al., 2011) and Chile (EPPO, 2011). A coordinated study on core genome SNPs and integrative conjugative elements (ICEs), combined with disease history, led to the hypothesis that the European, New Zealand, Chilean, and Korean strains of Psa3 are of Chinese origin (Butler et al., 2013, 2015; Marcelletti et al., 2011; McCann et al., 2013).
In Korea, the first bacterial canker by Psa2 was observed on Hayward in 1988 (Koh and Lee, 1992) and Hort16A in 2006 (Koh et al., 2010) on Jeju Island. Psa3 was first detected in 2011 on the yellow-fleshed kiwifruit cultivar Yellow-king and red-fleshed kiwifruit cultivar Hongyang in Goheung, Jeonnam province (Koh et al., 2012). Since then, frequent Psa3 incidences have occurred on yellow- and red-fleshed kiwifruit cultivars in major kiwifruit growing areas in Korea. Annual monitoring of Psa population enables estimation of various genetic variations in Psa3 strains currently distributed in Korea (Kim et al., 2106a, 2016b). In addition, there is the possibility of emergence of new biovars through evolutionary recombination between Psa2 existing only in Korea and Psa3 strains introduced from outside.
Recently, the fourth lineage biovar 4 designated Psa4 was detected in New Zealand, Australia, France, and Spain. Psa4 strains are substantially different from those of other biovars; they are less aggressive and cause only necrotic spots on leaves of green- and yellow-fleshed kiwifruit cultivars, whereas others also cause canker and shoot die-back. Therefore, Psa4, which showed no symptoms other than leaf spots, was transferred to the new pathovar actinidifoliorum (Psaf) (Abelleira et al., 2011; Cunty et al., 2015, 2016; Vanneste et al., 2013).
In Japan, Psa3 has been observed on yellow-fleshed kiwifruit cultivars nationwide since 2010, but different strains from other biovars (Psa1–Psa4) were detected in Saga prefecture in 2012. MLSA, additional PCR tests on pathogenic genes (argKtox cluster, cfl, and various effector genes), and biochemical and physiological characterization have shown that the fifth lineage biovar 5 designated Psa5 does not produce both phytotoxins phaseolotoxin and coronatine and is related to Psa2 more than major domestic Psa1 (Fujikawa and Sawada, 2016; Sawada et al., 2014). Although Psa5 has been found only in a limited local area in Japan until now, there is no guarantee it remains endemic in the future.
Disease Cycle
Bacterial canker caused by Psa develops in two phases (Serizawa et al., 1989). In its first phase during autumn and winter, the disease affects the main vine structure and overwintering canes. In its second phase during spring, the disease affects the new seasons’ growth of leaves, flowers, and canes.
Milky bacterial exudates typically flow out from natural openings or pruning injuries on canes or trunks from February in late winter or March in early spring. Bacterial exudates are very effective sources of inoculum dispersed by wind, splashing rain, wind-blown rain, etc. (Serizawa and Ichikawa, 1993d; Serizawa et al., 1989; Son et al., 2016). During spring, Psa enters through stomata, hydathodes, lenticels, wounds, etc. and moves systemically from leaves to young stems or from stems to blossoms through petioles (Ferrante et al., 2012; Serizawa and Ichikawa, 1993a; Son et al., 2016). Systemic displacement of the bacterium within young twigs most likely occurs via the xylem vessels (Spinelli et al., 2011).
High relative humidity and rain reduce the latent period between infection and symptom development of bacterial canker (Serizawa and Ichikawa, 1993d). Furthermore, strong winds and heavy rainfalls favor the disease, since injuries to vines and dispersal of bacterial exudates to wounds and/or natural openings are likely to occur (Serizawa et al., 1989). Psa causes disease at relatively low temperatures, since its optimum growth temperature on new canes is 12–18°C (Serizawa and Ichikawa, 1993b). At temperatures above 20°C and below 15°C, disease severity is reduced. Above 22°C, bacterial growth is abruptly inhibited by the development of wound-healing plant tissue surrounding the infected area, and only a few bacteria survive in diseased tissues during summer (Serizawa et al., 1994). Therefore, infection of vegetative and fruiting canes occurs mainly in spring.
