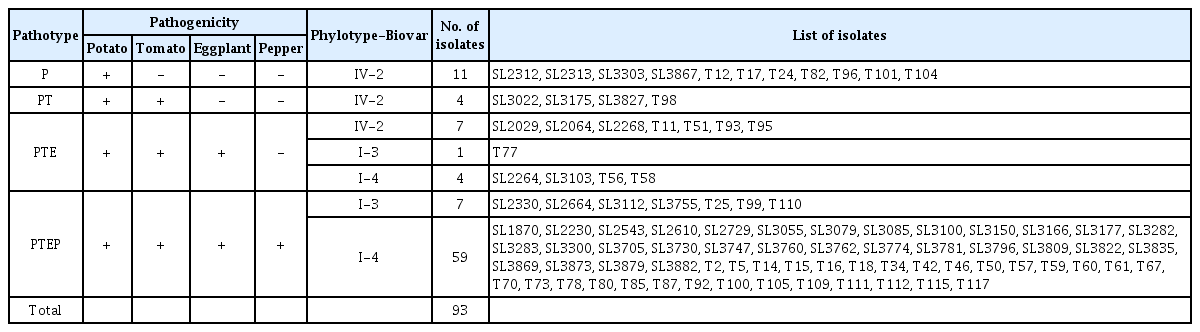
Analysis of Genetic and Pathogenic Diversity of Ralstonia solanacearum Causing Potato Bacterial Wilt in Korea
Article information
Abstract
The Ralstonia solanacearum species complex (RSSC) can be divided into four phylotypes, and includes phenotypically diverse bacterial strains that cause bacterial wilt on various host plants. This study used 93 RSSC isolates responsible for potato bacterial wilt in Korea, and investigated their phylogenetic relatedness based on the analysis of phylotype, biovar, and host range. Of the 93 isolates, twenty-two were identified as biovar 2, eight as biovar 3, and sixty-three as biovar 4. Applied to the phylotype scheme, biovar 3 and 4 isolates belonged to phylotype I, and biovar 2 isolates belonged to phylotype IV. This classification was consistent with phylogenetic trees based on 16S rRNA and egl gene sequences, in which biovar 3 and 4 isolates clustered to phylotype I, and biovar 2 isolates clustered to phylotype IV. Korean biovar 2 isolates were distinct from biovar 3 and 4 isolates pathologically as well as genetically - all biovar 2 isolates were nonpathogenic to peppers. Additionally, in host-determining assays, we found uncommon strains among biovar 2 of phylotype IV, which were the tomato-nonpathogenic strains. Since tomatoes are known to be highly susceptible to RSSC, to the best of our knowledge this is the first report of tomato-nonpathogenic potato strains. These results imply the potential prevalence of greater RSSC diversity in terms of host range than would be predicted based on phylogenetic analysis.
Introduction
Ralstonia solanacearum is a causal agent of bacterial wilt disease, and is one of the most destructive phytopathogenic bacteria worldwide (Hayward, 1991). A soil-borne pathogen, R. solanacearum infects host plants through wounds and natural openings, colonizes and blocks water in the xylem, and finally causes wilting and death of the host plant (Denny, 2006). When this bacterium infects potatoes, it causes brown rot on the tubers and aboveground symptoms including wilting, stunting, and yellowing of leaves (Kelman, 1954; Martin and French, 1985).