In summer with temperatures increasing above 25°C, degree of infection is drastically reduced, even though the bacterium can colonize the host through the stomata and hydathodes (Serizawa and Ichikawa, 1993c; Spinelli et al., 2011). Psa aestivates in infected trunks or roots or infested soils above 25°C (Son et al., 2016). During autumn, lenticels and buds are the main colonization sites for the pathogen (Serizawa et al., 1994). In autumn or early winter, the bacterium survives in latent form in the cortex tissue of infected branches and spreads throughout the whole vine from late winter to early spring. Recurrence of disease on branches after winter can be prevented by formation of wound tissue surrounding the infected parts. When the temperature drops from 22°C to 20°C, formation of wound tissue gradually decreases, and healing processes are very limited below 15°C.
In late winter or spring, bacterial sap mixes with pigments in bark tissue, creating dark-red ooze that give an appearance of bleeding trunks and leaders. Psa may cause girdling of whole limbs and kill entire vines. Thus, the spread and severity of disease in late winter to early spring depends primarily on the degree of infection of Psa in autumn to early winter (Donati et al., 2014). Psa does not induce ice nucleation (Takikawa et al., 1989), but winter frost and freeze/thawing promote migration of the pathogen within Actinidia twigs and between orchards (Ferrante and Scortichini, 2014). The disease cycle of bacterial canker caused by Psa in Korea is proposed in Fig. 4.
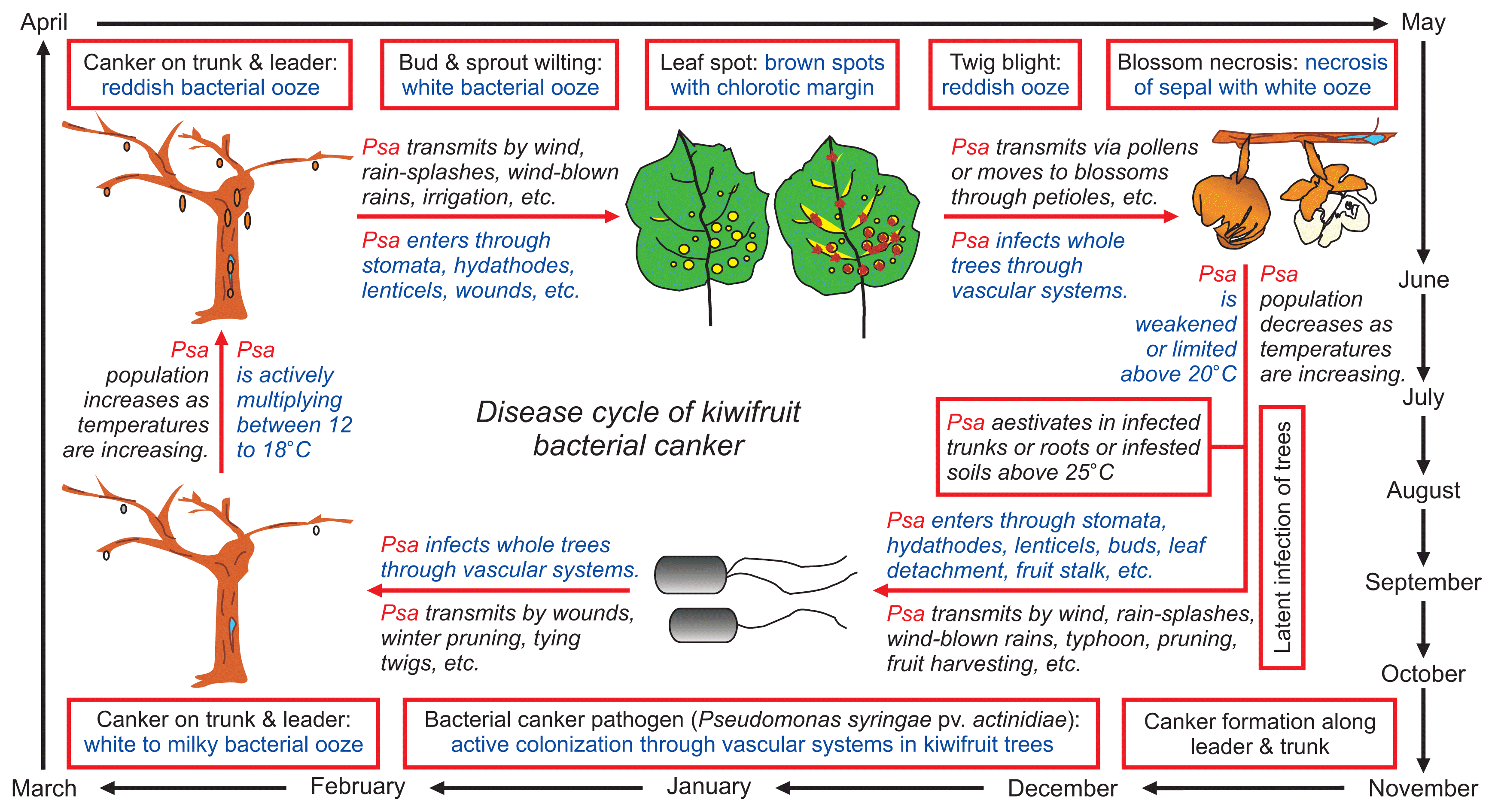
Disease cycle of bacterial canker on green-fleshed kiwifruit cultivar Hayward (Actinidia deliciosa) caused by Pseudomonas syringae pv. actinidiae biovar 2 (Psa2) in Korea.