The bacteria have an unusually broad host range of over 450 plant species, encompassing monocots and dicots (Hayward, 1991; Wicker et al., 2007). Phylogenetic analysis of R. solanacearum has revealed great diversity, and this group is known as the R. solanacearum species complex (RSSC) (Elphinstone, 2005; Genin and Denny, 2012). Previously, R. solanacearum has been classified into “races” based on host range (Buddenhagen et al., 1962; Hayward, 1964; He et al., 1983; Pegg and Moffett, 1971) and “biovars” based on carbohydrate utilization (Hayward, 1964, 1991). However, it has been difficult to define the correlation between races and biovars, with the exception of race 3/biovar 2. Recently, the RSSC has been divided into four phylogenetic groups (“phylotypes”) based on sequence analysis of the internal transcribed spacer (ITS) region of the 16S–23S rRNA gene (Poussier et al., 2000; Prior and Fegan, 2005). This scheme corresponds to geographic origin: phylotype I (Asia), phylotype II (America), phylotype III (Africa), and phylotype IV (Indonesia) (Fegan and Prior, 2005; Prior and Fegan, 2005). The scheme is also broadly consistent with various genetic typing analyses. The RSSC has recently undergone reclassification: R. solanacearum of phylotype II was reclassified as true R. solanacearum, phylotype I and III as R. pseudosolanacearum, R. syzygii of phylotype IV as R. syzygii supsp. syzygii, R. solanacearum of phylotype IV as R. syzygii supsp. indonesiensis, and the blood disease bacterium (BDB) of phylotype IV as R. syzygii supsp. celebesensis (Safni et al., 2014).
Korean agriculture has been severely affected by bacterial wilt. This disease has been observed not only in many economically important solanaceous crops, such as potato, tomato, and pepper plants, but also in paprika, sesame, peanut, sunflower, etc (Jeong et al., 2007; Lee and Kang, 2013; Lim et al., 2008; Seo et al., 2007; Yun et al., 2004). Therefore, there have been great efforts to overcome this disease by breeding resistant varieties or detecting pathogenic bacteria using PCR-based methods (Cho et al., 2011; Han et al., 2009; Jung et al., 2014; Kang et al., 2007; Kim et al., 2016; Lee et al., 2011).