Psa infects vines through stomata, hydathodes, lenticels, flowers, broken trychomes, leaf and fruit scars, and natural or artificial wounds (Spinelli et al., 2011). Although many aspects of the epiphytic life of Psa have not yet been fully clarified, infected pollens, asymptomatic flowers and leaves, leaf litter, and pruning debris may play important roles as inoculum sources of Psa (Marcelletti et al., 2011; Stefani and Giovanardi, 2011; Tyson et al., 2012; Vanneste et al., 2011a, 2011b).
In Korea, wound-inducing winter pruning practices using contaminated scissors are responsible for the rapid spread of Psa and outbreaks of bacterial canker (Kim et al., 2016a, 2016b; Koh et al., 2010). The role of contaminated fruits as inoculum sources of Psa is still debatable in other countries (Gallelli et al., 2011a; Minardi et al., 2011; Stefani and Giovanardi, 2011). Since Psa aestivates in infected trunks or roots or infested soils above 25°C during summer and autumn, epiphytic or endophytic Psa has never been detected from middle July until the harvest period until now in Korea (Son et al., 2016).
Diagnosis
The development of rapid and reliable diagnostic methods to screen the propagative material of kiwifruit circulating worldwide is a fundamental step to reduce further spread of the pathogen to Psa-free regions (Donati et al., 2014). Traditionally, Psa detection has been based on the isolation of bacteria in pure culture, followed by identification using rep-PCR (Ferrante and Scortichini, 2009) or sequencing of 16S rDNA (Balestra et al., 2009). A PCR-based method for identification of bacterial cultures was shown to be satisfactory for preliminary investigation of Psa, although sometimes non-specific bands and amplification from Pseudomonas syringae pv. theae can be obtained using both techniques (Gallelli et al., 2011b; Koh and Nou, 2002; Rees-George et al., 2010). Recently, other PCR-based techniques were developed in order to overcome these limitations, such as duplex-PCR method (Gallelli et al., 2011b) and nested PCR/RFLP assay (Biondi et al., 2013). A quantitative method based on real-time PCR, which allows quantification of the Psa population inside symptomless plant organs, was also developed (Gallelli et al., 2013).
Psa is highly heterogeneous and divided into biovars 1, 2, 3, and 5 with varying degrees of virulence against kiwifruit (Cunty et al., 2015; Sawada et al., 2014, 2015; Scortichini et al., 2012). Thus, clear distinction is required in the field and quarantine to implement control measures quickly and efficiently. Psa1-specific primers (Sawada et al., 2002), Psa3-specific primers (Ferrante and Scortichini, 2010; Sawada et al., 2015), all Psa-universal primers (Balestra et al., 2013), and Psa5-specific primers (Fujikawa and Sawada, 2016) have been developed.
In Korea, a multiplex PCR assay was developed for the specific detection of Psa3 (Koh et al., 2014). The primer pair designated as TacF and TacR specifically amplified a 545-bp fragment using genomic DNA of Psa3 strains. A multiplex PCR conducted using the TacF/TacR primer pair and universal primer pair for all Psa strains designed by Balestra et al. (2013) can be simultaneously applied for the detection of Psa and for differentiation of Psa3 strains.