In the present study, we collected bacteria from plants affected by potato bacterial wilt in Korea, conducted various genetic analyses, and determined host range. Our results demonstrate a relationship between genetic and pathogenic traits, and form the basis for comparative genomic analyses of the RSSC.
Materials and Methods
Collection of isolates and culture conditions
Ralstonia solanacearum isolates were collected by Dr. Young Kee Lee (T-numbered strains) and Dr. Seungdon Lee (SL-numbered strains) from 1998 to 2003 in Korea. Among the twenty-five locations surveyed, potato bacterial wilt was observed in twelve cities in six Korean provinces (Fig. 1). We analyzed ninety-three isolates, which are listed in Table 1. All isolates were identified to R. solanacearum by 16S rRNA sequence analysis. These bacteria were cultured on tetrazolium chloride (TZC) agar medium (peptone 10 g, glucose 2.5 g, casamino acid 1 g, agar 18 g, TZC 50 mg in 1 L distilled water) at 28°C for 48 h and frozen in 40% glycerol stock at −70°C (Kelman, 1954).
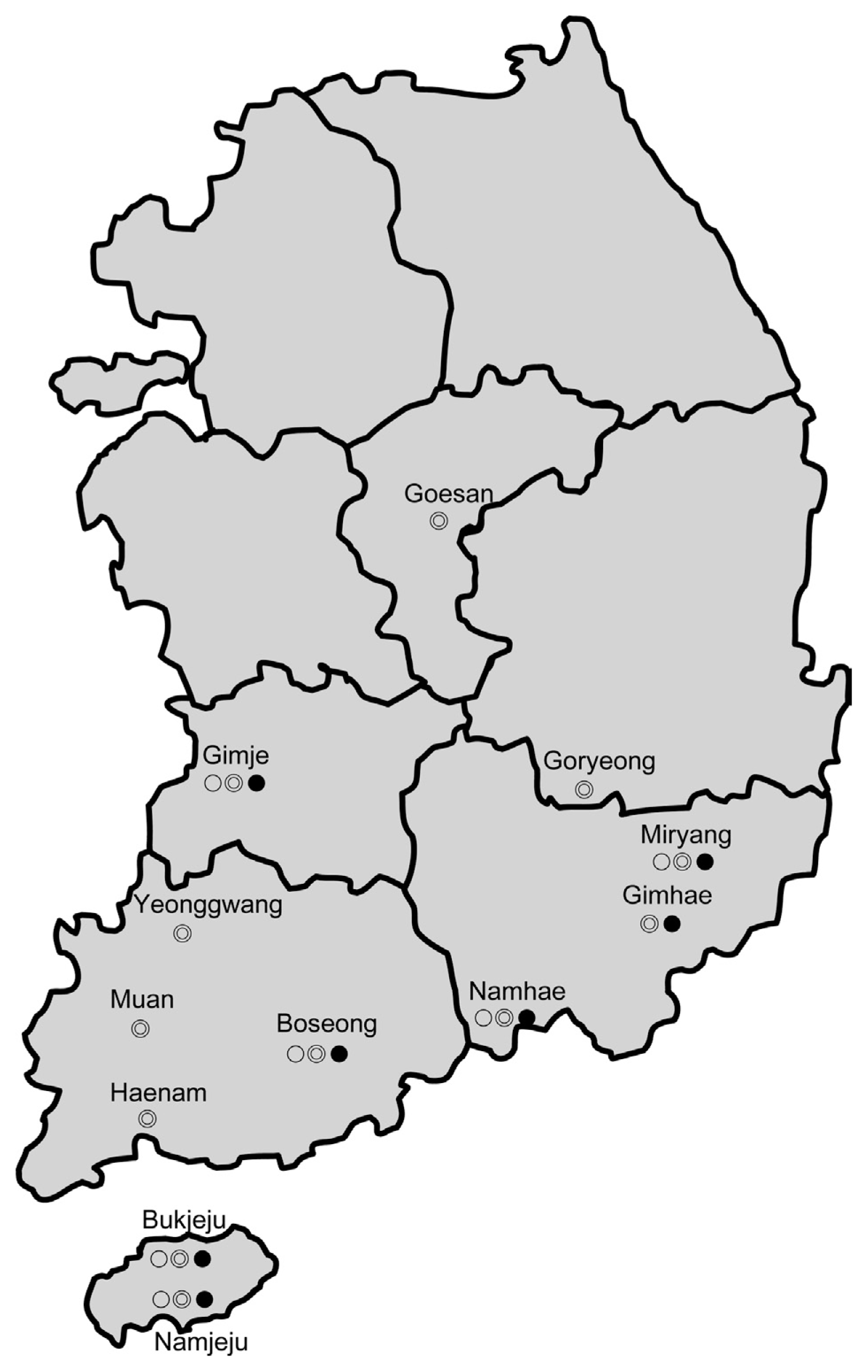
Potato-growing areas and geographic locations infected with potato bacterial wilt in Korea from 1998–2003. Potato-growing areas are colored gray, and symbols indicate isolates: ○, phylotype I-biovar 3; ⦾, phylotype I-biovar 4; ●, phylotype IV-biovar 2. Each symbol indicates representative locations for the same geographical origin, phylotype, and biovar.
Isolation of genomic DNA
To prepare genomic DNA, bacterial cells grown on TZC agar medium were subcultured in LB broth (peptone 10 g, yeast extract 5 g, sodium chloride 5 g in 1 l distilled water) in a 28°C shaking incubator for 16 h. Genomic DNA of all isolates was extracted using the Wizard Genomic DNA Purification Kit (Promega, Madison, WI, USA) according to the manufacturer’s instructions.
Phylotype identification
The ninety-three isolates were classified into phylotypes as previously described (Prior and Fegan, 2005; Sagar et al., 2014). We determined phylotype using the method of Sagar (2014) based on phylotype-specific multiplex PCR (Pmx-PCR) using the following primers: Nmult:21:1F, 5′-CGTTGATGAGGCGCGCAATTT-3′ (specific for phylotype I, amplicon size is 144 bp when paired with Nmult22:RR); Nmult:21:2F, 5′-AAGTTATGGACGGTGGAAGTC-3′ (phylotype II, 372 bp); Nmult:23:AF, 5′-ATTACGAGAGCAATCGAAAGATT-3′ (phylotype III, 91 bp); Nmult:22:InF, 5′-ATTGCCAAGACGAGAGAAGTA-3′ (phylotype IV, 213 bp), and Nmult:22:RR, 5′-TCGCTTGACCCTATAACGAGTA-3′ (reverse primer for all phylotypes). PCR reactions were carried out in a total volume of 20 μl with Profi-Premix (Bioneer) with primer sets and genomic DNA in an automated thermocycler (model PTC-200, MJ Research Inc., Waltham, MA, USA) as follows: initial denaturation at 96°C for 5 min, followed by 30 cycles of denaturation at 95°C for 15 sec, annealing at 59°C for 15 sec, and extension at 72°C for 30 sec, with a final extension at 72°C for 10 min. Seven microliters of each PCR product was examined by electrophoresis through 1% agarose gel, stained with ethidium bromide, and visualized on a UV-trans-illuminator.
Biovar determination
The ability of each isolate to oxidize three disaccharides (maltose, lactose, and cellobiose) and three hexose alcohols (mannitol, sorbitol, and dulcitol) was evaluated by inoculating the isolates on biovar plates following a modified Hayward method (Hayward, 1964). Along with the isolates, 1 ml of the basal medium (NH4H2PO4 1 g, KCl 0.2 g, MgSO4·7H2O 0.2 g, peptone 1 g, bromothymol blue 8 mg, agar 1.5 g in distilled water 1 l, pH 7.1) containing 1% sterilized carbon sources (maltose, lactose, cellobiose, mannitol, sorbitol, and dulcitol) was dispensed into the wells of 24-well plates (SPL Life Sciences, Seoul, Korea). All isolates were inoculated into individual wells with 5 μl of the 108 cfu/ml bacterial suspensions, with two replicates per isolate. The plates were incubated at 28°C for 14 days. The plates were observed every day and the color change of the medium was recorded two weeks after inoculation. The test was repeated twice.
Detection of rsa1 gene
We carried out PCR to validate the presence of rsa1, which has been reported to be a gene for avirulence of R. solanacearum on pepper hosts (Jeong et al., 2011). To eliminate errors, we prepared two rsa1 gene primer pairs. One pair produced a 720-bp fragment containing the full rsa1 gene including the promotor region (720-rsa1F; 5′-GCCGCTCGCCGCAATGCTGCC-3′, 720-rsa1R; 5′-TGGGCTGGGTGGGACTTAACC-3′), and the other produced a 315-bp fragment containing a partial rsa1 open reading frame (ORF) region (315-rsa1F; 5′-ATCACCAAGATTACCGGAAAG-3′, 315-rsa1R 5′-TGGGCTGGGTGGGACTTAACC-3′). The reactions were carried out as described previously for phylotype determination with a primer set of 720-rsa1F/720-rsa1R at an annealing temperature of 70°C, and with a primer set of 315-rsa1F/315-rsa1R at an annealing temperature of 59°C.