Another two sets of PCR primers specific to Psa1 and Psa2 strains, respectively, were developed based on random amplified polymorphic DNA analyses in Korea (Lee et al., 2016). Primers PsaJ-F and PsaJ-R produced a 481-bp region using genomic DNA of Psa1 strains, whereas primers PsaK-F and PsaK-R amplified a 413-bp region present only in the genome of Psa2 strains.
Epidemiology
Psa2 epidemics in Korea
In Korea, bacterial canker caused by Psa2 was detected for the first time on the green-fleshed kiwifruit cultivar Hayward on Jeju Island in 1988 (Koh and Lee, 1992). Since then, bacterial canker disease has rapidly spread to major cultivation regions of kiwifruit. Until 1990, the disease had only occurred in Jeju, Wando, Haenam, and Goheung but thereafter observed in Jindo, Bosseong, Suncheon, and Yeosu from 1991–2000 (Koh, 1995; Koh et al., 1994). Recently, the disease was detected in Jeju and Seogwipo in Jeju province, Jindo, Haenam, Wando, Gangjin, Boseong, Goheung, Suncheon, Yeosu, and Gwangyang in Jeonnam province, and Namhae in Gyeonnam province (Fig. 5).
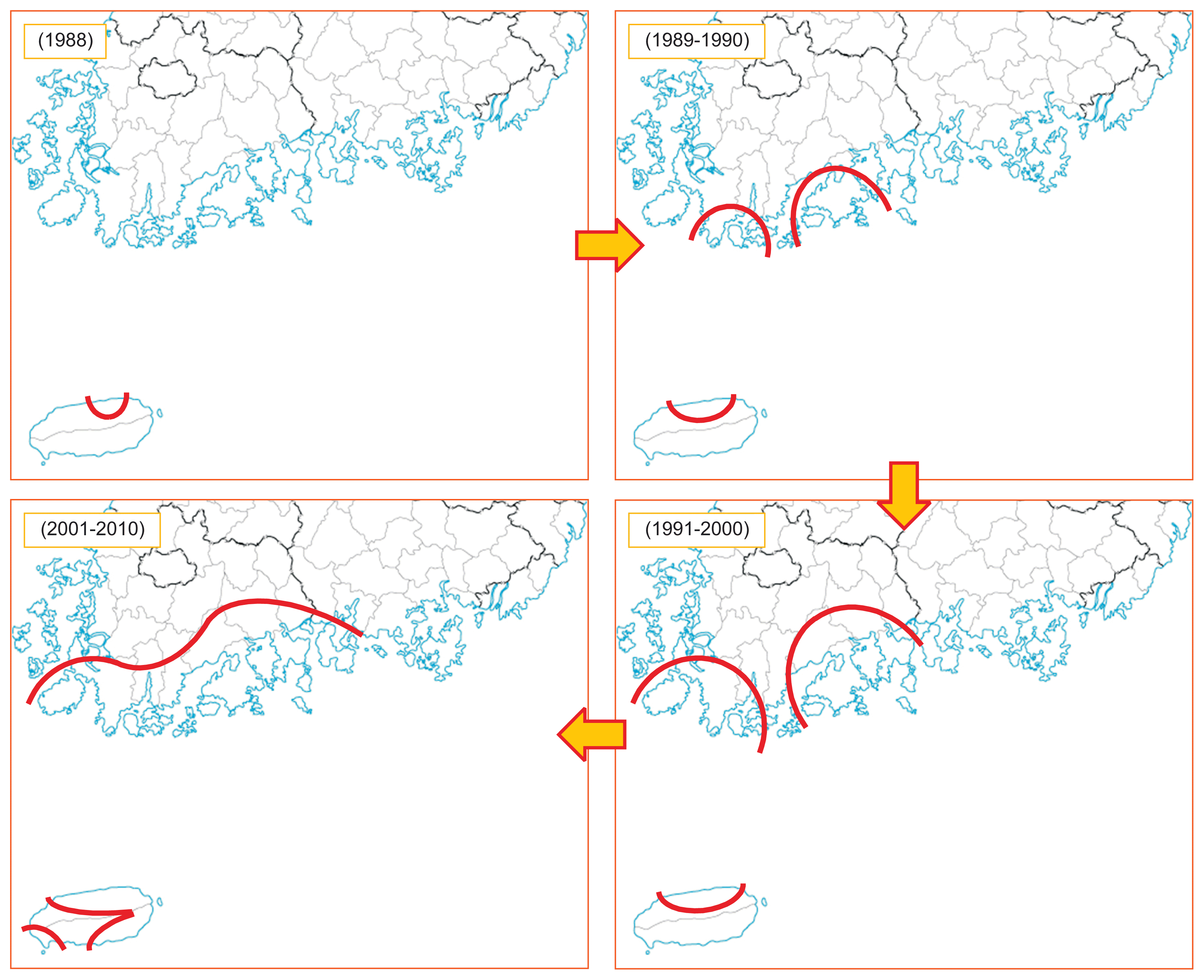
Distribution regions of kiwifruit orchards infected by Pseudomonas syringae pv. actinidiae biovar 2 (Psa2) from 1988–2010 in Korea.