Phylogenetic analysis of 16S rRNA and partial endoglucanase (egl) gene sequences
The 16S rRNA genes of the 93 isolates were amplified by PCR in 25 μl reaction volumes containing 1.25 U of Pfu Turbo DNA Polymerase (Stratagene), 2.5 μl of 10× Pfu polymerase buffer, 0.25 mM of each dNTP, 1 μl of 10 pmoles of each primer (9F, 5′-GAGTTTGATCCTGGCTCAG-3′; 1512R, 5′-ACGGCTACCTTGTTACGACTT-3′), and 50 ng of genomic DNA. The reaction was performed in an automated thermocycler (model PTC-200, MJ Research Inc.) with initial denaturation at 95°C for 5 min, followed by 30 cycles of denaturation at 95°C for 45 sec, annealing at 55°C for 45 sec, and extension at 72°C for 1 min, with a final extension at 72°C for 10 min. The 750-bp partial endoglucanase gene was amplified by PCR in the same reaction as the 16S rRNA gene with primer pairs of Endo-F (5′-ATGCATGCCGCTGGTCGCCGC-3′) and Endo-R (5′-GCGTTGCCCGGCACGAACACC-3′) (Poussier et al., 2000). After the PCR amplicons of 16S rRNA and endoglucanase genes were confirmed by electrophoresis, the sequences of the two genes were determined using an ABI BigDye Terminator v3.1 Cycle Sequencing Kit and an ABI3730XL automated DNA sequencer (Applied Biosystems Inc., Foster City, CA, USA) according to the manufacturer’s instructions. The 16S rRNA and endoglucanase gene sequences were confirmed and edited with BioEdit 7.2.5 software and trimmed with EditSeq software (DNASTAR Lasergene8, DNASTAR Inc., Madison, WI, USA). The analyzed 16S rRNA and partial endoglucanase gene sequences were deposited in the GenBank database, and the accession numbers are listed in Table 1. Reference sequences for the 16S rRNA gene and endoglucanase gene were obtained from the NCBI website with the following GenBank accession numbers: AL646052.1 (GMI1000), FP885891.2 (PSI07), FP885896.1 (CMR15), FP885897.1 (CFBP2957), and CP002820.1 (Po82). The ClustalW method was used for sequence alignment and the construction of phylogenetic trees using 1,000 bootstrap replicates in the MegAlign program (DNASTAR Lasergene8, DNASTAR Inc.).
Host range determination
For the pathogenicity test, tomato (Lycopersicon esculentum cv. Seokwang) (Lee and Kang, 2013), eggplant (Solanum melongena cv. Heukmajang) (Lee, 1999), and pepper (Capsicum annuum cv. Nokkwang) (Jeong et al., 2007) were grown in a greenhouse at 25–35°C under natural light conditions. For the positive control, potatoes (Solanum tuberosum cv. Sumi) (the original cultivar name: Superior) (Park et al., 2016) were grown in a greenhouse at 20–25°C. Two-week-old seedlings of eggplant and pepper, and 10-day-old seedlings of tomato were transplanted into plastic pots 7 cm in diameter containing commercial soil (Baroker, Seoul Agriculture Materials Co., Seoul, Korea) and grown in a greenhouse for 2–3 weeks. To prepare the inoculum, all isolates were grown on TZC plates for 48 h at 28°C. Bacterial cells were suspended in sterile distilled water and the concentration was adjusted to OD600 0.1. After wounding the root of each plant by stabbing with a 3-cm-wide scoop, 50 ml of bacterial suspension of the 93 isolates was poured into each pot. Three plants from each crop were inoculated with each isolate, and pathogenicity assays were repeated two or three times in a greenhouse under natural light conditions. Symptom development was observed every 3 days and recorded at 28 days post-inoculation using the following scale: −, no symptoms; +, one to three leaves wilted; ++, four to six leaves wilted; and +++, seven or more leaves or whole plant wilted.
The tomato plants were photographed at 12 days post-inoculation, and the eggplants and pepper plants were photographed at 19 days post-inoculation.
Results
Determination of biovar, phylotype, and rsa1 gene
The Korean potato bacterial wilt isolates were analyzed for biovar, phylotype, and presence of the rsa1 gene. Of the 93 isolates analyzed, twenty-two were of biovar 2 (about 24%), eight were of biovar 3 (less than 8%), and sixty-three were of biovar 4 (68%) (Table 1). The isolates of biovar 3 and biovar 4 were classified as phylotype I, and biovar 2 was classified as phylotype IV.