Psa2 epidemics were detected on the yellow-fleshed kiwifruit cultivar Hort16A in 2006, which was 2 years after the yellow-fleshed kiwifruit cultivar Hort16A was introduced to Jeju Island in 2004. The disease spread more quickly in Hort16A than Hayward and completely destroyed whole orchards on Jeju Island in one or several growing seasons (Koh et al., 2010). Over the past three decades, almost all the eradicated orchards were located in high mountain or seaside areas (Ko et al., 2000a, 2000b). Contaminated pruning shears and cold winter climatic conditions appear to have been responsible for the sudden outbreak and rapid spread of the epidemics.
Interestingly, there have been no Psa2 epidemics in Gyeonnam province except Namhae in 2015. Since Gyeonnam province is located in the southeast of the Korean peninsula, Gyeonnam province is warmer than Jeonnam province during winter. This suggests that warm winter climatic conditions are unfavorable to epidemics in Gyeonnam province.
Psa3 epidemics in Korea
Psa3 was first discovered on the yellow-fleshed kiwifruit cultivar Yellow-king and the red-fleshed kiwifruit cultivar Hongyang cultivated in Goheung, Jeonnam province in 2011 (Koh et al., 2012). Psa3-contaminated Yellow-king and Hongyang seedlings imported from China in 2006 were the first inoculum source of Psa3 epidemics in Goheung (Kim et al., 2016b). Genome analysis of Psa3 strains isolated from Yellow-king revealed that Psa3 strains are very closely related to those collected in China, New Zealand, Italy, and Chile (Butler et al., 2015). Similarly, Psa3 strains causing global pandemic outbreaks of bacterial canker of kiwifruit belong to the same genetic lineage originating from China (Mazzaglia et al., 2012).
In 2014, sudden Psa3 epidemics occurred on fruit-bearing branches and leaves of yellow-flesh kiwifruit cultivars such as Hort16A and red-fleshed kiwifruit cultivars such as Hongyang on Jeju Island, followed by Jeonnam and Gyeongnam provinces in 2015 and 2016 (Kim et al., 2016b). The imported pollens contaminated with Psa3 were the second inoculum source of Psa3 epidemics on Jeju Island (Kim et al., 2016b). Previous studies in other countries also support the possibility of Psa3-contaminated pollens being an inoculum source for canker (Gallelli et al., 2011a; Kim et al., 2016a; Stefani and Giovanardi, 2012; Vanneste et al., 2011a). Overall, Psa2 has been present in Korea since 1988, but Psa3 is very likely introduced to Korea through Psa3-infected seedlings imported from China in 2006 and Psa3-contaminated pollen imported from New Zealand and China around 2014. The possible domestic flow route of Psa3 is shown in Fig. 6. Bacterial canker caused by Psa3 became a pandemic in 2015 in Korea (Kim et al., 2016b). Among 202 diseased orchards, Psa2 and Psa3 were detected in 73 and 129 kiwifruit orchards, respectively. The number of kiwifruit orchards infected by Psa3 in 2016 increased nearly two times compared to that in 2015 (Fig. 7).

Possible domestic flow route of Psa3 strains of Pseudomonas syringae pv. actinidiae responsible for recent bacterial canker epidemics in Korea.

Distribution sites of kiwifruit orchards infected by Pseudomonas syringae pv. actinidiae biovar 2 (Psa2) and biovar 3 (Psa3) for the survey period of 2013–2016 in Korea.