It has been reported that the Rsa1 protein, which is an aspartic protease secreted through the type II secretion systems (T2SSs), is responsible for the loss of bacterial virulence to pepper (Jeong et al., 2011). To validate the presence of this gene, we carried out rsa1 detection PCR. For greater certainty, we used two sets of rsa1 gene primers. One pair was for the full rsa1 gene including the promotor (720-rsa1F/720-rsa1R), while the other was for a partial rsa1 ORF (315-rsa1F/315-rsa1R). From the ninety-three isolates of potato bacterial wilt, all phylotype IV-biovar 2 isolates produced a full 720-bp rsa1 gene fragment at an annealing temperature of 70°C. On the other hand, biovar 3 and biovar 4 isolates of phylotype I did not produce this fragment under these annealing conditions. Partial rsa1 gene PCR was carried out with a 315-rsa1F/315-rsa1R primer set at an annealing temperature of 59°C, and the results were the same as for the PCR of the full rsa1 gene, which was detected in all isolates of phylotype IV and not detected in any isolates of phylotype I.
Geographical distribution
Fig. 1 shows the geographical distribution of the potato bacterial wilt isolates in Korea from 1998 to 2003. Ralstonia solanacearum was first isolated in southern provinces (Gyeongsangnam-do, Jeollanam-do, and Jeju-do, Korea) with biovar 2 and biovar 4 isolates in 1998. The biovar 3 bacterium, which infects potato, was first identified in Gyeongsangnam-do in 1999, and was subsequently found in Jeollanam-do, Jeollabuk-do, and Jeju-do. Biovar 4 bacteria that infect potatoes have been isolated in all locations where potato bacterial wilt has been observed. R. solanacearum of biovar 2 has been isolated in seven locations, which are the same as the regions where biovar 3 bacteria have been isolated.
Phylogenetic analysis of 16S rRNA and partial endoglucanase (egl) gene sequences
To evaluate the genetic relationship of Korean potato isolates, we sequenced the 16S rRNA gene (about 1,453 nucleotides) and partial endoglucanase gene (760 and 766 nucleotides) of 93 isolates (Table 1) and compared the sequences using the ClustalW program (Fig. 2).

Phylogenetic trees of 16S rRNA (A) and partial endoglucanase (egl) gene sequences (B) analyzed by ClustalW with 1,000 bootstrap replicates using MegAlign of DNASTAR Lasergene 8 (DNASTAR Inc.). Roman numerals indicate phylotypes and symbols (★) indicate the representative biovar 4 strain.
The 16S rRNA sequences of biovar 2 isolates were identical with each other, and the sequences of biovar 3 and biovar 4 isolates were also identical. To downsize the phylogenetic tree, SL2729 was chosen to represent the biovar 4 strains (Fig. 2-A). In the 16S rRNA phylogenetic tree, Korean R. solanacearum isolates were separated into two groups following GMI1000 (phylotype I) and PSI07 (phylotype IV). Korean biovar 2 isolates, which were identified as phylotype IV, were clustered with PSI07 of a representative phylotype IV strain. The isolates of biovar 3 and 4, which were identified as phylotype I, were not only clustered with GMI1000, but also identical with GMI1000 in the sequenced 1,453 nucleotides.
Since the partial endoglucanase gene (egl) sequences were identical among biovar 4 isolates, we analyzed the egl sequence of SL2729 as the representative strain of biovar 4 (Fig. 2B). While the sequences of biovar 4 isolates were identical, the egl sequences of biovar 3 isolates showed subtle differences in nucleotides. In a phylogenetic tree based on the egl gene, biovar 3 and biovar 4 isolates were grouped to phylotype I without GMI1000, which was placed as the outgroup to Korean phylotype I.
Host range determination
To determine the host range of the bacteria that cause potato bacterial wilt, we assessed the pathogenicity of isolates on several solanaceous plants-potato, tomato, eggplant, and pepper. The patterns of pathogenicity were divided into four types: first, pathogenic only to potato, but nonpathogenic to tomato, eggplant, and pepper (P); second, pathogenic to potato and tomato, but nonpathogenic to eggplant and pepper (PT); third, pathogenic to potato, tomato, and eggplant, but nonpathogenic to pepper (PTE); and finally, pathogenic on all four tested crops (potato, tomato, eggplant, and pepper, PTEP) (Table 2).
The isolates of phylotype IV-biovar 2 showed various patterns of host pathogenicity, including P, PT, and PTE. Of the twenty-two biovar 2 isolates, eleven infected only potato (P), four infected only potato and tomato (PT), and seven infected potato, tomato, and eggplant (PTE). These results showed that none of the biovar 2 isolates could infect pepper. The isolates of phylotype I (including biovar 3 and 4) could be divided into two groups: pathogenic to potato, tomato, and eggplant, but nonpathogenic on pepper (PTE); and pathogenic to all test plants (PTEP). For phylotype I, one biovar 3 isolate and four biovar 4 isolates were classified as PTE (7% of phylotype I), and seven biovar 3 and fifty-nine biovar 4 isolates were classified as PTEP (93% of phylotype I). Fig. 3 shows the host range of representative pathotype strains.

Host range determination of Ralstonia solanacearum on tomato, eggplant, and pepper. SL2312 and T12 were not pathogenic to tomato, eggplant, and pepper (pathotype P); SL3175 was pathogenic to tomato, and not pathogenic to eggplant or pepper (PT); SL3103 was pathogenic to tomato and eggplant, and not pathogenic to pepper (PTE); and SL2729 was pathogenic to all tested plants (PTEP). IV-2, phylotype IV-biovar 2; I-4, phylotype I-biovar 4. Pictures of tomato were taken at 12 DPI, and pictures of eggplant and pepper were taken at 19 DPI.
Discussion
When potato bacterial wilt was first reported in Korea, we surveyed potato-growing regions from 1998 to 2003, isolated bacteria, and characterized them using various genetic and pathogenic tests. During this period, among the twenty-five potato cultivation areas, potato bacterial wilt was observed in twelve locations in southern region of Korea. By 16S rRNA sequence analysis and BLAST search, ninety-three bacteria were identified as R. solancearum. These isolates were analyzed to determine their phylotype, biovar, phylogenetic relationship of 16S rRNA and egl gene, presence of an rsa1 gene, and host range on major solanaceous crops-potato, tomato, eggplant, and pepper.
In Korea, the potato bacterial wilt isolates were classified into biovars 2, 3, and 4. When applied to the phylotype scheme, biovar 3 and 4 isolates belonged to phylotype I (Asian origin). All biovar 2 isolates belonged to phylotype IV (Indonesian origin), and none belonged to phylotype II (American origin).
The sequence variation of isolates from biovars 3 and 4 differed between the 16S rRNA and egl gene sequences. For 16S rRNA gene sequences, biovar 3 and 4 isolates were identical and built a phylotype I cluster with the representative strain GMI1000. For egl gene sequences, while all biovar 4 isolates were identical, biovar 3 isolates had variations in some nucleotides. Surprisingly, the representative phylotype I strain GMI1000 was distinguished from Korean biovar 3 and 4 isolates and was placed outside of the Korean phylotype I clade. On the other hand, Korean biovar 2 isolates were consistently located in the phylotype IV clade with the representative strain PSI07, either in the 16S rRNA tree or in the egl tree. These results may suggest that Korean biovar 2 strains were introduced from a foreign country relatively recently, and that Korean biovar 3 and 4 strains represent the Asian-origin phylotype I, which evolved differentially from GMI1000.
The egl gene was used for “sequevar” determination of the phylotype-sequevar classification, as discussed by Fegan and Prior (2005). The evolutionary dynamics of RSSC have previously been revealed by phylogenetic and statistical analysis of housekeeping and virulence-related genes by Castillo and Greenberg (2007). Among virulence-related genes, hrpB and fliC, which are essential for species survival, have undergone purifying selection, like essential housekeeping genes. On the other hand, egl, which is directly related to pathogenicity, has undergone diversifying selection with a high level of recombination. The divergence of egl between Korean phylotype I and GMI1000 was attributed by Castillo and Greenberg (2007) to geographic isolation.