Kiwifruit canker occurs not only on commercially grown kiwifruits but also on kiwifruit species selected from the wild and those grown in orchards (A. arguta, A. kolomikta, etc.) (McCann et al., 2013; Scortichini et al., 2012). Psa3 has infected most yellow-, red-, and green-fleshed kiwifruits that have been cultivated in Korea. More specifically, Psa3 infected not only the first reported kiwifruit cultivars Yellow-king and Hongyang but also other yellow- and red-fleshed kiwifruit cultivars such as Hort16A, Jecy-gold, Enza-gold, Enza-red, Haegeum, and Halla-gold as well as green-fleshed kiwifruit cultivars such as Hayward and Daeheung. In addition, Psa3 also infected the kiwiberry cultivar Skinny-green (Kim et al., 2016a, 2016b).
All plant parts can be affected by Psa3, which induces necrotic spots on leaves and canker as well as dieback symptoms on canes and trunks of yellow- and red-fleshed kiwifruit cultivars as opposed to leaf spots on green-fleshed kiwifruit cultivars (Kim et al., 2016a). Nevertheless, disease intensity differs between kiwifruit cultivars. In fact, Hayward is known to be more resistant to Psa3 infection than Hort16A (Vanneste et al., 2013). Further, Psa2 infection was shown to be relatively more prevalent on kiwifruit cultivars such as Hayward, Hort16A, and Hongyang than others, which is distinguishes Psa2 from Psa3.
Especially, sudden outbreaks of Psa3 epidemics were observed on the yellow-fleshed kiwifruit cultivar Jecygold in Gyeongnnam province in 2015 and 2016. Epidemiological and random amplification of polymorphic DNA analyses in Gyeongnam province strongly support the domestic flow route of Psa3 through imported pollens (Kim et al., 2016a). The first incidence of Psa3 on Hongyang in Sacheon, Gyeongnam province might be attributed to introduction of Psa3-contaminated pollens from China used for artificial pollination in 2014. Therefore, outbreaks of Psa3 in 2015 and 2016 might be due to rapid spread of bacterial canker by secondary infection of Psa3 from Hongyang orchards to neighboring Jecy-gold and Hayward orchards in Sacheon and Goseong, Gyeongnam province. Unusually, frequent rainfalls with strong winds in spring favored rapid spread of bacterial canker by secondary infection of Psa3 in Gyeongnam province in 2016.
As Psa3 was specified as a quarantine pathogen in 2014, import of infected kiwifruit seedlings or contaminated pollens with Psa3 has been prohibited in order to prevent additional introduction of Psa3 into Korea. In addition, developing technologies for healthy pollen production is essential to block the spread of canker, since it is now possible that pollen collected domestically might be infected with Psa3.
Concluding Remarks
Bacterial canker of kiwifruit caused by Psa2 has been prevalent since 1988 in Korea. Although some efforts have been made to control epidemics, sufficient research has not been conducted since Psa3 outbreaks in 2011 in Korea. Global outbreaks of Psa3 in 2010 forced researchers to study Psa3 in order to understand the kiwifurit-Psa pathosystem. Several aspects related to the detection, genomics, identification, and characterization of Psa pathovars have been examined in recent years. Deeper understanding of the pathogen and environmental conditions associated with Psa epidemics is still needed for the development of an effective strategy to reduce risk of Psa outbreaks worldwide.
It is necessary to monitor the dynamics of the Psa population in Korea in order to develop a systematic control strategy. When diagnosis of Psa strains in samples such as pollen or other plant organs is required, plant quarantine inspectors or researchers will be able to perform PCR by using PCR detection primer sets (Koh et al., 2014; Lee et al., 2016). PCR detection methods may serve as a useful tool for identification of Psa biovars and will be helpful in monitoring migration of Psa strains for development of canker-resistant cultivars of kiwifruit and a national early warning and surveillance system for canker epidemics.
Phytotoxin production ability is important to the differentiation of Psa biovars (Chapman et al., 2012). However, classification of Psa biovars needs to be reexamined in the future, as certain strains of Psa1 in Japan do not contain the argK-tox cluster (Fujikawa and Sawada, 2016). It is assumed that Psa keeps continuously evolving in order to adapt to the diverse kiwifruit cultivar-Psa pathosystem. Therefore, new virulent biovars or subgroups within distributed biovars Psa2 and Psa3 in Korea might appear eventually.
Acknowledgments
This work was supported by the Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, Forestry and Fisheries (IPET) through the Agri-Bio Industry Technology Development Program, funded by Ministry of Agriculture, Food, and Rural Affairs (MAFRA) (315019-2).