Regarding the recent introduction of Korean phylotype IV, Jeong et al. (2007) discussed the possibility of the import of phylotype IV strains from Japan. In phylogenetic analysis of 16S rDNA, egl, and hrpB, Korean phylotype IV isolate (SL2029) clustered with Japanese (MAFF301558, MAFF301559) and Indonesian (R142) phylotype IV strains. However, Korean SL2029 was closer to Japanese MAFF301558 and MAFF301559 than to Indonesian R142, which suggests that the Korean and Japanese lineages have diverged more recently than the Japanese and Indonesian lineages. Korean phylotype IV strains appeared after import of the potato cultivar Daeji (Japanese variety name: Dejima) from Japan in the 1990s. It is reasonable to infer that phylotype IV ingressed with the import of Daeji, since the timing of Daeji cultivation in southern region of Korea is consistent with the emergence of R. solanacearum phylotype IV.
Among biovars 2, 3, and 4, biovar 4 is the most common in Korea and was found in all regions of potato bacterial wilt. Most biovar 4 isolates (93.6%) were pathogenic to all of the tested solanaceous plants (potato, tomato, eggplant, and pepper). The distribution and pathogenicity results are consistent with a previous report that biovar 4 was predominant in other crops in Korea (Jeong et al., 2007). It seems that the high humidity and temperatures of the Korean summer are suitable for biovar 4 outbreaks, and the cultivation season of susceptible crops permits the spread of the pathogens. In light of these results, biovar 4 is considered the main and most destructive pathovar; hence, we should monitor biovar 4 to predict and prevent the spread of bacterial wilt from potato to other crops, or vice versa.
Korean biovar 2 isolates were genetically and pathologically distinct from biovar 3 and 4. All biovar 2 isolates were classified as phylotype IV, and clustered with phylotype IV reference strain PSI07 in 16S rRNA and in egl phylogenetic trees. Furthermore, only biovar 2 isolates contained the rsa1 gene, which confers avirulence to pepper-infecting strains (Jeong et al., 2011). This result is consistent with our host-determining pathogenicity assays, which showed that all biovar 2 isolates were nonpathogenic to pepper. We also identified tomato-nonpathogenic biovar 2 isolates. In the course of pathogenicity assays, we observed that tomato plants were the most susceptible among all hosts, which is consistent with previous reports (Ramesh et al., 2014; Sakthivel et al., 2016). However, some biovar 2 isolates could not infect all tested plants (tomato, eggplant, and pepper), but only potato, which was the original host plant. The nonpathogenic strains on tomato, eggplant, and pepper were reported previously: R288 (phylotype I) isolated from Morus alba in China and MAFF211266 (phylotype I) and MAFF301558 (phylotype IV) isolated from Solanum lycopersicum in Japan (Lebeau et al., 2011). However, the tested cultivars (tomato L390, eggplant Florida Market, and pepper Yolo Wonder) in these earlier reports differed from those in the present study (tomato Seokwang, eggplant Heukmajang, and pepper Nokkwang), and did not include potato. Therefore, the tomato-nonpathogenic isolates used in the present study are important as a genetic resource.
From this study, we also obtained the groups of eggplant-pathogenic and nonpathogenic isolates and pepper-pathogenic and nonpathogenic isolates. These groups could be the materials for investigating of eggplant-specific (or pepper-specific) infection factors. The genomic difference between the tomato-pathogenic and tomato-nonpathogenic biovar 2 groups may provide a clue to host specificity on tomato. Therefore we aim to conduct further comparative analyses of the tomato pathogenic and nonpathogenic genomes.
Acknowledgments
This work was supported by a grant from the National Institute of Agricultural Sciences, Rural Development Administration (PJ010085 and PJ012466), Republic of Korea.